- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Analyzing the Translatome of Lymphatic and Venous Endothelial Cells In Vivo via Translating Ribosome Affinity Purification (TRAP)
(*contributed equally to this work) Published: Vol 15, Iss 23, Dec 5, 2025 DOI: 10.21769/BioProtoc.5516 Views: 1713
Reviewed by: Alberto RissoneAmr Galal Abdelraheem IbrahimAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Visualization of Actin Cytoskeleton in Cellular Protrusions in Medaka Embryos
Toru Kawanishi [...] Hiroyuki Takeda
Jul 5, 2023 1917 Views

Metabolic RNA Labeling and Translating Ribosome Affinity Purification for Measurement of Nascent RNA Translation
Hirotatsu Imai and Akio Yamashita
Oct 20, 2024 2544 Views

SunTag-Based Single-Molecule Translation Imaging in Caenorhabditis elegans
Elise van der Salm [...] Suzan Ruijtenberg
Oct 20, 2025 2323 Views
Abstract
Zebrafish are a powerful model for investigating vascular and lymphatic biology due to their genetic tractability and optical transparency. While translating ribosome affinity purification (TRAP) has been widely applied in other systems, its application in zebrafish has remained limited. Here, we present an optimized TRAP protocol for isolating ribosome-associated mRNAs from endothelial cells in vivo, without the need for cell dissociation or sorting. Using a novel transgenic zebrafish line, which expresses HA-tagged Rpl10a under the mrc1a promoter, we enriched actively translating endothelial transcripts. Differential expression analysis revealed robust upregulation of vascular and lymphatic genes including flt4, kdrl, and lyve1b. This approach captures the endothelial cell translatome with high specificity and offers a robust platform for investigating the molecular mechanisms of endothelial biology under genetic, environmental, or toxicological perturbations.
Key features
• Permits in vivo isolation of the endothelial ribosome without cell sorting.
• Optimized TRAP protocol to analyze lymphatic and venous endothelial gene expression in zebrafish.
• Utilizes a stable transgenic zebrafish line.
• Compatible with real-time quantitative PCR (qPCR) and next-generation sequencing (RNA-seq).
Keywords: TRAPGraphical overview
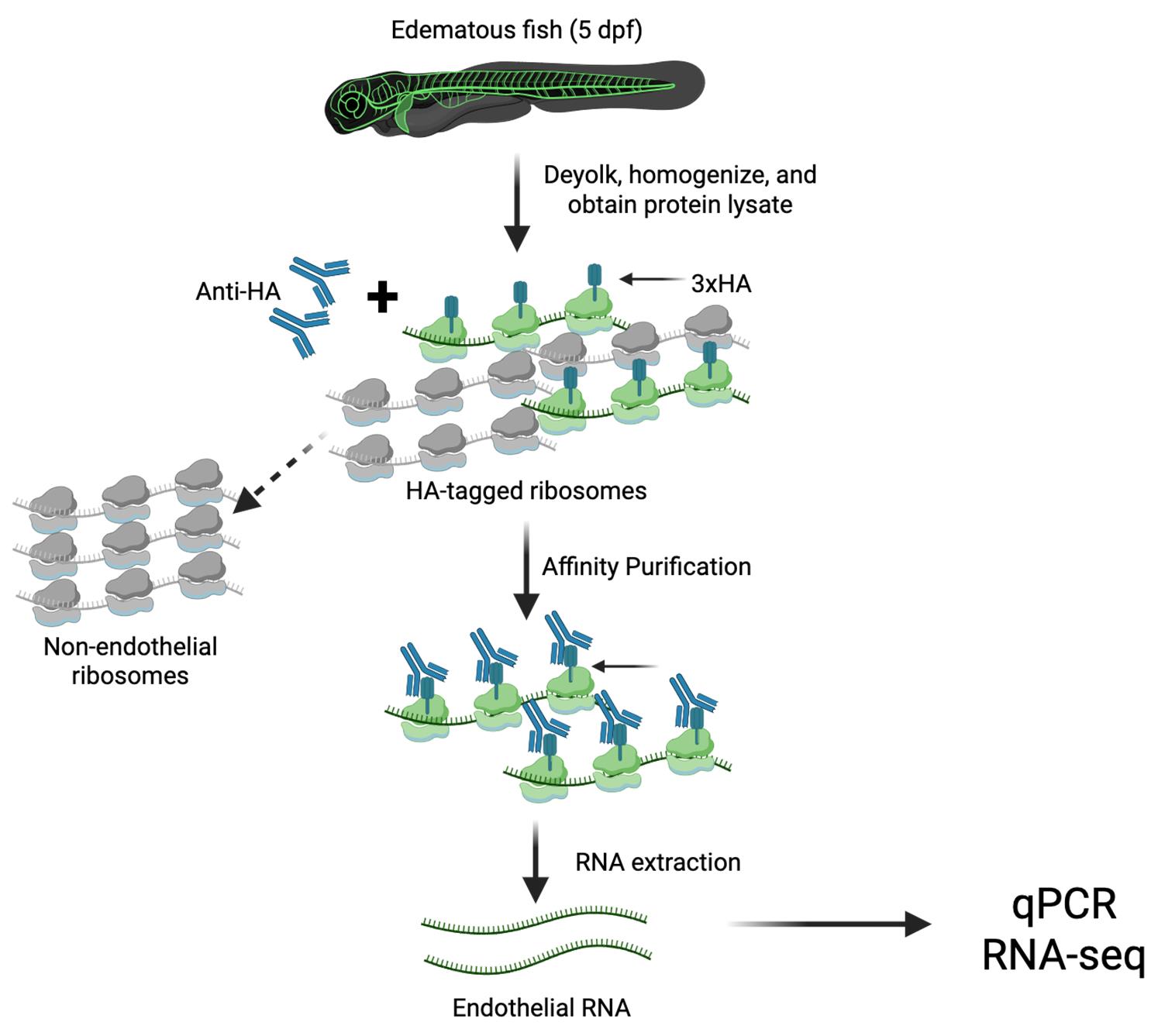
Graphical overview of the translating ribosome affinity purification (TRAP) protocol. Transgenic RiboTag zebrafish are deyolked and homogenized to generate whole-tissue lysates. Anti-HA antibodies are added to the lysate to selectively bind HA-tagged endothelial ribosomes. Magnetic Dynabeads conjugated to the antibody Fc domain enable affinity purification of the HA-tagged ribosomes using a magnetic stand. Unbound, non-endothelial ribosomes remain in the supernatant, while the Dynabead–αHA-ribosome–mRNA complex is subjected to RNA extraction to isolate actively translated endothelial RNA. This protocol provides an in vivo snapshot of cell type–specific translational activity.
Background
The blood and lymphatic systems form an integrated vascular network that delivers oxygen and nutrients, removes metabolic waste, and maintains fluid balance and homeostasis. These networks are composed of endothelial cells, whose functions are tightly regulated under both physiological and pathological conditions [1,2]. Disruption of endothelial regulation can impair fluid balance, resulting in fluid accumulation and tissue edema. In response to edemagenic signals in the microenvironment, lymphatic and venous endothelial cells activate molecular programs that promote edema clearance [3].
Zebrafish have emerged as a valuable animal model for studying vascular development due to their optical transparency, genetic tractability, and evolutionary conservation of endothelial gene function [4–7]. In our recent study, we utilized live imaging of early-stage zebrafish larvae exposed to abrupt salinity changes to induce osmotic stress and tissue edema. This model revealed increased lymphangiogenesis and remodeling of the lymphatic vasculature during edema resolution, characterized by proliferation of lymphatic progenitors and expansion of the primary lymphatic network [3].
Although endothelial responses to edema are generally initiated at the transcriptional level, post-transcriptional mechanisms, including translational regulation, likely contribute to dynamic changes in gene expression [8,9]. Transcriptome profiling tools such as microarrays and RNA sequencing from total RNA provide insights into RNA abundance, but they do not reflect the state of translation activity [10,11]. Since protein synthesis is determined by mRNAs engaged with actively translating ribosomes, profiling the translatome offers a more accurate representation of functional gene expression.
Conventional transcriptome analyses rely on fluorescence-activated cell sorting (FACS) of labeled endothelial cells, which requires mechanical and chemical dissociation into single-cell suspensions [12,13]. However, this process can disrupt native cell–cell and cell–matrix interactions, potentially altering gene expression and introducing artifacts. Translating ribosome affinity purification (TRAP) using the RiboTag strategy provides a powerful alternative for cell type–specific isolation of ribosome-associated mRNAs in vivo, bypassing the need for cell sorting [14–20]. The rapid, non-disruptive nature of TRAP minimizes ex vivo artifacts and preserves in vivo gene expression profiles.
To enable endothelial-specific TRAP in zebrafish, we developed a transgenic line Tg(mrc1a:egfp-2a-rpl10a-3xHA)y723, expressing hemagglutinin (HA)-tagged Rpl10a and eGFP via a viral 2A peptide under control of the endothelial-specific mrc1a promoter [3]. Following osmotic challenge, larvae display vascular remodeling and enable the isolation of actively translating mRNAs from lymphatic and venous endothelial cells. Translatome analysis revealed upregulation of key endothelial genes such as flt4, kdrl, and lyve1b [3]. Coupling TRAP with RNA sequencing allows for comprehensive identification of genes involved in endothelial responses to edema.
This protocol describes an optimized TRAP workflow for profiling the endothelial translatome in edematous zebrafish. It provides a robust, reproducible method for capturing dynamic gene expression changes during edema formation and resolution.
Materials and reagents
Biological materials
1. Stable transgenic zebrafish line: Tg(mrc1a:egfp-2a-rpl10a-3xHA)y723
Reagents
1. Invitrogen DynabeadsTM Protein G (ThermoFisher, catalog number: 10004D)
2. Direct-zolTM RNA MicroPrep (ZymoResearch, catalog number: R2060)
3. 100 μg/mL cycloheximide (Sigma, catalog number: C7698)
4. Protease inhibitor cocktail (Sigma, catalog number: P8340)
5. PierceTM Bradford Protein Assay kit (ThermoFisher, catalog number: 23200)
6. 1 mM DTT (Sigma, catalog number: 646563)
7. 200 units/mL RNAsin (Promega, catalog number: N2115)
8. 1 mg/mL heparin (Sigma, catalog number: H3393-10KU)
9. 10% NP-40 (Roche, catalog number: 11-332-473-001)
10. Anti-HA antibody (Abcam, catalog number: ab9110)
11. TRIzol (Ambion Life Technologies, catalog number: 15596026)
12. Glycogen RNA-grade (ThermoFisher, catalog number: R0551)
13. High-Capacity cDNA Reverse Transcription kit (Applied Biosystem, catalog number: 4374966)
14. TaqMan Fast Advanced Master Mix (Applied Biosystem, catalog number: 4444557)
15. Phosphate-buffered saline (PBS), pH 7.4 (Fisher, catalog number: BP2438)
16. Pronase from Streptomyces griseus (20 mg/mL) (Sigma, catalog number: 10165921001)
17. Ethyl alcohol 200 proof (Sigma, catalog number: E7023-1L)
18. Chloroform HPLC-grade (Fisher, catalog number: C606SK-1)
19. Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S3014-1KG)
20. Potassium chloride (KCl) (Fisher Chemical, catalog number: P217-500)
21. Sodium bicarbonate (NaHCO3) (Fisher Chemical, catalog number: S233-500)
22. Calcium chloride (CaCl2) (MP Biomedicals, catalog number: 193819)
23. Magnesium chloride (MgCl2) (Invitrogen, catalog number: AM9530G)
24. Calcium nitrate hydrate [Ca(NO3)2·xH2O] (Alfa Aesar, catalog number: 44515-14)
25. Magnesium sulfate heptahydrate (MgSO4·7H2O) (Sigma-Aldrich, catalog number: 230391-25G)
26. Tris pH 8.5 (Teknova, catalog number: 21327)
27. Tris-HCl pH 7.4 (Teknova, catalog number: T1074)
28. Molecular grade water (H2O) (Corning, catalog number: 46-000-CM)
29. HEPES (Fisher Bioreagents, catalog number: BP310-500)
Solutions
1. Deyolking buffer (see Recipes)
2. Deyolking wash buffer (see Recipes)
3. Homogenization buffer (HB) (see Recipes)
4. Homogenization buffer+ (HB+) (see Recipes)
5. High salt buffer (HSB) (see Recipes)
6. High salt buffer+ (HSB+) (see Recipes)
7. Hypertonic salt solution (3× Danieau buffer) obtained from 10× Danieau buffer stock (see Recipes)
8. High-capacity RT mix for cDNA synthesis (see Recipes)
9. TaqMan Real-Time PCR (see Recipes)
Recipes
1. Deyolking buffer
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 1 M NaCl | 55 mM | 5.50 mL |
| 1 M KCl | 1.9 mM | 0.19 mL |
| 1 M NaHCO3 | 1.25 mM | 0.125 mL |
| H2O | n/a | 94.185 mL |
| Total | n/a | 100 mL |
Store at 4 °C.
2. Deyolking wash buffer
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 1 M NaCl | 110 mM | 11.00 mL |
| 1 M KCl | 3.5 mM | 0.35 mL |
| 1 M CaCl2 | 2.7 mM | 0.27 mL |
| 1 M Tris (pH 8.5) | 10 mM | 1.00 mL |
| H2O | n/a | 87.38 mL |
| Total | n/a | 100 mL |
Store at 4 °C.
3. Homogenization buffer (HB)
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 1 M Tris-HCl (pH 7.4) | 50 mM | 5.00 mL |
| 1 M KCl | 100 mM | 10.00 mL |
| 1 M MgCl2 | 12 mM | 1.20 mL |
| 10% NP-40 | 1% | 10 mL |
| H2O | n/a | 73.80 mL |
| Total | n/a | 100 mL |
Store at 4 °C.
4. Homogenization buffer+ (HB+)
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| DTT | 1 mM | 10 μL |
| Protease inhibitor cocktail | 0.01× | 100 μL |
| RNAsin | 200 units/mL | 50 μL |
| Cycloheximide | 100 μg/mL | 50 μL |
| Heparin | 1 mg/mL | 100 μL |
| HB (see Recipe 3) | n/a | 9.69 mL |
| Total | n/a | 10 mL |
Freshly prepared on ice.
5. High salt buffer (HSB)
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 1 M Tris (pH 7.4) | 50 mM | 5.00 mL |
| 1 M KCl | 300 mM | 30.00 mL |
| 1 M MgCl2 | 12 mM | 1.20 mL |
| 10% NP-40 | 1% | 10.00 mL |
| H2O | n/a | 53.80 mL |
| Total | n/a | 100 mL |
Store at 4 °C.
6. High salt buffer+ (HSB+)
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| DTT | 1 mM | 10 μL |
| Protease inhibitor cocktail | 0.01× | 100 μL |
| RNAsin | 200 units/mL | 50 μL |
| Cycloheximide | 100 μg/mL | 50 μL |
| Heparin | 1 mg/mL | 100 μL |
| HSB (see Recipe 5) | n/a | 9.69 mL |
| Total | n/a | 10 mL |
Freshly prepared on ice.
7. Hypertonic salt solution (3× Danieau buffer) obtained from 10× Danieau buffer stock
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 580 mM NaCl | 58 mM | 10.00 mL |
| 6.71 mM KCl | 0.7 mM | 10.43 mL |
| 8.22 mM MgSO4 | 0.4 mM | 4.87 mL |
| 8.53 mM Ca(NO3)2 | 0.6 mM | 7.03 mL |
| 50 mM HEPES | 10 mM | 20.0 mL |
| H2O | n/a | 47.67 mL |
| Total | n/a | 100 mL |
Store at 4 °C.
8. High-capacity RT mix for cDNA synthesis
| Component | Quantity or volume |
|---|---|
| 10× RT buffer | 2.0 µL |
| 25× dNTP Mix | 0.8 µL |
| 10× RT random primers | 2.0 µL |
| MultiScribeTM reverse transcriptase | 1.0 µL |
| RNase inhibitor | 1.0 µL |
| Nuclease-free H2O | 3.2 µL |
| RNA template (100 ng) | 10 µL |
| Total per reaction | 20 µL |
Volumes per reaction. Store at -20 °C.
9. TaqMan real-time PCR
| Component | Quantity or volume |
|---|---|
| TaqMan Fast Advanced Master Mix (2×) | 5.0 µL |
| TaqMan target gene primers FAM (20×) | 0.5 µL |
| TaqMan reference gene primers VIC (20×) | 0.5 µL |
| Nuclease-free H2O | 2.0 µL |
| cDNA (1:5) | 2.0 µL |
| Total per reaction | 10 µL |
Volumes per reaction. Store at -20 °C.
Laboratory supplies
1. 1.5 mL microcentrifuge tubes (ABDOS, catalog number: P10202)
2. RNase-free elution tubes (Invitrogen, catalog number: AM12480)
3. VWR disposable pestles (VWR, catalog number: 47747-358)
4. SureOne sterile pipette tips (FisherBrand, catalog number: 02-707-442)
Equipment
1. Magnetic Eppendorf stand (ThermoFisher, catalog number: 12321D)
2. SorvallTM LegendTM Micro 21R Microcentrifuge 4 °C microfuge (ThermoFisher, catalog number: 75002445)
3. 4 °C Gentle Eppendorf tube rotator (Fisher, catalog number: 11-676-342)
4. Zymo-DR Duet kit (ZymoResearch, catalog number: R2060)
5. Labnet centrifuge (Thomas Scientific, catalog number: 1160W29)
6. Vortex (Fisher, catalog number: 88-882-011)
7. Zeiss Stemi 305 microscope (Zeiss, catalog number: 435063-9020-100)
8. Biovortexer (BioSpec, model: 1083MC)
9. NanoDrop (Thermo Scientific, catalog number: 13400518)
10. Confocal laser scanning microscope (Zeiss, model: LSM880)
11. Quantitative PCR machine (Applied Biosystems, model: Quant Studio 7 Flex)
Software and datasets
1. GraphPad Prism Software, version 9.2.0
Procedure
Day 1
A. Experimental animal setup
1. Set up breeding tanks with an equal number of male and female Tg(mrc1a:egfp-2a-rpl10a-3xHA)y723 zebrafish. To synchronize embryo collection at a defined developmental stage, place a divider in the tank to separate males and females overnight. Although both homozygous and heterozygous transgenic embryos can be used, homozygous transgenic animals allow for better yield.
Day 2
B. Collect embryos/larvae
1. In the morning, collect fertilized eggs and maintain them at 28.5 °C under standard zebrafish husbandry conditions, in accordance with institutional animal care guidelines.
2. Collect embryos at the desired developmental stage. The mrc1a promoter drives vascular transgene expression prior to 24 h post-fertilization (hpf) and continues to adulthood [6].
Note: This protocol was optimized using embryos at 3 days post-fertilization (dpf).
C. Dechorionate embryos
1. Add pronase (20 mg/mL stock solution) to 0.3× Danieau solution at a final concentration of 2 mg/mL.
2. Incubate embryos at 28.5 °C until the chorions are digested and begin to disintegrate.
3. Wash three times with 0.3× Danieau solution and collect the dechorionated embryos.
D. Induce edema
1. Subject dechorionated 3 dpf embryos to hypertonic conditions by incubating them in 3× Danieau solution for 24 h at 28.5 °C (Figure 1A).
2. Maintain control embryos in 0.3× Danieau solution under the same conditions.
3. After 24 h, transfer the treated embryos to 0.3× Danieau solution and incubate for an additional 24 h. Induced edema is visible with a brightfield microscope at 4 dpf (Figure 1B).
4. Collect both edematous and control larvae at desired developmental stages and proceed with TRAP protocol. This protocol used 5 dpf larvae. Samples are processed simultaneously to minimize batch effects.
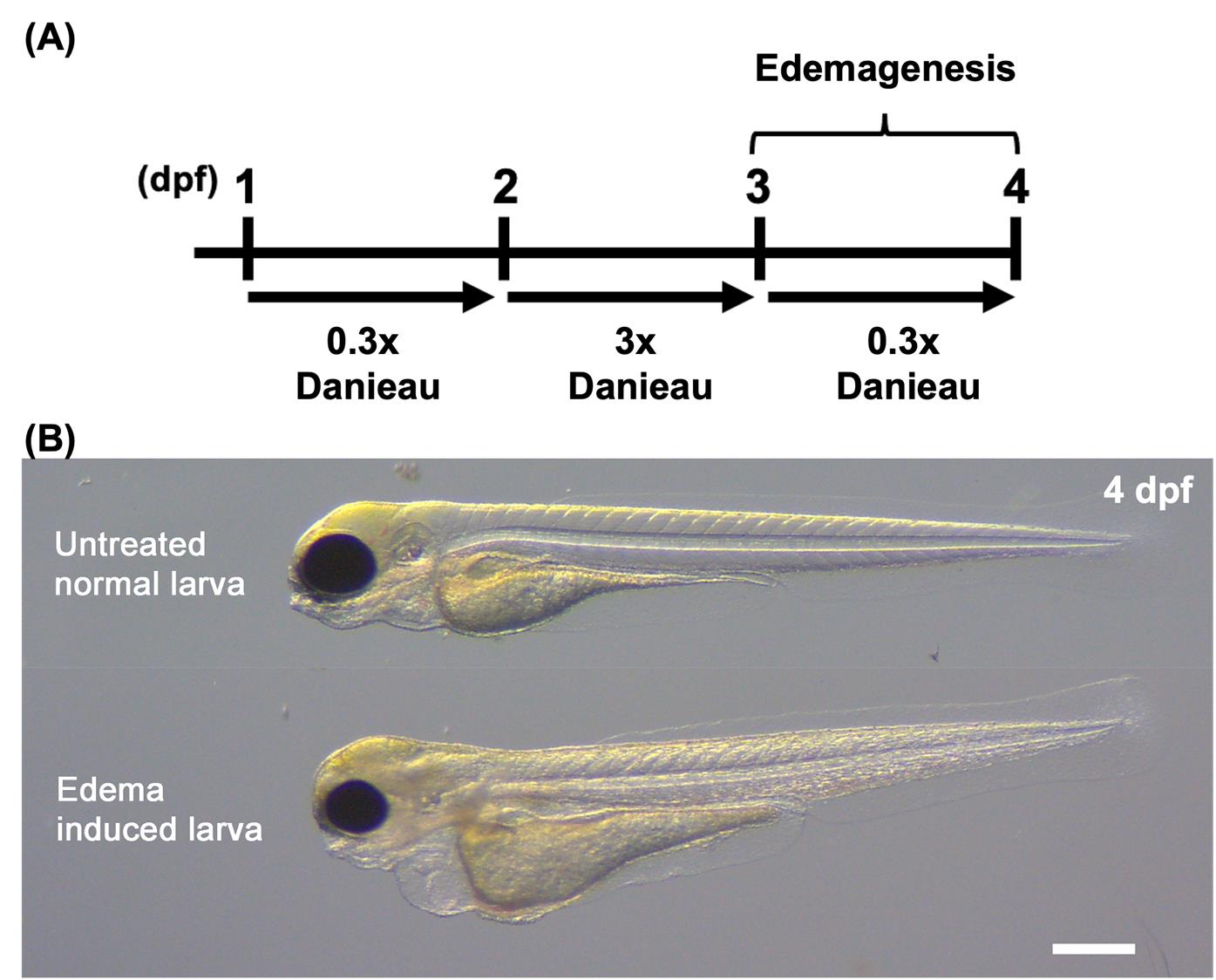
Figure 1. Schematic of edema induction. (A) Timeline of the edema induction protocol. Larvae were treated with 3× Danieau solution from 2 to 3 dpf. Edemagenesis was induced by replacing the medium with 0.3× Danieau solution at 3 dpf to impose osmotic stress. (B) Brightfield image of a control (untreated) larva and edema-induced larva at 4 dpf. Scale bar, 500 µm. More detailed information on the edema induction protocol is available in Olayinka et al. [3].
E. Buffer preparation
1. Chill all buffers on ice or at 4 °C prior to use.
2. Thaw all necessary inhibitors for HB+ buffer preparation.
Note: Cycloheximide may require warming to 30 °C to thaw completely.
3. Prepare HB+ buffer by adding inhibitors to the homogenization buffer (HB) immediately before use. Keep the HB+ buffer on ice.
F. Deyolk embryos
1. Transfer 200–500 embryos from each embryo group (edematous and control) into separate 1.5 mL microcentrifuge tubes and remove all remaining 0.3× Danieau solution.
2. Add 1 mL of deyolking buffer supplemented with 10 μL of protease inhibitor cocktail.
3. Gently disrupt the yolk by pipetting up and down using a 200 μL pipette tip. Confirm yolk separation under a stereomicroscope.
4. Centrifuge at 300× g for 30 s at 4 °C and discard the supernatant.
5. Wash the embryos three times with 1 mL of cold deyolking wash buffer, centrifuging at 300× g for 30 s at 4 °C each time. Repeat until the supernatant is clear.
6. Perform a final wash with cold PBS. Centrifuge at 300× g for 30 s, then discard the supernatant.
G. Embryo homogenization
1. Add 2 μL of HB+ buffer per embryo to the deyolked pellet for each group (edematous and control).
2. Homogenize thoroughly using a Biovortexer with pestles. Avoid introducing bubbles during homogenization.
3. Incubate the homogenate on ice for 10 min to ensure complete cell lysis.
4. Centrifuge at 3,000× g for 10 min at 4 °C. Transfer the supernatant to a new tube and discard the pellet.
5. Repeat the centrifugation of the supernatant phase at 10,000× g for 10 min at 4 °C to remove cellular debris.
6. Collect the final supernatant in a new 1.5 mL tube and keep it on ice before proceeding to immunoprecipitation.
H. Protein quantification
1. Quantify the total protein concentration from each embryo lysate (edematous and control) using the Bradford protein assay, following the manufacturer’s instructions.
Note: Use BSA as a standard and perform measurements in triplicate to ensure accuracy.
I. Set up indirect immunoprecipitation (IP)
1. Add 1 μL of anti-HA antibody per 400 μg of protein into the lysate supernatant collected from section G.
2. Incubate the mixture on a tube rotator at 4 °C for 4 h to allow antibody-ribosome complex formation.
J. Prepare Dynabeads Protein G
1. Vigorously shake the Dynabeads Protein G tube to resuspend the beads evenly in the storage buffer.
2. Dispense 57 μL of Dynabeads per 400 μg of protein to a 1.5 mL tube.
3. Place the tube on a magnetic stand and allow the beads to collect on the tube wall; carefully remove and discard the storage buffer (Figure 2).
4. Take out the tube from the magnetic stand and add 800 μL of HB+ buffer to the beads.
5. Use a tube rotator to equilibrate beads to HB+ buffer by rotating at 4 °C for 1 h.
K. Affinity purification
1. Remove the HB+ buffer from the equilibrated Dynabeads using a magnetic stand (Figure 2).
2. Add 800 μL of the HA-antibody and lysate mixture to the beads.
3. Incubate the mixture overnight (~16 h) at 4 °C on a tube rotator to allow affinity binding.

Figure 2. Image illustrating Dynabead-bound endothelial ribosome lysates and unbound protein lysates on a magnetic Eppendorf stand
Day 3
L. Wash beads
1. Remove and save the supernatant (unbound protein lysate) from each sample on ice (Figure 2).
2. Wash the beads three times with 800 μL of HSB+.
a. Rotate at 4 °C for 5 min.
b. Use a magnetic stand to collect the beads for 3 min and carefully remove the wash buffer (Figure 2).
3. After the second wash, transfer the beads to a fresh tube to reduce background before the final wash.
M. RNA isolation from the bead–antibody complex
1. Add 500 μL of TRIzol reagent directly to the antibody-bound beads.
2. Incubate on ice for 5 min to ensure thorough lysis.
3. Add 100 μL of chloroform and then vortex vigorously for 30 s.
4. Incubate the mixture at room temperature for 3 min.
5. Centrifuge at 12,000× g for 15 min at 4 °C. The mixture separates into a lower phenol-chloroform phase, an interphase, and a colorless upper aqueous phase.
6. Carefully transfer the upper aqueous phase to a fresh RNase-free tube.
7. Add 1.5× volume of 100% ethanol and 1 μL of glycogen as a carrier.
8. Purify RNA using the Direct-zolTM RNA MicroPrep kit according to the manufacturer’s instructions.
9. Quantify RNA concentration by nanodrop.
10. Store RNA at -80 °C or continue immediately.
Note: This protocol used 200–500 larvae at 5 dpf, collecting an RNA yield ranging from 400 to 600 ng. Samples should be pooled for RNA isolation to achieve sufficient yield for certain downstream applications (e.g., RNA-seq).
Data analysis
qPCR amplification curves and threshold cycle data were exported in .xlsx format for processing in Microsoft Excel. For each vascular gene, the ΔCT value was calculated by subtracting the cycle threshold (CT) of the reference gene (eef1a1) from the corresponding CT of the target (Ct_target–Ct_eef1a1). The ΔΔCT values were then obtained by subtracting the mean ΔCT of the supernatant control sample from each ΔCT value in both supernatant (sup) and purified endothelial (Endo) RNA. Fold changes in gene expression were calculated using the 2-ΔΔCT method. Replicate fold changes were averaged for each vascular gene and compared between endothelial and supernatant samples (Figure 3). The results show that the TRAP protocol, using the RiboTag strategy, efficiently enriches for specific endothelial gene markers such as lyve1b, flt4, and cdh5, while not enriching for ubiquitous genes like n4bp1 (Figure 3A). This technique enables targeted analysis of gene expression for selected cell types within complex tissue samples and can be applied in a range of clinical and research contexts to profile cell type–specific gene expression (Figure 3B).
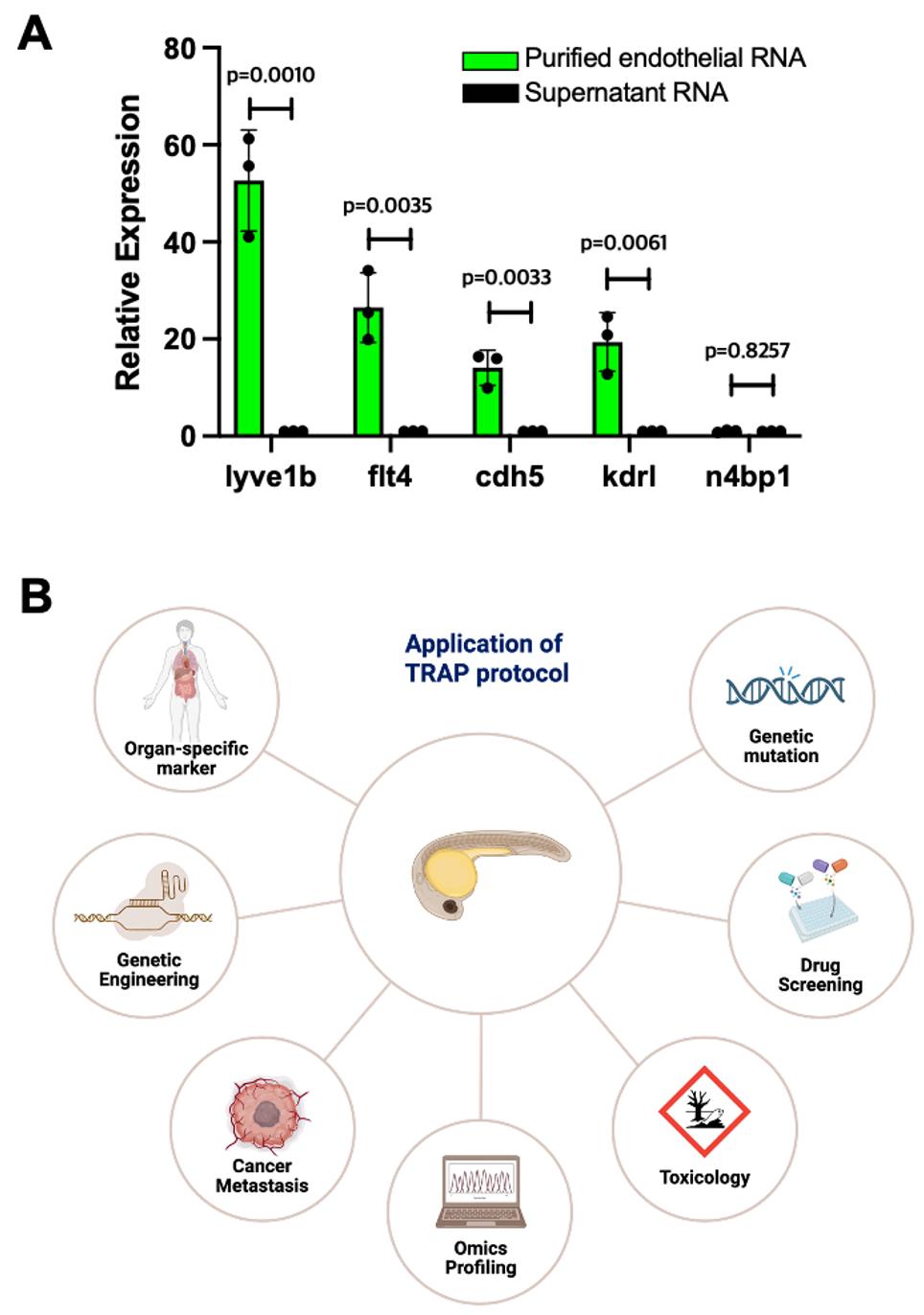
Figure 3. Analyzing endothelial translatome using the translating ribosome affinity purification (TRAP) assay. (A) Real-time qPCR showing specific enrichment of endothelial genes lyve1b, flt4, cdh5, and kdrl in the purified endothelial RNA, compared to non-endothelial RNA from the supernatant. n4bp1, a ubiquitous gene, served as a negative control. Fold change values were imported into GraphPad Prism version 9.2.0 for visualization and statistical analysis. Data were plotted as mean ± SD. An unpaired two-tailed Student’s t-test was used to compare the groups. Significance was defined as p-value < 0.05. (B) Diverse clinical and biomedical research themes where the TRAP protocol can be employed.
Validation of protocol
Purified endothelial and supernatant RNA samples were normalized to equal concentrations and reverse transcribed using the High-Capacity cDNA Reverse Transcription Kit following the manufacturer’s instructions. Each reaction contained an equal volume (10 µL) of master mix (Recipe 6) and normalized RNA in a PCR tube and was incubated in a thermal cycler under the following conditions: 25 °C for 10 min, 37 °C for 120 min, 85 °C for 5 min, and then held at 4 °C. The resulting cDNA was stored at -20 °C or used immediately for qPCR.
cDNA samples were diluted 1:5 with nuclease-free water and kept on ice. Real-time multiplex PCR reactions were performed using the TaqMan Fast Advanced Master Mix (Recipe 9), with reaction volumes adjusted according to the number of target genes and the number of technical replicates. Gene-specific FAM-labeled TaqMan probes were used for vascular targets, while a VIC-labeled probe targeting the reference gene eef1a1α served as the internal control. Multiplex reactions were prepared in a 96-well fast optical plate, sealed, and briefly centrifuged to ensure homogeneity. Plates were loaded into the Quant Studio 7 Flex Real-Time PCR System and amplified under the following conditions: hold at 95 °C for 2 min, followed by 40 cycles of denaturation at 95 °C for 15 s and annealing/extension at 60 °C for 30 s.
This protocol (or parts of it) has been used and validated in the following research article:
Olayinka et al. [3]. Compensatory lymphangiogenesis is required for edema resolution in zebrafish. Scientific Reports.
General notes and troubleshooting
General notes
1. Screen all embryos for transgenic expression prior to TRAP to ensure sample consistency. Both homozygous and heterozygous embryos can be used; however, a larger sample pool is recommended when embryos are heterozygous.
2. Perform all steps following edema induction on ice to preserve RNA integrity and protein complexes.
3. Prepare all supplemented buffers fresh on the day of the experiment to maintain reagent activity.
4. Handle samples gently throughout the protocol to minimize material loss and maximize yield.
Troubleshooting
| Step | Problem | Possible cause | Solution |
|---|---|---|---|
| Embryo collection | Low fertilization or embryo yield | Suboptimal breeding conditions, unhealthy or aged fish | Use healthy breeding pairs; optimize tank setup and timing; separate breeding pairs overnight; avoid overcrowding. |
| Edema formation | Mild edema or normal appearance | Incorrect hypertonic solution composition or timing | Verify hypertonic solution composition; shake well before use; induce edema by 3 dpf at the latest. |
| Deyolking | Embryo damage or lysis | Aggressive pipetting or narrow pipette tips | Use wide-bore pipette tips (200 µL) and pipette gently to minimize mechanical stress. |
| Homogenization | Incomplete tissue lysis | Insufficient grinding or short homogenization and incubation time | Use a motorized pestle; homogenize thoroughly without air bubbles; incubate lysate on ice for at least 10 min. |
| Lysate | Low protein yield | Too few embryos or inadequate lysis | Use approximately 2 μL of HB+ buffer per embryo; verify lysis visually under a microscope. |
| IP setup | Nonspecific binding | Poor antibody specificity or inadequate washing | Use validated anti-HA antibody; prepare fresh cold HSB+ for washes; increase washes to 5 times. |
| IP setup | Bead clumping or loss | Prolonged incubation or harsh pipetting | Use gentle rotation; avoid foam formation; minimize bead handling during pipetting. |
| IP setup | Weak binding to HA-Rpl10a | Insufficient antibody concentration or low transgene expression | Confirm antibody activity; use 1 μL of antibody per 400 μg of protein; verify transgene expression at 2 dpf. |
| Washing | High background in eluate | Incomplete wash steps | Perform all washes with cold HSB+; transfer beads to a fresh tube after the second wash. |
| RNA isolation | Poor RNA yield | Loss of aqueous phase during phase separation | Use phase-lock gel tubes and pipette carefully to collect the aqueous phase. |
| RNA integrity | RNA degradation characterized by a RIN score under 7 or smearing | RNase contamination | Use RNase-free tubes, tips, and reagents; clean surfaces with RNaseZap; include DNase treatment. Avoid multiple freeze-thawing of RNA samples. |
| RNA quality | Ineffective inhibitors | Inhibitors degraded or thawed improperly | Always thaw inhibitors on ice; prepare HB+ fresh; avoid repeated freeze-thaw cycles. |
| RNA precipitation | Poor RNA yield | Missing carrier or low RNA concentration | Add 1 μL of glycogen and 1.5× volume 100% ethanol for precipitation; concentrate RNA using cleanup kits. |
Acknowledgments
The specific contributions of each author are as follows: Conceptualization, H.M.J.; Investigation, O.O. and L.Z.; Writing—original draft, O.O. and L.Z.; Writing—review and editing, H.M.J. This work was supported by the American Heart Association (Grant #24PRE1199881 to O.O.) and the UIC College of Medicine start-up fund to H.M.J. This protocol was initially described in Miller et al. (2019) and was subsequently modified and optimized in Olayinka et al. (2025).
The following figures were created using BioRender: Graphical overview, https://BioRender.com/vs4grkh; Figure 3B, https://BioRender.com/44gaqew.
Competing interests
The authors declare no conflicts of interest.
Ethical considerations
All procedures followed the required guidelines and regulations, conforming to the ARRIVE guidelines. Zebrafish husbandry and research protocols were approved by the University of Illinois Animal Care and Institutional Biosafety Committee (Animal Care and Use Protocol 23–112).
References
- Oliver, G., Kipnis, J., Randolph, G. J. and Harvey, N. L. (2020). The Lymphatic Vasculature in the 21st Century: Novel Functional Roles in Homeostasis and Disease. Cell. 182(2): 270–296. https://doi.org/10.1016/j.cell.2020.06.039
- Montenegro-Navarro, N., García-Báez, C. and García-Caballero, M. (2023). Molecular and metabolic orchestration of the lymphatic vasculature in physiology and pathology. Nat Commun. 14(1): e1038/s41467–023–44133–x. https://doi.org/10.1038/s41467-023-44133-x
- Olayinka, O., Ryu, H., Wang, X., Malik, A. B. and Jung, H. M. (2025). Compensatory lymphangiogenesis is required for edema resolution in zebrafish. Sci Rep. 15(1): e1038/s41598–025–92970–1. https://doi.org/10.1038/s41598-025-92970-1
- Yaniv, K., Isogai, S., Castranova, D., Dye, L., Hitomi, J. and Weinstein, B. M. (2006). Live imaging of lymphatic development in the zebrafish. Nat Med. 12(6): 711–716. https://doi.org/10.1038/nm1427
- Küchler, A. M., Gjini, E., Peterson-Maduro, J., Cancilla, B., Wolburg, H. and Schulte-Merker, S. (2006). Development of the Zebrafish Lymphatic System Requires Vegfc Signaling. Curr Biol. 16(12): 1244–1248. https://doi.org/10.1016/j.cub.2006.05.026
- Jung, H. M., Castranova, D., Swift, M. R., Pham, V. N., Galanternik, M. V., Isogai, S., Butler, M. G., Mulligan, T. S. and Weinstein, B. M. (2017). Development of the larval lymphatic system in the zebrafish. Development. 144: e145755. https://doi.org/10.1242/dev.145755
- Shin, M., Nozaki, T., Idrizi, F., Isogai, S., Ogasawara, K., Ishida, K., Yuge, S., Roscoe, B., Wolfe, S. A., Fukuhara, S., et al. (2019). Valves Are a Conserved Feature of the Zebrafish Lymphatic System. Dev Cell. 51(3): 374–386.e5. https://doi.org/10.1016/j.devcel.2019.08.019
- Zhao, B. S., Roundtree, I. A. and He, C. (2016). Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 18(1): 31–42. https://doi.org/10.1038/nrm.2016.132
- Reynaud, K., McGeachy, A. M., Noble, D., Meacham, Z. A. and Ingolia, N. T. (2023). Surveying the global landscape of post-transcriptional regulators. Nat Struct Mol Biol. 30(6): 740–752. https://doi.org/10.1038/s41594-023-00999-5
- Becker, K., Bluhm, A., Casas-Vila, N., Dinges, N., Dejung, M., Sayols, S., Kreutz, C., Roignant, J. Y., Butter, F., Legewie, S., et al. (2018). Quantifying post-transcriptional regulation in the development of Drosophila melanogaster. Nat Commun. 9(1): e1038/s41467–018–07455–9. https://doi.org/10.1038/s41467-018-07455-9
- Buccitelli, C. and Selbach, M. (2020). mRNAs, proteins and the emerging principles of gene expression control. Nat Rev Genet. 21(10): 630–644. https://doi.org/10.1038/s41576-020-0258-4
- Gurung, S., Restrepo, N. K., Chestnut, B., Klimkaite, L. and Sumanas, S. (2022). Single-cell transcriptomic analysis of vascular endothelial cells in zebrafish embryos. Sci Rep. 12(1): e1038/s41598–022–17127–w. https://doi.org/10.1038/s41598-022-17127-w
- Li, Z., Ross Stewart, K. M., Bruton, F. A., Denvir, M. A. and Brittan, M. (2022). Isolation of Cardiac Endothelial Cells for Transcriptomic Analysis of the Zebrafish and Mouse Heart. Methods Mol Biol. 2441: 297–309. https://doi.org/10.1007/978-1-0716-2059-5_23
- Sanz, E., Yang, L., Su, T., Morris, D. R., McKnight, G. S. and Amieux, P. S. (2009). Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci USA. 106(33): 13939–13944. https://doi.org/10.1073/pnas.0907143106
- Tryon, R. C., Pisat, N., Johnson, S. L. and Dougherty, J. D. (2013). Development of translating ribosome affinity purification for zebrafish. Genesis. 51(3): 187–192. https://doi.org/10.1002/dvg.22363
- Heiman, M., Kulicke, R., Fenster, R. J., Greengard, P. and Heintz, N. (2014). Cell type–specific mRNA purification by translating ribosome affinity purification (TRAP). Nat Protoc. 9(6): 1282–1291. https://doi.org/10.1038/nprot.2014.085
- Reynoso, M. A., Juntawong, P., Lancia, M., Blanco, F. A., Bailey-Serres, J. and Zanetti, M. E. (2015). Translating Ribosome Affinity Purification (TRAP) Followed by RNA Sequencing Technology (TRAP-SEQ) for Quantitative Assessment of Plant Translatomes. Methods Mol Biol. 1284: 185–207. https://doi.org/10.1007/978-1-4939-2444-8_9
- Miller, M. F., Greenspan, L. J., Gildea, D. E., Monzo, K., Margolin, G., Pham, V. N., Ameyaw, K., Price, L., Aloi, N., Stratman, A. N., et al. (2019). Profiling the endothelial translatome in vivo using ‘AngioTag’ zebrafish. bioRxiv. e1101/815696. https://doi.org/10.1101/815696
- Miller, M. F., Greenspan, L. J., Gildea, D. E., Monzo, K., Margolin, G., Pham, V. N., Ameyaw, K., Price, L., Aloi, N., Stratman, A. N., et al. (2025). In vivo profiling of the endothelium using ‘AngioTag’ zebrafish. Angiogenesis. 28(3):40. https://doi.org/10.1007/s10456-025-09990-8
- Salussolia, C., Winden, K. and Sahin, M. (2022). Translating Ribosome Affinity Purification (TRAP) of Cell Type-specific mRNA from Mouse Brain Lysates. Bio Protoc. 12(9): e4407. https://doi.org/10.21769/bioprotoc.4407
Article Information
Publication history
Received: Aug 22, 2025
Accepted: Oct 22, 2025
Available online: Nov 3, 2025
Published: Dec 5, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Olayinka, O., Zarinebaf, L. and Jung, H. M. (2025). Analyzing the Translatome of Lymphatic and Venous Endothelial Cells In Vivo via Translating Ribosome Affinity Purification (TRAP). Bio-protocol 15(23): e5516. DOI: 10.21769/BioProtoc.5516.
Category
Molecular Biology > RNA > mRNA translation
Cell Biology > Organelle isolation > Polyribosome
Developmental Biology > Cell growth and fate > Lymphangiogenesis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








