- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2024
Volume: 14, Issue: 21
Biological Sciences
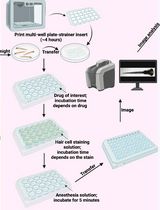
PEPITA: Parallelized High-Throughput Quantification of Ototoxicity and Otoprotection in Zebrafish Larvae
Biophysics
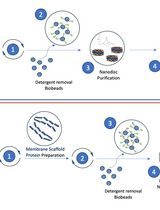
Optimizing Transmembrane Protein Assemblies in Nanodiscs for Structural Studies: A Comprehensive Manual
Cancer Biology
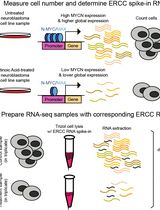
Accurate Measurement of Cell Number–Normalized Differential Gene Expression in Cells Treated With Retinoic Acid
Developmental Biology
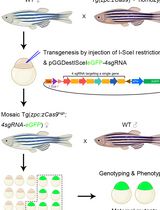
Generation of Zebrafish Maternal Mutants via Oocyte-Specific Knockout System
An In Vitro Model of Murine Osteoclast-Mediated Bone Resorption
Environmental science
The on-Site Monitoring and Specimen-Making of Ectoparasites on Rodents and Other Small Mammals
Neuroscience
Assessment and Quantification of Foam Cells and Lipid Droplet–Accumulating Microglia in Mouse Brain Tissue Using BODIPY Staining
A Real-Time Approach for Assessing Rodent Engagement in a Nose-Poking Go/No-Go Behavioral Task Using ArUco Markers
Semi-Automated Assessment of Long-Term Olfactory Habituation in Drosophila melanogaster Using the Olfactory Arena
Plant Science
Fluorescent Staining and Quantification of Starch Granules in Chloroplasts of Live Plant Cells Using Fluorescein
Stem Cell
Efficient Gene-Editing in Human Pluripotent Stem Cells Through Simplified Assembly of Adeno-Associated Viral (AAV) Donor Templates
Novel Cross-Species Salivary Gland-Parasympathetic Neuron Coculture System