- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2022
Volume: 12, Issue: 19
Biological Engineering
Dual-target Bridging ELISA for Bispecific Antibodies
LIST: A Newly Developed Laser-assisted Cell Bioprinting Technology
Cell Biology
CRISPR/Cas9-mediated LRP10 Knockout in HuTu-80 and HEK 293T Cell Lines
Enhanced Ribonucleoprotein Immunoprecipitation (RIP) Technique for the Identification of mRNA Species in Ribonucleoprotein Complexes
Drug Discovery
Gastrulation Screening to Identify Anti-metastasis Drugs in Zebrafish Embryos
Immunology
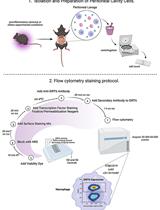
Flow Cytometry Analysis of SIRT6 Expression in Peritoneal Macrophages
Microbiology
Assay for Protealysin-like Protease Inhibitor Activity
Neuroscience
A Simplified Paradigm of Late Gestation Transient Prenatal Hypoxia to Investigate Functional and Structural Outcomes from a Developmental Hypoxic Insult
Plant Science
Extraction and Quantification of Plant Hormones and RNA from Pea Axillary Buds
Collection of Xylem Exudates from the Model Plant Arabidopsis and the Crop Plant Soybean