- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2023
Volume: 13, Issue: 1
Cell Biology
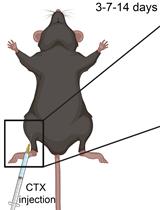
Myonecrosis Induction by Intramuscular Injection of CTX
Developmental Biology
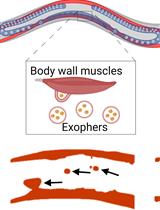
Preparation of Caenorhabditis elegans for Scoring of Muscle-derived Exophers
Immunology
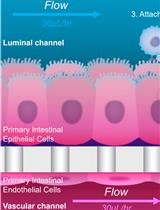
Safety Profiling of Tumor-targeted T Cell–Bispecific Antibodies with Alveolus Lung- and Colon-on-Chip
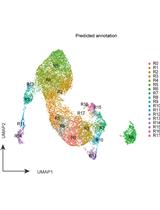
Sample Preparation and Integrative Data Analysis of a Droplet-based Single-Cell ATAC-sequencing Using Murine Thymic Epithelial Cells
Microbiology
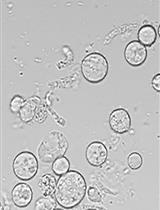
Sclerotinia sclerotiorum Protoplast Preparation and Transformation
Molecular Biology
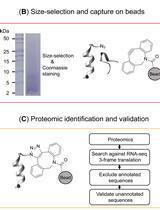
BONCAT-based Profiling of Nascent Small and Alternative Open Reading Frame-encoded Proteins
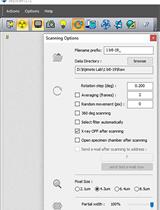
Staining and Scanning Protocol for Micro-Computed Tomography to Observe the Morphology of Soft Tissues in Ambrosia Beetles
Neuroscience
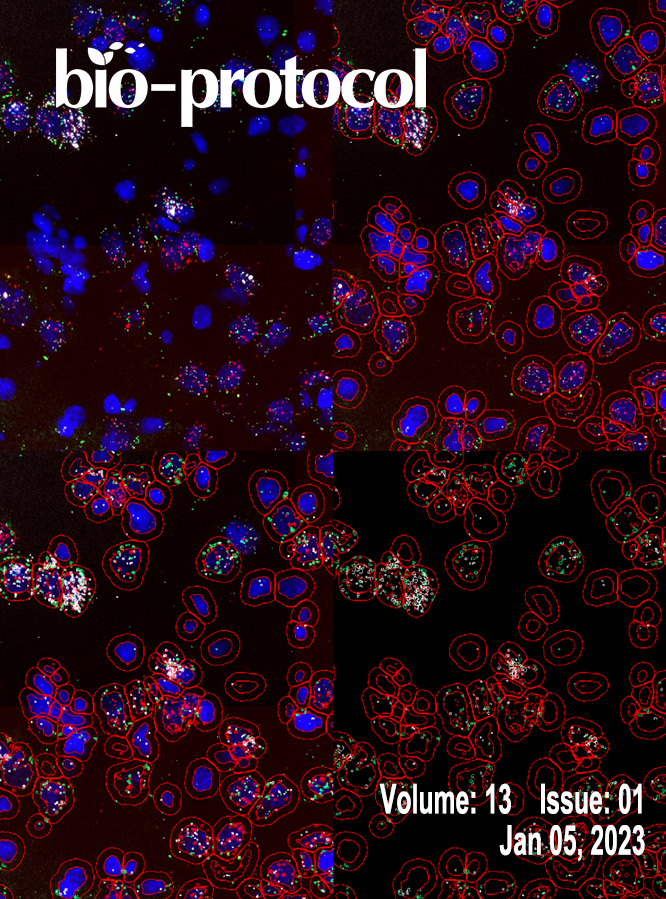
Quantitative Analysis of Gene Expression in RNAscope-processed Brain Tissue
Parasitoid Wasp Culturing and Assay to Study Parasitoid-induced Reproductive Modifications in Drosophila
Systems Biology
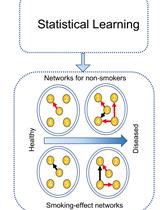
A Cartographic Tool to Predict Disease Risk-associated Pseudo-Dynamic Networks from Tissue-specific Gene Expression