- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2023
Volume: 13, Issue: 13
Biochemistry
Human-rabbit Hybrid Translation System to Explore the Function of Modified Ribosomes
Cuticular Hydrocarbon Profiling by Fractionation and GC-MS in Socially Parasitic Ants
Biological Engineering
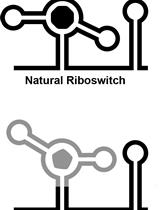
In vitro Selection and in vivo Testing of Riboswitch-inspired Aptamers
Biophysics
Dual-color Colocalization in Single-molecule Localization Microscopy to Determine the Oligomeric State of Proteins in the Plasma Membrane
Cancer Biology
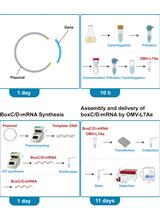
mRNA Delivery Platform Based on Bacterial Outer Membrane Vesicles for Tumor Vaccine
3D Ultrastructural Visualization of Mitosis Fidelity in Human Cells Using Serial Block Face Scanning Electron Microscopy (SBF-SEM)
Cell Biology
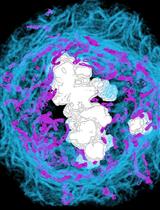
Iterative Indirect Immunofluorescence Imaging (4i) on Adherent Cells and Tissue Sections
Visualizing NBD-lipid Uptake in Mammalian Cells by Confocal Microscopy
Developmental Biology
Visualization of Actin Cytoskeleton in Cellular Protrusions in Medaka Embryos
Systematic Analysis of Smooth Muscle and Cartilage Ring Formation during Mouse Tracheal Tubulogenesis
Immunology
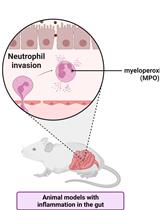
Measuring Myeloperoxidase Activity as a Marker of Inflammation in Gut Tissue Samples of Mice and Rat
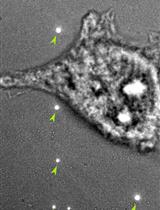
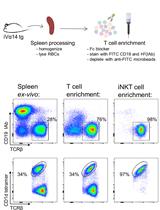
Primary Mouse Invariant Natural Killer T (iNKT) Cell Purification and Transduction
Medicine
Ex vivo Culture and Contractile Force Measurements of Non-human Primate Heart Slices
Microbiology
Qualitative and Quantitative Methods to Measure Antibacterial Activity Resulting from Bacterial Competition
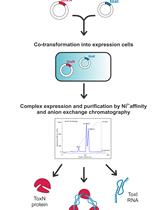
Large-scale Purification of Type III Toxin-antitoxin Ribonucleoprotein Complex and its Components from Escherichia coli for Biophysical Studies
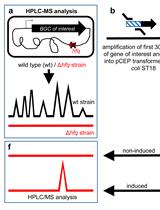
easyPACId, a Simple Method for Induced Production, Isolation, Identification, and Testing of Natural Products from Proteobacteria
A Simple and Reproducible Stereomicroscopic Method to Assess Fungal Biofilms: Application to Antifungal Susceptibility Testing
Molecular Biology
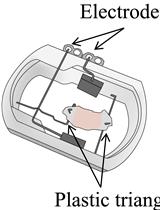
In vivo Electroporation of Skeletal Muscle Fibers in Mice
Plant Science
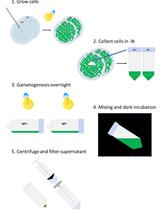
Autolysin Production from Chlamydomonas reinhardtii
Systems Biology
Lipidomics Workflow for Analyzing Lipid Profiles Using Multiple Reaction Monitoring (MRM) in Liver Homogenate of Mice with Non-alcoholic Steatohepatitis (NASH)