- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Trehalase Activity in Arabidopsis thaliana Optimized for 96-well Plates
Published: Vol 3, Iss 20, Oct 20, 2013 DOI: 10.21769/BioProtoc.946 Views: 12695
Reviewed by: Tie Liu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Semi-throughput Procedure for Assaying Plant NADP-malate Dehydrogenase Activity Using a Plate Reader
Kevin Baudry and Emmanuelle Issakidis-Bourguet
Aug 20, 2023 1465 Views

An in vitro Assay to Probe the Formation of Biomolecular Condensates
Yu Zhang and Shen Lisha
Sep 5, 2023 3196 Views

Immunofluorescence for Detection of TOR Kinase Activity In Situ in Photosynthetic Organisms
Ana P. Lando [...] Giselle M. A. Martínez-Noël
Dec 20, 2024 1811 Views
Abstract
Trehalose is a nonreducing disaccharide. It is a common sugar in bacteria, fungi and yeast, where it functions as a carbon source and stress protectant. In contrast, plants, although encoding large trehalose biosynthesis gene families, contain only small amounts of trehalose. The intermediate compound of trehalose, trehalose-6-phosphate (T6P), is a signaling molecule in plants, regulating metabolism, growth, and development. Most plants contain only a single trehalase, the enzyme that specifically hydrolyzes trehalose into two glucose molecules. High trehalase activity has been suggested to be part of the defense mechanism in plants hosting mycorrhizal fungi, rhizobia, and the plant pathogen Plasmodiophora brassica. Recently, it was shown in Arabidopsis thaliana that high trehalase activity is associated with an increase in drought stress tolerance and that trehalase fulfills an important role in stomatal regulation. Here we describe a protocol for measuring trehalase activity in Arabidopsis tissues, optimized for 96-well plates. Dialyzed protein extracts will be incubated with trehalose, followed by the quantitation of the released glucose using glucose oxidase-peroxidase.
Keywords: Arabidopsis thalianaMaterials and Reagents
- Plant tissues
- Liquid nitrogen
- MES
- Phenylmethylsulfonyl fluoride (PMSF)
- EDTA
- Polyvinylpyrrolidone (PVP)
- Dithiothreitol (DTT)
- CaCl2
- Glucose
- Bovine serum albumin (BSA)
- Na2CO3
- K-Na-tartrate
- CuSO4.5H2O
- KOH
- NaOH
- Folin & Ciocalteu's phenol reagent (Sigma-Aldrich, catalog number: 47641 )
- Trehalose (Sigma-Aldrich, catalog number: T9531 )
- Glucose, GOD-PAP (DIALAB GmbH, catalog number: D95218B )
- Extraction buffer (see Recipes)
- Dialysis buffer (see Recipes)
- Trehalose buffer (see Recipes)
- Glucose standards (see Recipes)
- BSA standards (see Recipes)
- Lowry buffers (see Recipes)
Equipment
- Mortars and pestles
- Spectra/Por® 1 Dialysis Membrane (IEEE, catalog number: 132660 )
Note: 96-well dialysis systems can be used for the dialysis of multiple samples.
- Transparent 96-well plates with flat bottom
- Rocker
- Refrigerated microcentrifuge
- Pre-chilled microcentrifuge tubes
- 100 ml cylinder
- Magnetic stir plate and magnet
- Cold room
- Spectrophotometric plate reader
Procedure
- Protein extraction
Note: Work always on ice unless stated differently.
- Grind plant tissues in liquid nitrogen with mortar and pestle.
- Aliquot 100 mg tissue powder in chilled microcentrifuge tubes. Prepare at least 3 replicates per sample.
- Add 1 ml of ice cold extraction buffer to each sample. Homogenize samples by pipetting up and down. Centrifuge at 18,000 x g, 4 °C, 10 min.
- Transfer the supernatant to a new, chilled microcentrifuge tube. The obtained protein extracts can be stored at -80 °C.
- Grind plant tissues in liquid nitrogen with mortar and pestle.
- Dialysis
- Wet a piece of Spectra/Por?1 Dialysis Membrane with water and tie a knot at the bottom (Figure 1).
- Transfer 500 μl of the protein extract into the tubing and tie a knot at the top (Figure 1).

Figure 1. Tying a knot at the bottom and top of a dialysis membrane
- Place tubing in a 100 ml cylinder filled with ice cold dialysis buffer. Dialyze the extract at 4 °C for 2-3 h under continuous stirring on a magnetic stir plate.
- Replace dialysis buffer and continue the dialysis at 4 °C overnight.
- Transfer the dialyzed extracts to new, chilled microcentrifuge tubes. The dialyzed extracts can be stored at -80 °C.
- Wet a piece of Spectra/Por?1 Dialysis Membrane with water and tie a knot at the bottom (Figure 1).
- Trehalase activity adapted for 96-well plates
Note: Work always on ice unless stated differently.
- Prepare a water bath at 95 °C.
- Prepare a water bath at 95 °C.
For samples
- Transfer 10 μl of the dialysis product in a 96-well plate with flat bottom (= plate S).
- Transfer 10 μl of each respective glucose standard to plate S.
For blanks
- Transfer 10 μl of the dialysis product in a 96-well plate with flat bottom (= plate B).
- Place plate B at 95 °C temperature for 5 minutes to denature the trehalase enzyme present in the blanks.
- Place plate B on ice for 2 min.
For samples and blanks
- Add 50 μl of trehalose buffer to plates S and B. Mix by pipetting up and down.
- Incubate plates S and B for 30 min on a rocker at 30 °C.
- Stop the reaction by boiling for 5 min at 95 °C (denaturation of the trehalase).
- Place plates S and B on ice for 2 min.
- Add 200 μl of Glucose, GOD-PAP to plates S and B for determining the glucose concentration of the samples and blanks by colorimetry. Mix by pipetting up and down.
- Incubate plates S and B for 15 min at 30 °C on a rocker.
- Measure the absorbance of plates S and B at 505 nm with a spectrophotometric plate reader (30 °C).
- Determine the glucose standard curve to calculate the glucose concentration (nmol) present in the samples and blanks. Subtract the glucose concentration of the blanks from the samples (see Example: Calculating the trehalase activity in excel).
For proteins (Lowry procedure, Van Houtte et al., 2013)
- Transfer 10 μl of the dialysis product in a 96-well plate with flat bottom (= plate P) and add 30 μl water to these samples.
- Transfer 40 μl of each respective BSA standard to plate P.
- Add 200 μl of Reagent C (Lowry buffers) to plate P and incubate for 10 min at room temperature.
- Add 20 μl of Reagent D (Lowry buffers) to plate P and incubate for 30 min at 30 °C.
- Measure the absorbance of plate P at 546 nm with a spectrophotometric plate reader (30 °C).
- Use the BSA standard curve to determine the amount of protein (μg) present in the extracts (see Example: Calculating the trehalase activity in excel).
- Express the trehalase specific activity as nmol of glucose released per min per mg protein (see Example: Calculating the trehalase activity in excel).
Data analysis
Here we show an example how to calculate the trehalase activity from a protein extract of Arabidopsis Col-0 seedlings.
- Glucose standard curve
Using excel, plot the glucose concentration of the respective glucose standards on the X axis and the corresponding absorbances (505 nm) on the Y axis (Table 1; Figure 2). Add a linear trendline to the glucose standard curve and diplay its equation (Figure 2; [1]).
[1] y = 0.0157x + 0.0858
Table 1. Absorbances at 505 nm (Plates S and B)Glucose standards
Absorbance (505 nm) 0 nmol glucose
0.08
20 nmol glucose
0.3721
40 nmol glucose
0.7421
60 nmol glucose
1.0355
80 nmol glucose
1.3792
100 nmol glucose
1.6143
Arabidopsis Col-0 tissues
Absorbance (505 nm)
Sample
0.1483
Blank
0.0898
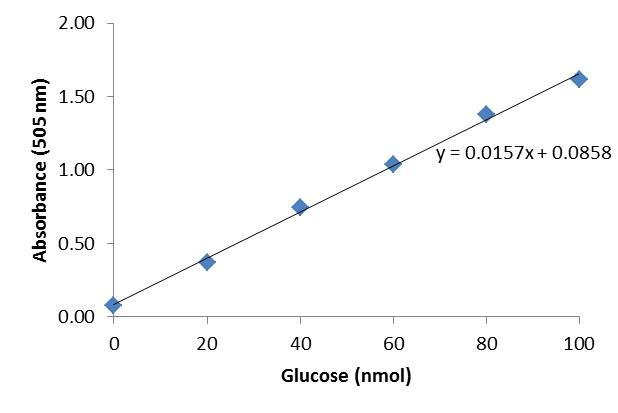
Figure 2. Glucose standard curve
- Determination of the glucose concentration
Equation [1] and the absorbances (505 nm) of the sample and blank (Table 1) can be used to calculate the glucose concentration present in the sample and blank.
[2] Nmol glucose in sample = (0.1483-0.0858)/0.0157 = 3.9809
[3] Nmol glucose in blank = (0.0898-0.0858)/0.0157 = 0.2548
In order to know how many glucose is released per min in the extract, subtract [3] from [2], and divide by the duration of the incubation time (min).
[4] Nmol glucose released per min = (3.9809 - 0.2548)/30 = 0.1242
- BSA standard curve
Since trehalase activity is expressed per unit of protein, we need to determine the amount of protein present in the extract. Using excel, plot the protein content of the respective BSA standards on the X axis and the corresponding absorbances (546 nm) on the Y axis (Table 2; Figure 3). Add a linear trendline to the BSA standard curve and display its equation (Figure 3; [5]).
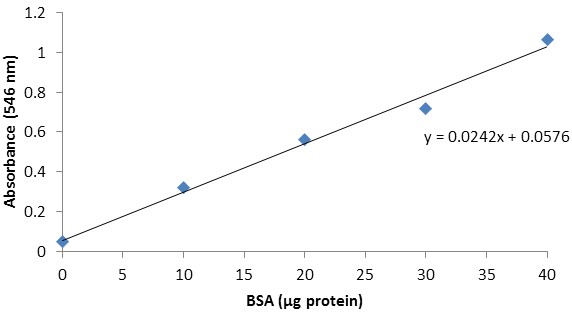
Figure 3. BSA standard curve
[5] y = 0.0242x + 0.0576
- Determination of the protein content
Equation [5] and the absorbance (546 nm) of the protein extract (Table 2) can be used to calculate the protein content.
Table 2. Absorbances at 546 nm (Plate P)BSA standards
Absorbance (546 nm)
0 μg protein
0.0488
10 μg protein
0.3224
20 μg protein
0.5603
30 μg protein
0.7159
40 μg protein
1.0637
Arabidopsis Col-0 tissues Absorbance (505 nm)
Protein 0.4786
[6] μg protein in extract = (0.4786 - 0.0576)/0.0242 = 17.3967
- Trehalase specific activity of Arabidopsis Col-0 seedlings
Since the specific trehalase activity is expressed as nmol glucose produced per min per mg protein, we need to divide [4] by [6], and multiply by 1,000.
[7] Trehalase specific activity in nmol glucose per min per mg protein = 0.1242/17.3967*1,000 = 7.1393
Recipes
- Extraction buffer
0.1 M MES-KOH, pH 6
1 mM PMSF
1 mM EDTA
1% (w/v) PVP
1 mM DTT
Stored at 4 °C
- Dialysis buffer
10 mM MES-KOH, pH 7
50 μM CaCl2
Stored at 4 °C
- Trehalose buffer
250 mM trehalose
62.5 mM MES-KOH, pH 7
125 μM CaCl2
Stored at 4 °C
- Glucose standards
Make standards with 10, 8, 6, 4, 2 and 0 μl of a 10 mM glucose solution in a total V of 10 μl
Fresh prepared or store 10 μl aliquots at -20 °C
- BSA standards
Make standards with 40, 30, 20, 10 and 0 μl of a 1 mg/ml BSA solution in a total V of 40 μl
Fresh prepared and keep on ice
- Lowry buffers
Reagent A:
2% (w/v) Na2CO3, 0.02% (w/v) K-Na-tartrate in 0.1 M NaOH
Stored at room temperature
Reagent B:
1% (w/v) CuSO4.5H2O
Stored at room temperature
Reagent C:
Mix solution A and B (100:1, [v/v])
Fresh prepared
Reagent D:
Mix Folin & Ciocalteu's phenol reagent with MilliQ water (1:2, [v/v])
Fresh prepared
Acknowledgments
This protocol was developed in the framework of the following paper: Van Houtte et al. (2013). It was developed based on two previous publications: Brodmann et al. (2002) and Pernambuco et al. (1996). Hilde Van Houtte was supported by the KU Leuven industrial research fund (IOF/KP/08/001). This work was supported by a grant from the FWO (G.0859.10).
References
- Brodmann, A., Schuller, A., Ludwig-Muller, J., Aeschbacher, R. A., Wiemken, A., Boller, T. and Wingler, A. (2002). Induction of trehalase in Arabidopsis plants infected with the trehalose-producing pathogen Plasmodiophora brassicae. Mol Plant Microbe Interact 15(7): 693-700.
- Lowry, O. H., Rosebrough, N. J., Farr, A. L. and Randall, R. J. (1951). Protein measurement with the Folin phenol reagent.J Biol Chem 193(1): 265-275.
- Pernambuco, M. B., Winderickx, J., Crauwels, M., Griffioen, G., Mager, W. H. and Thevelein, J. M. (1996). Glucose-triggered signaling in Saccharomyces cerevisiae: different requirements for sugar phosphorylation between cells grown on glucose and those grown on non-fermentable carbon sources. Microbiology 142(7): 1775-1782.
- Van Houtte, H., Vandesteene, L., Lopez-Galvis, L., Lemmens, L., Kissel, E., Carpentier, S., Feil, R., Avonce, N., Beeckman, T., Lunn, J. E. and Van Dijck, P. (2013). Overexpression of the trehalase gene AtTRE1 leads to increased drought stress tolerance in Arabidopsis and is involved in abscisic acid-induced stomatal closure. Plant Physiol 161(3): 1158-1171.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Houtte, H. V. and Dijck, P. V. (2013). Trehalase Activity in Arabidopsis thaliana Optimized for 96-well Plates. Bio-protocol 3(20): e946. DOI: 10.21769/BioProtoc.946.
- Van Houtte, H., Vandesteene, L., Lopez-Galvis, L., Lemmens, L., Kissel, E., Carpentier, S., Feil, R., Avonce, N., Beeckman, T., Lunn, J. E. and Van Dijck, P. (2013). Overexpression of the trehalase gene AtTRE1 leads to increased drought stress tolerance in Arabidopsis and is involved in abscisic acid-induced stomatal closure. Plant Physiol 161(3): 1158-1171.
Category
Plant Science > Plant biochemistry > Protein > Activity
Biochemistry > Protein > Isolation and purification
Biochemistry > Carbohydrate > Disaccharide
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











