- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Detailed Method for the Purification of Rhamnogalacturonan-I (RG-I) in Arabidopsis thaliana
Published: Vol 16, Iss 3, Feb 5, 2026 DOI: 10.21769/BioProtoc.5579 Views: 217
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Simple, Rapid, and Cost-Effective Method for Assessing Carbohydrate Partitioning in Microalgae and Arabidopsis thaliana
Araceli N. Bader [...] Leonardo Curatti
Dec 5, 2024 2381 Views

Enzymatic Starch Quantification in Developing Flower Primordia of Sweet Cherry
Nestor Santolaria [...] Afif Hedhly
Apr 5, 2025 1926 Views

New Approach to Detect and Isolate Rhamnogalacturonan-II in Arabidopsis thaliana Seed Mucilage
Dayan Sanhueza and Susana Saez-Aguayo
Sep 5, 2025 1278 Views
Abstract
The plant cell wall is a dynamic and complex extracellular matrix that not only provides structural integrity and determines cell shape but also mediates intercellular communication. Among its major components, pectins play essential roles in cell adhesion, wall porosity, hydration, and flexibility. Rhamnogalacturonan-I (RG-I), a structurally diverse pectic polysaccharide, remains one of the least understood components of the plant cell wall. Its backbone is substituted with arabinan, galactan, and arabinogalactan side chains that vary in length, branching, and composition across tissues, species, and developmental stages. In addition, RG-I can undergo modifications such as backbone acetylation, further contributing to its structural complexity and functional diversity. To advance understanding of RG-I, we present a detailed method for isolating RG-I from the model plant Arabidopsis thaliana. Leveraging Arabidopsis as a model system provides major advantages owing to its well-characterized genome and powerful molecular toolkit, enabling deeper investigation into the roles of RG-I in plant development and responses to environmental stress. Our method consists of two major steps: an initial chemical extraction using oxalate, followed by endo-polygalacturonase (EPG) digestion to fragment the pectic domains. An advantage of this approach is that it produces a dry material that can be stored at room temperature without special handling and does not introduce chemicals that may interfere with downstream analyses. The purified RG-I can be used for detailed compositional and structural analyses, as well as for functional studies of enzymes involved in pectin biosynthesis, modification, and degradation. Although this protocol was developed for isolating RG-I from Arabidopsis rosette leaves, it is also applicable to other Arabidopsis organs and other plant species.
Key features
• This protocol provides a detailed description of RG-I isolation from Arabidopsis rosette leaves.
• The isolated RG-I can be used for compositional and structural analyses and serves as a substrate for functional studies of enzymes.
• This protocol is also applicable for isolating RG-I from other Arabidopsis organs and from different plant species.
Keywords: Plant cell wallGraphical overview
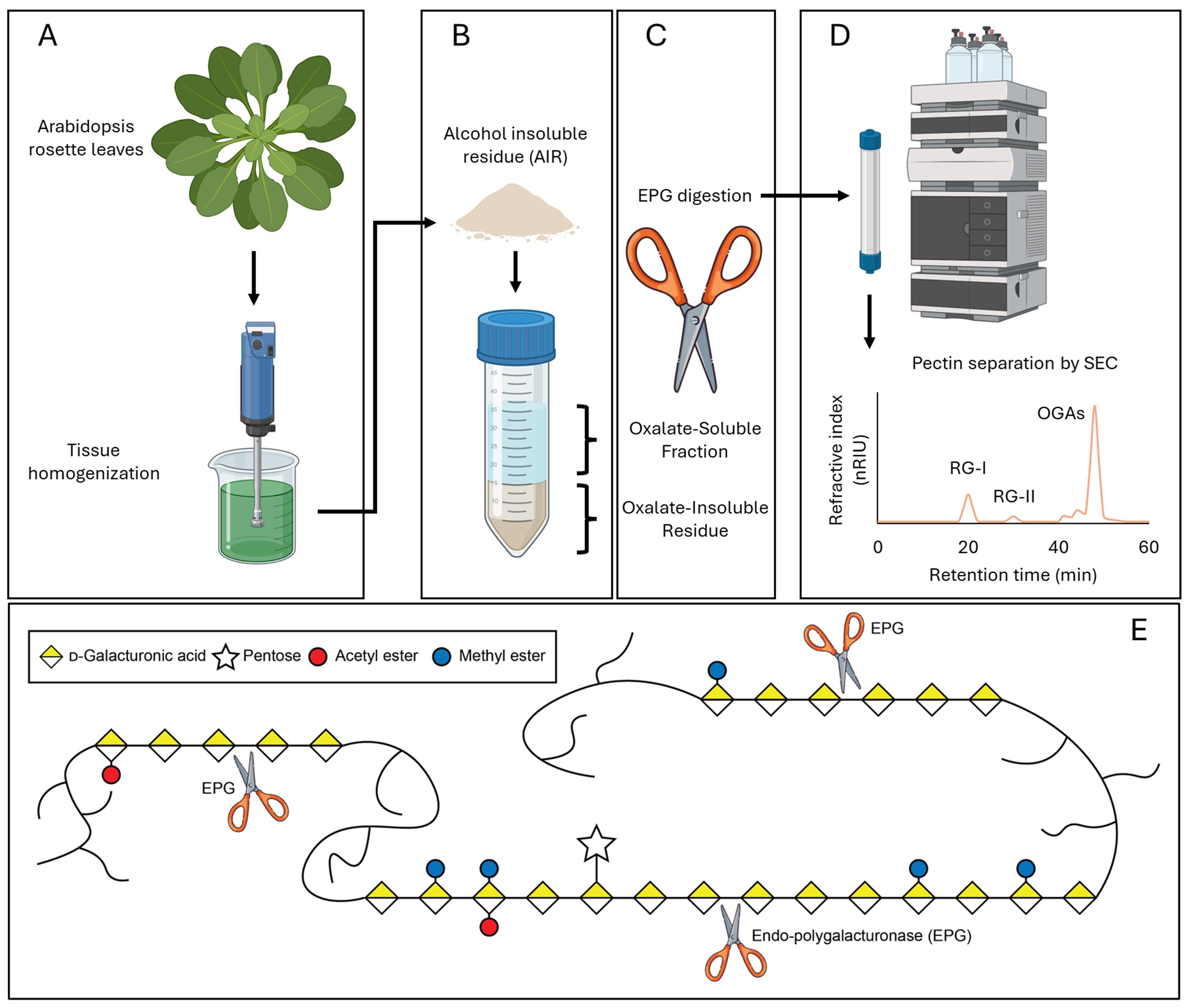
An outline of the steps for extracting rhamnogalacturonan-I (RG-I) from Arabidopsis rosette leaves. (A) Arabidopsis leaves are ground into a homogenate. (B) The resulting grounded plant material is treated to produce alcohol-insoluble residue (AIR), which is then treated with ammonium oxalate, resulting in an oxalate-soluble fraction and oxalate-insoluble residue. (C) Endo-polygalacturonase (EPG) digestion of the oxalate-soluble fraction and oxalate-insoluble residue releases RG-I. (D) RG-I is purified by size exclusion chromatography (SEC), and the resulting hypothetical spectrum is shown. (E) Schematic depicting a simplified mechanism of the activity of EPG used herein. For simplicity, the backbones and side chains of RG-I and rhamnogalacturonan II (RG-II) are represented as curved lines. EPG cleaves the α-1,4 glycosidic linkages between galacturonic acid residues within the homogalacturonan (HG) backbone. Pectin domains, including HG, RG-I, and RG-II, are believed to be covalently linked to one another via HG in the wall. Cleavage of HG by EPG releases RG-I from the pectin network [1,2].
Background
The plant cell wall is a dynamic and complex extracellular structure that surrounds all plant cells, providing mechanical support, determining cell shape, mediating intercellular communication, and regulating growth. It is primarily composed of cellulose microfibrils embedded in a matrix of hemicelluloses, pectins, proteins, and other polysaccharides [3]. Pectins are a diverse group of polysaccharides that are abundant in the middle lamella, as well as in the primary walls of dicots and nongraminaceous monocots. They play crucial roles in numerous biological processes contributing to cell–cell adhesion, cell wall porosity, hydration, and flexibility. Mutant plants with defects in pectin composition or modification often show developmental abnormalities, such as smaller rosette leaves, irregular organ formation, uneven cell expansion, and defects in pollen tube growth [1,4,5]. Rhamnogalacturonan-I (RG-I) is a major type of pectin characterized by a backbone of alternating rhamnose and galacturonic acid residues, often decorated with arabinan, galactan, and arabinogalactan side chains, that may also be modified by backbone acetylation (Figure 1).
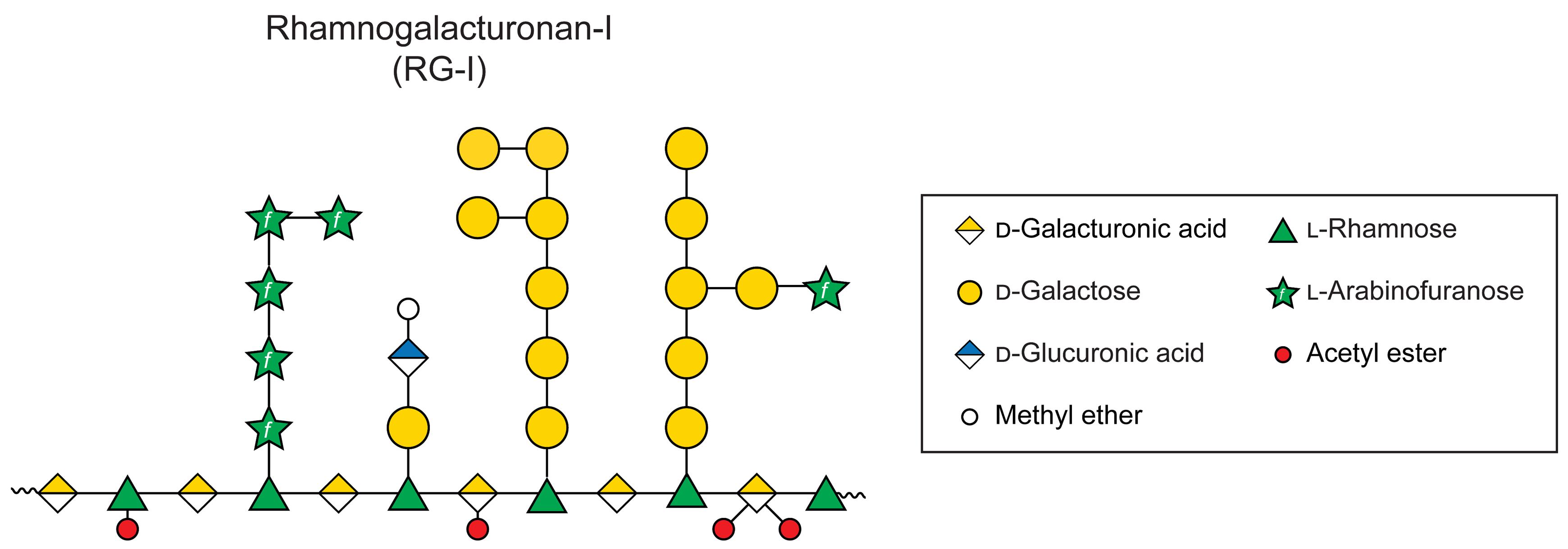
Figure 1. Representative model structure of rhamnogalacturonan-I (RG-I) sidechains. The specific structure of RG-I side chains is not known and is representative. Monosaccharide symbols used in the representative schematic structure are taken from the Symbol Nomenclature for Glycans (SNFG) from the Consortium for Functional Glycomics.
Most research on pectin has focused on homogalacturonan (HG), while the structure and function of RG-I have remained poorly understood, especially in model species. Indeed, much of our knowledge of RG-I largely comes from non-model species, which limits genetic studies of its synthesis and biological functions. RG-I is highly diverse, varying in structure by tissue, species, and developmental stage [6], and its biosynthesis likely involves a multitude of enzymes with distinct characteristics. Arabidopsis has long served as a model for studying plant cell wall structure, biosynthesis, remodeling, and function [7,8]. Thus, foundational knowledge about the structural features of RG-I in Arabidopsis is necessary for designing future experiments to gain insights into its biosynthesis and roles in plant development.
To better understand the structure and function of RG-I, it is essential to isolate this polysaccharide in sufficient quantity and purity. Isolated RG-I can be analyzed for its monosaccharide composition, glycosyl linkages, and structural features, providing insights into its diversity and biological roles. Furthermore, purified RG-I serves as a valuable substrate for enzymatic studies, enabling the characterization of pectin-synthesizing [9] and pectin-degrading [10] enzymes and the investigation of cell wall remodeling processes. This makes the development of reliable isolation protocols a critical step for advancing both fundamental and applied plant cell wall research. Here, we present a detailed protocol for isolating RG-I from Arabidopsis rosette leaves. Our protocol comprises two main steps: solubilization of pectin using ammonium oxalate and the release of RG-I through cleavage of pectin with endo-polygalacturonase (EPG) (Graphical overview). The oxalate functions as a chelating agent, sequestering divalent cations and thereby solubilizing pectin that is stabilized in the cell wall through calcium-mediated cross-links. Other chelators, such as ethylenediaminetetraacetic acid (EDTA) and 1,2-cyclohexylenedinitrilotetraacetic acid (CDTA), have also been employed for pectin extraction [11]. However, these compounds are difficult to remove by dialysis, which can interfere with downstream analyses. Following the solubilization step, pectin is digested with EPG, producing a mixture of RG-I, rhamnogalacturonan II (RG-II), and oligogalacturonides (OGAs), which can subsequently be separated by size-exclusion chromatography (SEC). Although this protocol is demonstrated using Arabidopsis rosette leaves, it has also been successfully applied to other Arabidopsis tissues as well as to various plant species [12,13].
Materials and reagents
Biological materials
1. Arabidopsis thaliana ecotype Columbia-0 (Col-0)
2. Spirizyme Excel (Novozymes, catalog number: NAPFM084)
3. Liquozyme SC DS (Novozymes, catalog number: AUP61163)
4. Endo-polygalacturonase M2 (EPG, Megazyme, catalog number: 700004232); an alternative can be found from Neogen [Megazyme endo-Polygalacturonanase (Pectobacterium carotovorum), catalog number: E-PGALPC]
Reagents
1. 190 proof ethanol (KOPTEC, catalog number: V1105HC)
2. Chloroform (VWR, catalog number: BDH83626.4)
3. Methanol (VWR, catalog number: BDH85800400)
4. Acetone (Tedia, catalog number: AS1112_001)
5. Sodium acetate (Fisher, catalog number: S220.1)
6. Ammonium oxalate (Sigma-Aldrich, catalog number: 32304)
7. Ammonium formate (Sigma-Aldrich, catalog number: 156264)
8. Sodium formate (Sigma-Aldrich, catalog number: 247596)
9. ACS-grade hydrochloric acid (HCl) 36.5%–38% (VWR, catalog number: BDH3028)
10. Miracle-Gro 20-20-20 water-soluble all-purpose food fertilizer (Jacks Classic, catalog number: 52024)
11. Metromix 830 soil (Hummert International, Berger BM7 35% Bark HP, catalog number: 10121500)
12. Vermiculite (PALMETTO VERMICULITE, A-1 Super Fine)
13. Formic acid (Fisher Scientific, catalog number: A117-50)
Solutions
1. 80% ethanol (see Recipes)
2. Chloroform:methanol (1:1) (see Recipes)
3. 100 mM sodium acetate buffer, pH 5 (see Recipes)
4. 0.5% ammonium oxalate buffer, pH 5 (see Recipes)
5. 50 mM ammonium formate buffer, pH 5 (see Recipes)
Recipes
1. 80% ethanol
| Reagent | Final concentration | Volume |
|---|---|---|
| Ethanol | n/a | 800 mL |
| ddH2O | n/a | 200 mL |
| Total | n/a | 1,000 mL |
Store at room temperature (RT).
2. Chloroform:methanol (1:1)
| Reagent | Final concentration | Volume |
|---|---|---|
| Chloroform | n/a | 500 mL |
| Methanol | n/a | 500 mL |
| Total | n/a | 1,000 mL |
Store at RT.
3. 100 mM sodium acetate buffer, pH 5
| Reagent | Final concentration | Quantity |
|---|---|---|
| Sodium acetate | 100 mM | 13.6 g |
| ddH2O | n/a | 1,000 mL |
| Total | n/a | 1,000 mL |
Adjust the pH to approximately 5 with 1 M HCl. Store at RT.
4. 0.5% (w/v) ammonium oxalate buffer, pH 5
| Reagent | Final concentration | Quantity |
|---|---|---|
| Ammonium oxalate | 35.18 mM | 5 g |
| ddH2O | n/a | 1,000 mL |
| Total | n/a | 1,000 mL |
Adjust the pH to approximately 5 with 1 M HCl. Store at RT.
5. 50 mM ammonium formate buffer, pH 5
| Reagent | Final concentration | Quantity |
|---|---|---|
| Ammonium formate | 50 mM | 3.153 g |
| ddH2O | n/a | 1,000 mL |
| Total | n/a | 1,000 mL |
Adjust the pH to approximately 5 with formic acid. Store at RT.
Laboratory supplies
1. 1.5 mL Eppendorf tubes (AXYGEN, catalog number: MCT-150-C)
2. 250 mL Erlenmeyer flask with sidearm tabulation (Corning, catalog number: 10-180D)
3. 2 mL Eppendorf tubes (Fisher Scientific, FisherbrandTM, catalog number: 13-698-792)
4. 0.2 mL PCR tubes (ASI alkali scientific ProCycle, catalog number: 25141)
5. 0.45 μm nylon filter (Thermo Scientific, catalog number: F2517-4)
6. Rotary salad spinner (Mainstays, catalog number: MS563617582S1)
7. GF/B glass microfiber filter (GE Healthcare Life Sciences, catalog number: 1820-070)
8. Bel-ArtTM plain spinbar magnetic stirring bar (Fisher Scientific, catalog number: 03-411-782)
9. SnakeSkinTM dialysis clips (Thermo ScientificTM, catalog number: PI68011PM)
10. Nylon filter mesh fabric 0.2 mm thick (Amazon OTGMGCB, catalog number: 765015439381)
11. Büchner funnel with 90 mm pore plate (GSC International, catalog number: BFP90)
12. 250 mL, 500 mL, 1 L, and 5 L beakers (Fisher Scientific, FisherbrandTM, catalog number: FB100250)
Equipment
1. Benchtop centrifuge (Eppendorf, model: 5810R)
2. Swing bucket rotor (Eppendorf, 5810/5810R, catalog number: S-4-104)
3. Freeze dry system (Labconco Corporation, model: 4.5 FreeZone, catalog number: 7960040)
4. Water bath (VWR, model: WB05, serial no. W113C0864)
5. Superdex 75 Increase 10/300 GL column (Cytiva, catalog number: 17517401)
6. Agilent 1260 Infinity II LC system (Agilent)
7. SM208 Screen Brightness Meter Screen Luminance Meter (M&A Instruments)
8. POLYTRON homogenizer (Brinkmann)
Software and datasets
1. Agilent OpenLab CDs ChemStation Rev. C.01.10 [201]
Procedure
A. Cultivation of Arabidopsis thaliana
1. Sterilize soil (Metromix 830) by autoclaving using a liquid cycle at 121 °C, 18 psi, for 40 min. Allow the soil to cool to room temperature.
2. Mix autoclaved Metromix 830 soil and vermiculite in a 1:1 ratio. Add tap water until the soil is evenly moistened.
3. Dissolve 5.3 g of Miracle-Gro 20-20-20 water-soluble all-purpose food fertilizer in 1 L of tap water. Add the prepared solution to the cooled soil until evenly moistened, but not wet, and mix thoroughly by hand.
4. Add the soil mixture to 48-cell garden tray inserts sitting in plastic garden trays with no drain holes. Place one Arabidopsis seed per cell on the soil surface.
5. Cover the seeded trays with plastic humidity domes or plastic wrap. Place trays at 4 °C in the dark for 2 days. This step is crucial for breaking seed dormancy and ensuring uniform, high-rate germination.
6. Move the trays to a walk-in growth chamber. Set growth conditions to an 18/6 h light/dark cycle at 21 °C, with a light intensity of 120 μmol photons·m-2·s-1.
Optional: Confirm light intensity using a luminance meter at the height of the pot and record this information.
7. Open the plastic humidity domes once the seedlings have developed two true leaves that are similar in size to the cotyledons (Figure 2), allowing them to gradually transition from a high-humidity environment to a lower one. After 2 days, completely remove humidity domes.

Figure 2. An Arabidopsis seedling with two true leaves (white arrowheads). Scale bar = 1 cm.
8. Water the plants every two days by filling the plastic garden trays with water. Avoid flooding the soil.
B. Harvesting plant material
1. Four weeks after Arabidopsis germination, use scissors to cut at the base of the rosette and collect the rosette leaves.
2. Wash leaves thoroughly with tap water to remove any soil, then dry them using a salad spinner according to the manufacturer’s instructions.
Notes:
1. Plant material can also be dried using a tissue towel or another method. Make sure no water is visible on the tissue.
2. Leaves can be stored directly at -80 °C before further processing.
C. Preparation of alcohol-insoluble residue (AIR)
1. Weigh 20 g of fresh or frozen leaves into a 1 L beaker. Add 500 mL of 80% ethanol and homogenize using a POLYTRON homogenizer at speed 8 until no large tissue fragments remain. (Optional: A household blender can also be used, but ensure that the material is sufficiently ground.)
Notes:
1. The speed setting and homogenization time may vary depending on the tissue type, sample amount, container size, and volume of 80% ethanol. Hard tissues or larger sample amounts require longer homogenization, and higher speeds may be needed to achieve complete disruption. The container should not be filled more than half full, as overfilling can cause the sample to spill during homogenization, particularly at high speeds.
2. Homogenization is performed in a cold room (4 °C) in our lab to prevent endogenous plant enzymes from altering the cell wall. Overheating was not observed in our experiments, even when using Arabidopsis inflorescence stems. However, for certain plant materials that are more sensitive or more difficult to handle, a pulse-and-go approach may be useful to maintain temperature control during extraction.
3. Before and after homogenization, wash the POLYTRON tip thoroughly with running water. Carefully inspect the tip and use tweezers to remove any plant material that may be stuck. Dry the tip completely using a paper towel or a hair dryer.
2. Assemble an Erlenmeyer flask vacuum filtration setup connected to the lab vacuum line (Figure 3). Filter the homogenate through a 50 μm nylon mesh. A gentle vacuum can be applied to aid filtration.
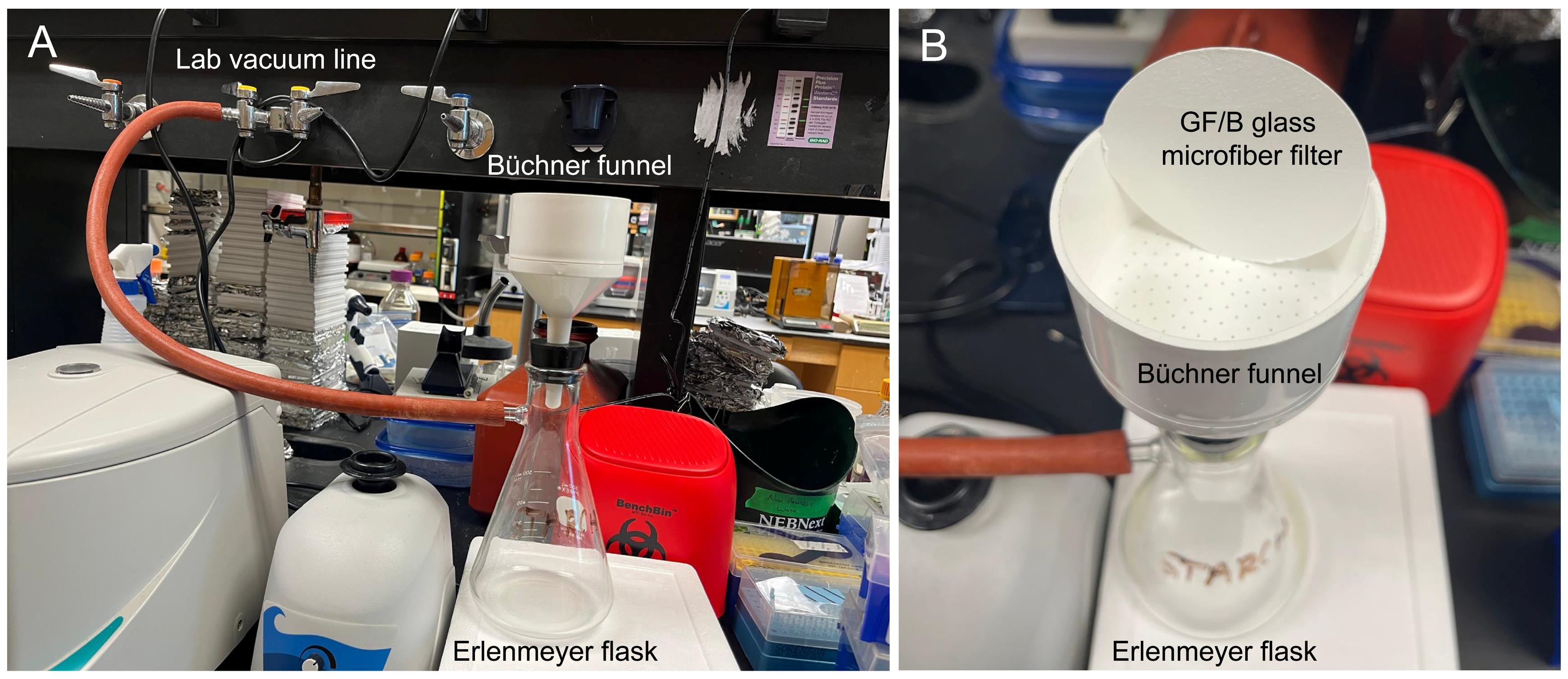
Figure 3. Vacuum filtration setup used in this protocol. (A) A Büchner funnel is assembled onto an Erlenmeyer flask, which is connected to a lab vacuum system. (B) A closer look at the Büchner funnel from the top and the GF/B glass microfiber filter.
3. Wash the residue retained on the nylon mesh by slowly adding 300 mL of 80% ethanol.
Note: 80% ethanol in the filtrate should be disposed of in appropriate hazardous waste containers.
4. Transfer the residue to a clean 500 mL beaker using a metal spatula. Add 300 mL of a 1:1 (v/v) chloroform:methanol solution. Place the beaker on a stir plate in a chemical fume hood, cover it with aluminum foil, and stir with a magnetic stir bar at 300 rpm at room temperature overnight. The next day, filter the mixture through a 50 μm nylon mesh using the setup described in step C2.
Note: The solution of 1:1 (v/v) chloroform:methanol should be disposed of in appropriate hazardous waste containers.
5. Wash the residue retained on the nylon mesh by slowly adding 300 mL of the 1:1 (v/v) chloroform:methanol solution, followed by 300 mL of acetone.
Notes:
1. Continue washing the residue with a chloroform:methanol mixture if you see a green color in the residue.
2. Acetone from the wash step should be disposed of in appropriate hazardous waste containers.
6. Transfer the washed residue using a metal spatula to a clean 500 mL beaker. Place the beaker in a chemical fume hood and cover with aluminum foil. Use a needle or sharp forceps to poke holes in the foil to facilitate evaporation and allow the sample to air-dry.
Note: Completely dried AIR appears white, whereas incompletely dried AIR is darker in color and retains the odor of acetone (Figure 4). Overnight (~16 h) is typically sufficient time to air-dry a sample from 20g of leaves, but check that the sample is completely dry prior to moving on to wall fractionation.
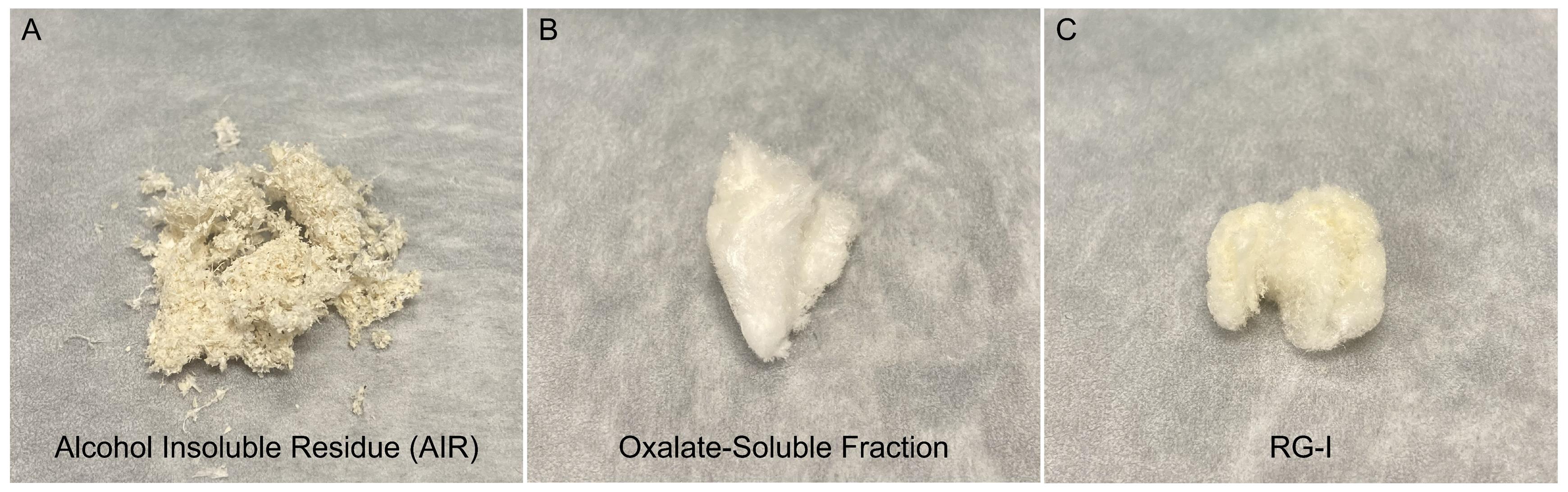
Figure 4. Representative samples from this protocol. (A) Alcohol insoluble residue (AIR) prepared from Arabidopsis rosette leaves. (B) Freeze-dried oxalate-soluble fraction. (C) Final RG-I product prepared using this protocol.
7. The dried sample is considered to be the alcohol-insoluble residue (AIR) (Figure 4). Transfer the AIR to labeled 50 mL conical tubes or Ziplock bags.
Note: AIR can be stored at room temperature and ambient humidity for years prior to downstream analyses.
8. Waste disposal: Used 80% ethanol, 1:1 (v/v) chloroform:methanol, and acetone should be disposed of in appropriate hazardous waste containers.
Note: In addition to ethanol-based AIR preparation, cell wall material (CWM) can also be prepared using methods such as 1.5% SDS extraction or phenol/acetic acid/water (PAW) treatment [14,15]. These procedures rapidly inactivate endogenous enzymes, thereby preventing unwanted structural modifications to cell wall polysaccharides during CWM preparation. The ethanol-based AIR preparation offers several advantages. It efficiently removes inorganic salts, low-molecular-weight metabolites, lipids, chlorophyll, and other pigments that could interfere with downstream analyses. The resulting material is dry and stable, allowing long-term storage at room temperature without special handling. In addition, the procedure does not introduce detergents such as SDS, which could compromise downstream experiments that are sensitive to residual SDS.
D. Preparation of destarched AIR
1. Weigh 1 g of AIR into a 500 mL flask. Add 100 mL of 0.1 M sodium acetate buffer (pH 5). We use a cocktail of two glucoamylase enzymes to hydrolyze starch into glucose, which can be easily removed by dialysis. For 1 g of AIR, add 120 μL of Spirizyme Excel and 600 μL of Liquozyme SC DS to the resuspended AIR slurry. Cover the flask with parafilm and incubate in a shaking incubator at 50 °C and 200 rpm for 24 h.
2. Filter the destarched AIR through 50 μm nylon mesh using the setup described in step C2.
3. Wash the residue retained on the nylon mesh by slowly adding 300 mL of 0.1 M sodium acetate buffer (pH 5). The resulting residue is considered destarched AIR. For pectin extraction, the destarched AIR is used directly in the next step without drying. (Optional: For long-term storage of destarched AIR, wash it with water followed by acetone as described in step C5, then air-dry and store at room temperature, same as for AIR detailed in step C7.)
Notes:
1. The filtrate from steps D2 and D3 can be collected and dialyzed, as it is enriched in arabinogalactan proteins (AGPs) and loosely bound pectins [16].
2. Liquozyme SC DS and Spirizyme Excel are commercial enzyme products containing α-amylase and amyloglucosidase, respectively. The specific activity for Spirizyme Excel is 750 amyloglucosidase units (AGU) per gram. The standard declared activity for Liquozyme SC DS is 240 Kilo Novo alpha-amylase units (KNU) per gram. In our lab, the combination of these two has been shown to efficiently remove starch. However, other commercially available α-amylases can also be used for destarching [14].
E. Pectin extraction
Note: The homogalacturonan domains of pectin are highly crosslinked by Ca2+ ions. Ammonium oxalate chelates Ca2+ ions, breaking these calcium bridges and solubilizing or releasing pectins from the cell wall.
1. Resuspend the destarched AIR from step D3 (or weigh out 1 g of dry destarched AIR) in 100 mL of 0.5% (w/v) ammonium oxalate (pH 5) in a clean 500 mL flask. Cover the flask with aluminum foil, place it in a shaking incubator, and incubate at room temperature with shaking at 200 rpm for 24 h.
2. Divide the slurry from step E1 evenly between four 50 mL conical centrifuge tubes and use 0.5% (w/v) ammonium oxalate (pH 5) to balance them if necessary. Centrifuge at 2,683× g for 10 min at room temperature. Carefully decant the supernatant into a 500 mL beaker.
3. For each tube, add 12.5 mL of 0.5% (w/v) ammonium oxalate (pH 5) and vortex the tubes for 30 s to wash the slurry. Balance the tubes using 0.5% (w/v) ammonium oxalate (pH 5) and centrifuge at 2,683× g for 10 min at room temperature. Transfer the supernatant into the same beaker.
4. Repeat step E3 two more times.
5. Filter the combined supernatants through a GF/B glass microfiber filter using the setup described in step C2.
Note: The GF/B glass microfiber filter is used in this protocol to avoid introducing contaminants, such as cellulose, that are present in regular cellulose-based filter paper. Such contaminants could interfere with the detection of pectin or other cell wall polysaccharides from plant tissue during downstream analyses.
6. The filtrate is considered to be the oxalate-soluble fraction and is enriched in pectins, including RG-I, RG-II, and HG. The final pellet is referred to as the oxalate-insoluble residue, which also contains a substantial amount of pectins.
Note: Both the oxalate-soluble fraction and the oxalate-insoluble residue contain RG-I and can be stored at -20 °C before further processing.
F. RG-I isolation from the oxalate-soluble fraction
1. Reduce the volume of the oxalate-soluble fraction to ~70 mL using a rotary evaporator with the water bath set at 37 °C.
Note: This step is optional, but it can help reduce the amount of dialysis tubing and water required.
2. Preparation of dialysis tubing: Cut the dialysis tubing (3.5 kDa MWCO) to the desired length and soak it in deionized water (DI) for at least 5 min. Rinse thoroughly with DI water.
Note: During the washing step, pinch the bottom of the tubing between two fingers, then fill it with water until taut to ensure there is no damage or punctures.
3. To set up dialysis, fill a 15 L bucket with 10 L of DI water. Secure one end of the washed dialysis tubing using a dialysis clip, placing the clip approximately 1 cm from the bottom of the tubing. (Optional: Fill the clipped tubing with DI water to check that the clip is secure and then discard water.) Over a clean secondary container or beaker, use a serological pipette to carefully transfer the concentrated oxalate-soluble fraction into the prepared dialysis tubing. Leave several centimeters of unfilled tubing at the top of the liquid for expansion during dialysis. Gently smooth out the air by closing/smoothing out the tubing between two fingers, then clip the top of the tubing with a labeled dialysis clip to seal it approximately 1 cm from the top. Place the tubing directly in the prepared bucket set upon a stir plate. Dialyze against 10 L of DI water for 24 h at room temperature while stirring gently with a magnetic stir bar at 100 rpm (Figure 5). Replace with 10 L of fresh DI water and continue dialysis for an additional 24 h.
Notes:
1. Longer dialysis times may be necessary to completely remove salts and glucose from the sample.
2. Dialysis is performed at room temperature in our lab. If temperature sensitivity is a concern, dialysis can also be carried out at 4 °C, although achieving complete buffer exchange requires more time.
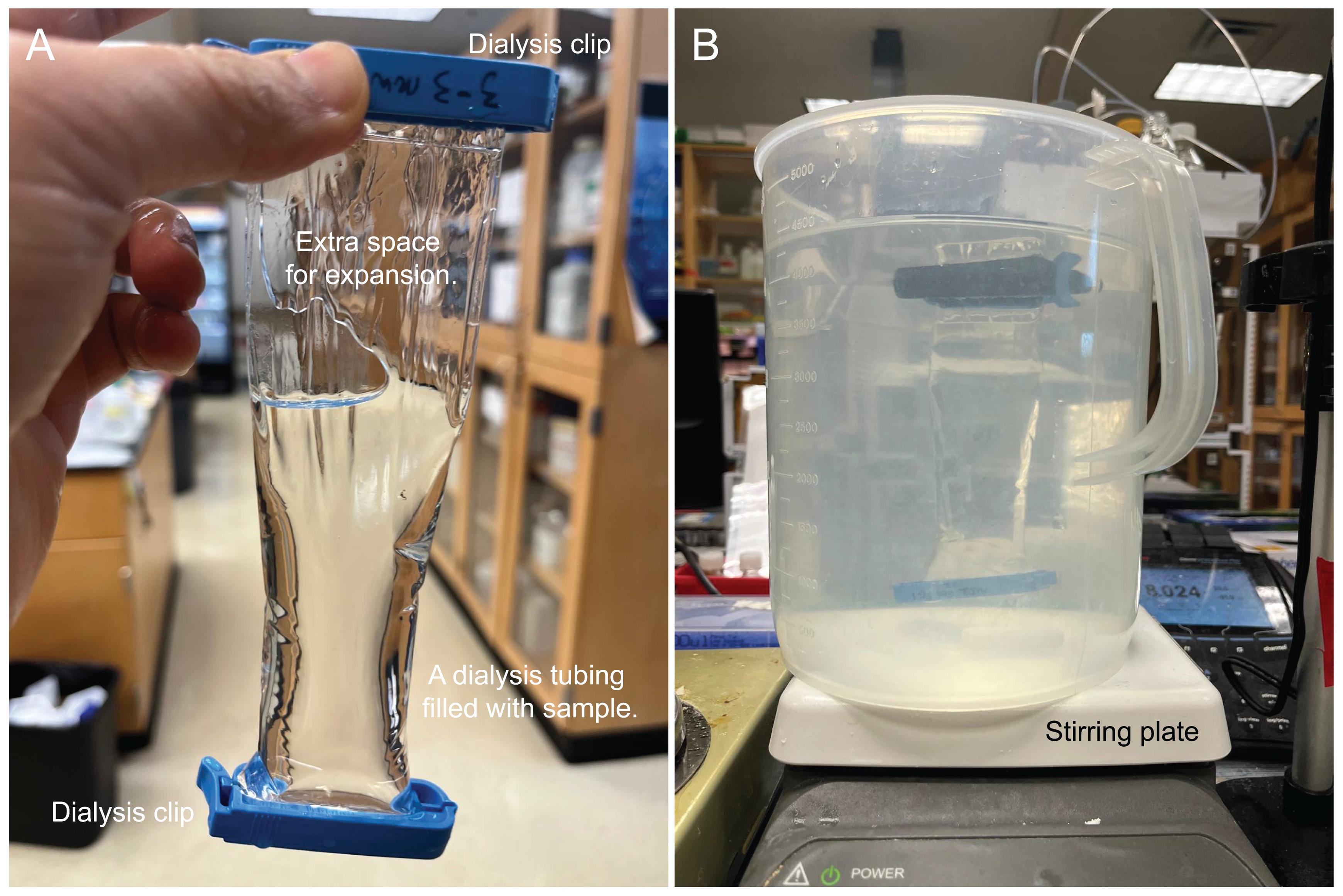
Figure 5. Dialysis setup used in this protocol. (A) Extra space is left at the top of the sample to allow for expansion during dialysis. (B) A 5 L beaker is shown here for demonstration.
4. Collect the dialyzed oxalate-soluble fraction by gently opening one dialysis clip and pipetting or pouring the solution into a 250 mL beaker.
Note: Make sure to carry out this step in a secondary container to recover any spilled sample.
5. Lyophilization: Divide the sample into multiple 50 mL conical centrifuge tubes, adding no more than 25 mL per tube. Freeze the tubes at -80 °C. Once frozen, remove the caps and cover each tube with aluminum foil or parafilm. Use a needle or sharp forceps to poke holes in the foil or parafilm to allow water to evaporate. Place the tubes in lyophilization jars and lyophilize until dry. It will take 24–48 h to fully dry the samples. A white, fluffy material should be obtained after lyophilization, and no ice should remain (Figure 4).
Notes:
1. Do not fill the centrifuge tubes more than 50% full, as the tubes tend to crack as liquids expand during freezing.
2. Pectin obtained after lyophilization can be stored at room temperature and ambient humidity for years prior to downstream analyses.
6. Endo-polygalacturonanase (EPG) digestion: Weigh 10 mg of the lyophilized oxalate-soluble fraction from step F5 into a 15 mL conical tube. Add 4 mL of 100 mM ammonium formate (pH 5.5). To release RG-I, add 4 μL of endo-polygalacturonanase M2 (approximately 0.4 U of enzyme per oxalate-soluble fraction). Place the tube in a shaking incubator and incubate at 40 °C while shaking at 200 rpm for 24 h. Filter the EPG digest through a 0.45 μm nylon spin filter.
G. Size exclusion chromatography (SEC)
1. Column preparation: Connect a Superdex 75 Increase 10/300 GL column on an Agilent 1260 Infinity II LC system equipped with a refractive index (RI) detector (or equivalent system). Equilibrate the column by washing with at least two column volumes of 50 mM ammonium formate buffer (pH 5) until the RI baseline is stable.
2. Manually inject 500 μL of the filtered EPG digest onto the column. Separate RG-I using 50 mM ammonium formate buffer (pH 5) as the mobile phase, with isocratic elution at a flow rate of 0.4 mL/min. Collect fractions corresponding to the RG-I peak, which typically elutes at ~20 min.
3. Lyophilize the RG-I eluate as described in step F5. The resulting white, fluffy material is enriched in RG-I (Figure 4).
Note: The RG-I enriched material obtained after lyophilization can be stored at room temperature and ambient humidity for years prior to downstream analyses.
H. RG-I isolation from the oxalate-insoluble residue
1. Resuspend all the oxalate-insoluble residue in 90 mL of 100 mM ammonium formate buffer (pH 5.5). To release RG-I, add 12 μL of endo-polygalacturonanase M2. Place the tube in a shaking incubator and incubate at 40 °C while shaking at 200 rpm for 24 h.
2. Filter the digest sequentially through a 50 μm nylon mesh and a GF/B glass microfiber filter. Transfer the filtrate to a 250 mL beaker and store covered at 4 °C.
3. Performing a second EPG digestion may help to increase RG-I yield. Resuspend the residue retained on the filter in 90 mL of 100 mM ammonium formate buffer (pH 5.5), add 12 μL of endo-polygalacturonanase M2, and incubate at 40 °C while shaking at 200 rpm for 24 h in a shaking incubator. Filter the digest as described in step G2.
4. Combine the filtrate from steps G2 and G3. Reduce the volume of the combined filtrate to ~40 mL using a rotary evaporator as described in step F1.
5. Dialyze the concentrated filtrate as described in steps F2–3.
6. Lyophilize the dialyzed filtrate as described in steps F4–5. A white, fluffy material should be obtained after lyophilization.
7. Isolate RG-I as described in step F7. The resulting white, fluffy material is enriched in RG-I (Figure 4).
Note: The RG-I-enriched material obtained after lyophilization can be stored at room temperature and ambient humidity for years prior to downstream analyses.
Data analysis
Chromatogram analysis is conducted with Agilent OpenLab CDs ChemStation Rev. C.01.10 [201]. Figure 6 shows representative chromatographs of the isolation of RG-I from the oxalate-soluble fraction and the oxalate-insoluble residue using size exclusion chromatography (SEC). RI signal data can be exported as a CSV file and further analyzed in Microsoft Office Excel. The RG-I peak is identified as the first eluting peak, appearing at approximately 20 min under our SEC conditions. Peak integration and normalization can be performed in ChemStation (Agilent). Automatic peak integration is available under the Integrate tab. If manual adjustments are required, such as correcting the baseline or modifying peak start and stop times, these can be made by editing the integration events in Edit Integration Events under the Method tab. Normalization can be performed in two ways. First, the RG-I peak area can be normalized to the amount of plant material used, using the following formula:
Normalized RG-I area = Area of RG-I/mg plant material
Alternatively, if an internal standard is used, the RG-I peak area can be normalized to that of the internal standard using the following formula:
Normalized RG-I area = Area of RG-I/Area of internal standard
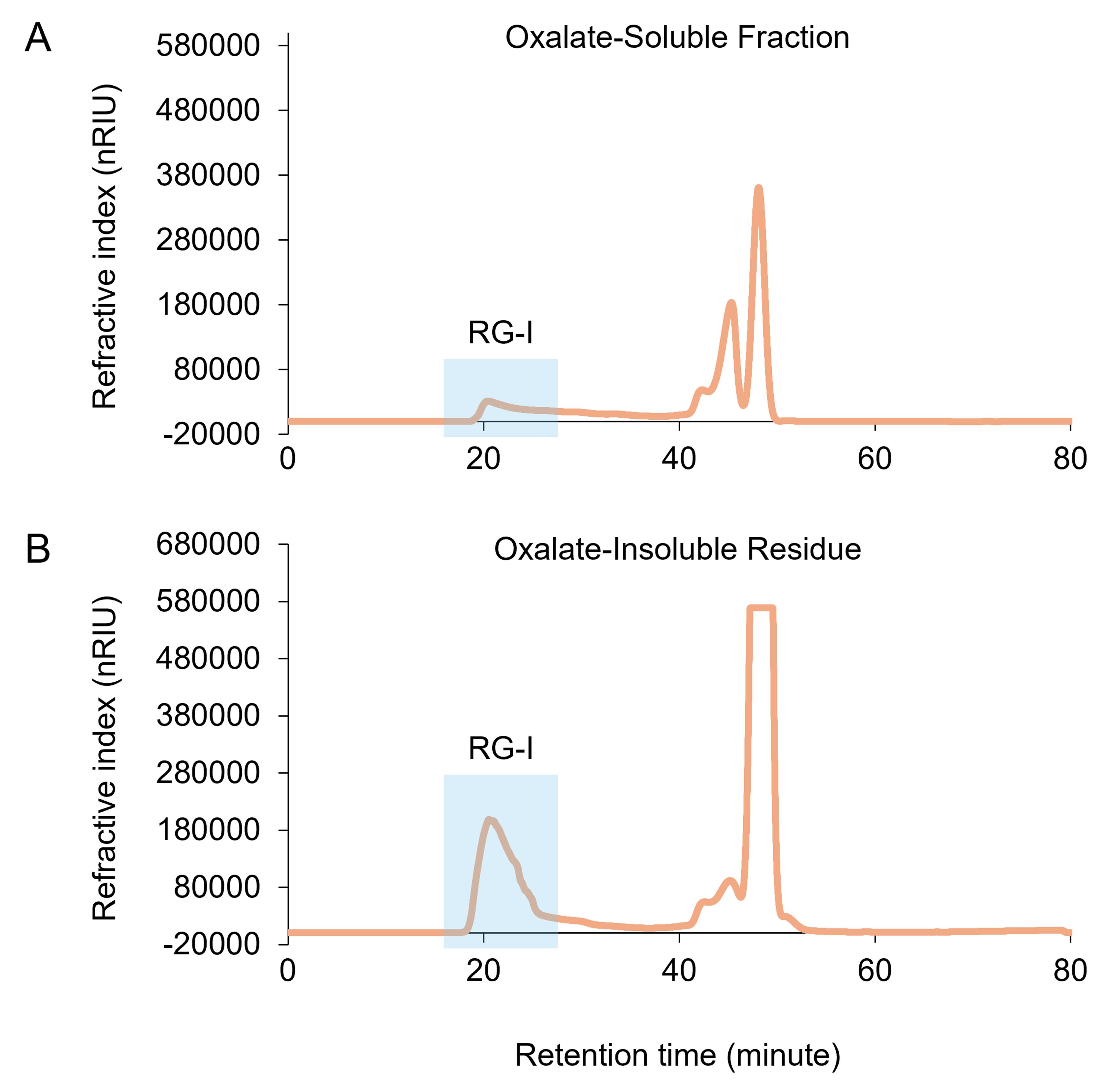
Figure 6. Isolation of RG-I from the oxalate-soluble fraction and the oxalate-insoluble residue using size exclusion chromatography (SEC). The oxalate-soluble fraction (A) and the oxalate-insoluble residue (B) were prepared from Arabidopsis rosette leaves. Peaks that elute beginning at ~20 min correspond to RG-I.
Validation of protocol
This protocol has been used and validated in the following research article [12].
• Liang Zhang, Jiri Vlach, Ian M. Black, Stephanie Archer-Hartmann, Christian Heiss, Parastoo Azadi, Breeanna R. Urbanowicz. (The pectin puzzle: Decoding the fine structure of rhamnogalacturonan-I (RG-I) in Arabidopsis thaliana uncovers new pectin features), Carbohydrate Polymers, Volume 368, Part 2, 2025, 124161, ISSN 0144-8617, https://doi.org/10.1016/j.carbpol.2025.124161.
General notes and troubleshooting
General notes
1. This protocol has been validated for the isolation of RG-I from various Arabidopsis organs, including rosette leaves, inflorescence stems, flowers, and siliques. It can also be applied to the isolation of RG-I from other plant species.
2. Endo-polygalacturonase (EPG) cleaves homogalacturonan (HG) regions within the pectin network, thereby releasing RG-I, rhamnogalacturonan-II (RG-II), and oligogalacturonides (OGAs). All three fractions can be collected using this protocol. The OGA fraction is usually the most abundant, while the abundance of RG-I and RG-II depends on the plant species and organ type.
3. We found that the oxalate-insoluble residue yields more RG-I per unit of EPG enzyme activity than the oxalate-soluble fraction.
4. Alcohol insoluble residue (AIR), oxalate-soluble fraction, oxalate-insoluble residue, and RG-I can be stored in conical centrifuge tubes or sealed containers at room temperature for years.
5. Use of sterile supplies is not required for this protocol.
6. The organic solvents used in this protocol, including ethanol, methanol, and acetone, are flammable. Chloroform and methanol are highly toxic. All steps involving these solvents should be carried out carefully in a chemical fume hood.
7. All vacuum steps in this protocol are performed using the laboratory vacuum line, operating at −25 to −26 in Hg (approximately −847 to −880 mbar) (Figure 3). Room temperature is defined as 22 °C.
8. The chamber pressure during lyophilization in this protocol is maintained at ~0.12 mbar, with shelf/sample temperature at approximately -86 °C.
Troubleshooting
Problem 1: RG-I is not pure.
Possible cause: Endo-polygalacturonase (EPG) is unable to completely degrade homogalacturonan (HG); therefore, residual HG remains attached to RG-I.
Solution: A monosaccharide composition assay can be performed to compare the ratio of galacturonic acid (GalA) to rhamnose (Rha). A pure RG-I is expected to have a GalA:Rha ratio close to 1:1. If the GalA content is higher than Rha, an additional digestion using EPG can be performed on the isolated RG-I. Highly methylesterified HG is resistant to digestion by this Megazyme EPG. Pectin methylesterase (PME) can be added during EPG digestion to remove methyl esters and improve HG hydrolysis.
Problem 2: The yield of RG-I is low.
Possible cause 1: Low abundance of pectin or RG-I in the specific tissue of interest.
Solution: A monosaccharide composition assay or a glycosyl linkage assay of the AIR sample is recommended prior to pectin isolation. If the GalA and Rha content is low, ion-exchange chromatography can be used to enrich the negatively charged pectic polysaccharides. Alternatively, the amount of plant material can be increased. However, a proportional increase in reagents, buffer, enzyme, and suitable laboratory vessels (tubes, bottles, flasks, etc.) is recommended.
Possible cause 2: The efficiency of EPG digestion is low on the oxalate-soluble fraction or the oxalate-insoluble residue derived from the specific tissue of interest.
Solution: Different HG hydrolases or lyases can be employed.
Acknowledgments
Authors’ contribution: Conceptualization, T.J. and B.U.; Investigation, L.Z.; Writing—Original Draft, L.Z.; Writing—Review & Editing, L.Z., T.J., and B.U.; Funding acquisition, B.U.; Supervision, B.U.
L.Z. and B.U. would like to acknowledge the U.S. Department of Energy, Office of Science, Basic Energy Sciences, under grant numbers DE-SC0015662 and DESC0008472, for support of research on complex pectins. B.U. would like to acknowledge the U.S. Department of Energy, Office of Science, Biological and Environmental Research, Genomic Science Program grant no. DE-SC0023223 and DE-SC0026057 for support of T.J. This protocol is adapted from Zhang et al. [12].
Items in the graphical overview figure (panels A–D), including Arabidopsis rosette leaves, a homogenizer, beaker, ash, Falcon tube, scissors, HPLC instrument, and column, were obtained from BioRender.com. The figure was assembled in Microsoft PowerPoint with the inclusion of the HPLC chromatogram and labels.
Competing interests
The authors declare no competing interests.
References
- Atmodjo, M. A., Hao, Z. and Mohnen, D. (2013). Evolving Views of Pectin Biosynthesis. Annu Rev Plant Biol. 64(1): 747–779. https://doi.org/10.1146/annurev-arplant-042811-105534
- O'Neill, M. A., Eberhard, S., Albersheim, P. and Darvill, A. G. (2001). Requirement of Borate Cross-Linking of Cell Wall Rhamnogalacturonan II for Arabidopsis Growth. Science. 294(5543): 846–849. https://doi.org/10.1126/science.1062319
- Cosgrove, D. J. (2024). Structure and growth of plant cell walls. Nat Rev Mol Cell Biol. 25(5): 340–358. https://doi.org/10.1038/s41580-023-00691-y
- Bou Daher, F., Chen, Y., Bozorg, B., Clough, J., Jönsson, H. and Braybrook, S. (2018). Anisotropic growth is achieved through the additive mechanical effect of material anisotropy and elastic asymmetry. eLife. 7: e38161. https://doi.org/10.7554/eLife.38161
- Jiang, L., Yang, S. L., Xie, L. F., Puah, C. S., Zhang, X. Q., Yang, W. C., Sundaresan, V. and Ye, D. (2005). VANGUARD1 Encodes a Pectin Methylesterase That Enhances Pollen Tube Growth in the Arabidopsis Style and Transmitting Tract. Plant Cell. 17(2): 584–596. https://doi.org/10.1105/tpc.104.027631
- Kaczmarska, A., Pieczywek, P. M., Cybulska, J. and Zdunek, A. (2022). Structure and functionality of Rhamnogalacturonan I in the cell wall and in solution: A review. Carbohydr Polym. 278: 118909. https://doi.org/10.1016/j.carbpol.2021.118909
- Reiter, W., Chapple, C. and Somerville, C. R. (1997). Mutants of Arabidopsis thaliana with altered cell wall polysaccharide composition. Plant J. 12(2): 335–345. https://doi.org/10.1046/j.1365-313x.1997.12020335.x
- Turner, S. R. and Somerville, C. R. (1997). Collapsed Xylem Phenotype of Arabidopsis Identifies Mutants Deficient in Cellulose Deposition in the Secondary Cell Wall. Plant Cell. 9(5): 689. https://doi.org/10.2307/3870425
- Prabhakar, P. K., Pereira, J. H., Taujale, R., Shao, W., Bharadwaj, V. S., Chapla, D., Yang, J. Y., Bomble, Y. J., Moremen, K. W., Kannan, N., et al. (2023). Structural and biochemical insight into a modular β-1,4-galactan synthase in plants. Nat Plants. 9(3): 486–500. https://doi.org/10.1038/s41477-023-01358-4
- Plouhinec, L., Zhang, L., Pillon, A., Haon, M., Grisel, S., Navarro, D., Black, I., Neugnot, V., Azadi, P., Urbanowicz, B., et al. (2025). Unlocking soybean meal pectin recalcitrance using a multi-enzyme cocktail approach. Sci Rep. 15(1): 1716. https://doi.org/10.1038/s41598-024-83289-4
- Renard, C. M. and Thibault, J. F. (1993). Structure and properties of apple and sugar-beet pectins extracted by chelating agents. Carbohydr Res. 244(1): 99–114. https://doi.org/10.1016/0008-6215(93)80007-2
- Zhang, L., Vlach, J., Black, I. M., Archer-Hartmann, S., Heiss, C., Azadi, P. and Urbanowicz, B. R. (2025). The pectin puzzle: Decoding the fine structure of rhamnogalacturonan-I (RG-I) in Arabidopsis thaliana uncovers new pectin features. Carbohydr Polym. 368: 124161. https://doi.org/10.1016/j.carbpol.2025.124161
- Barnes, W. J., Koj, S., Black, I. M., Archer-Hartmann, S. A., Azadi, P., Urbanowicz, B. R., Peña, M. J. and O’Neill, M. A. (2021). Protocols for isolating and characterizing polysaccharides from plant cell walls: a case study using rhamnogalacturonan-II. Biotechnol Biofuels. 14(1): 142. https://doi.org/10.1186/s13068-021-01992-0
- Gibeaut, D. M., Pauly, M., Bacic, A. and Fincher, G. B. (2005). Changes in cell wall polysaccharides in developing barley (Hordeum vulgare) coleoptiles. Planta. 221(5): 729–738. https://doi.org/10.1007/s00425-005-1481-0
- Ring, S. G. and Selvendran, R. R. (1980). Isolation and analysis of cell wall material from beeswing wheat bran (Triticum aestivum). Phytochemistry. 19(8): 1723–1730. https://doi.org/10.1016/s0031-9422(00)83802-9
- Tan, L., Cheng, J., Zhang, L., Backe, J., Urbanowicz, B., Heiss, C. and Azadi, P. (2024). Pectic-AGP is a major form of Arabidopsis AGPs. Carbohydr Polym. 330: 121838. https://doi.org/10.1016/j.carbpol.2024.121838
Article Information
Publication history
Received: Nov 7, 2025
Accepted: Dec 18, 2025
Available online: Jan 5, 2026
Published: Feb 5, 2026
Copyright
© 2026 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Zhang, L., Javaid, T. and Urbanowicz, B. R. (2026). Detailed Method for the Purification of Rhamnogalacturonan-I (RG-I) in Arabidopsis thaliana. Bio-protocol 16(3): e5579. DOI: 10.21769/BioProtoc.5579.
Category
Plant Science > Plant biochemistry > Carbohydrate
Plant Science > Plant cell biology > Cell wall
Biochemistry > Carbohydrate > Polysaccharide
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








