- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Rapid and Simplified Induction of Spinal Motor Neurons From Human Induced Pluripotent Stem Cells
(*contributed equally to this work, § Technical contact) Published: Vol 15, Iss 20, Oct 20, 2025 DOI: 10.21769/BioProtoc.5478 Views: 1667
Reviewed by: Marion HoggAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Quantitative, Dynamic Detection of Neuronal Na+ Transients Using Multi-photon Excitation and Fluorescence Lifetime Imaging (FLIM) in Acute Mouse Brain Slices
Sara Eitelmann [...] Jan Meyer
Feb 5, 2025 1738 Views

Differentiation, Maintenance, and Contraction Profiling of Human Induced Pluripotent Stem Cell–Derived Cardiomyocytes
Matthijs Snelders [...] Jeroen Essers
Mar 5, 2025 3951 Views

Human iPSC-Derived Neuron and Oligodendrocyte Co-culture as a Small-Molecule Screening Assay for Myelination
Stefanie Elke Chie [...] Maria Consolata Miletta
May 5, 2025 3446 Views
Abstract
Human induced pluripotent stem cell (hiPSC)-derived motor neurons (MNs) provide a critical source for the study of motor neuron diseases (MNDs), which has been hindered by the lack of appropriate disease models for many years. Although many spinal MN differentiation protocols have been established by mimicking in vivo neurogenesis using extrinsic signaling molecules, substantial variations in the duration and efficiency persist due to inconsistencies in concentrations, timing, and delivery methods of these molecules. Here, we present an efficient monolayer culture differentiation strategy that enables the generation of enriched CHAT+ spinal MNs (sMNs) in 18 days and functional sMNs exhibiting extensive network activities, as confirmed by multielectrode array (MEA), within 28 days. Therefore, this optimized MN differentiation protocol facilitates the production of mature sMNs for MND research, high-throughput drug screening, and potential cell replacement therapies.
Key features
• This protocol provides a rapid and simple monolayer culture strategy for differentiating hiPSCs into sMNs in 18 days.
• Early administration of the Notch inhibitor Compound E accelerates the generation of hiPSC–sMNs 10 days in advance compared to a previous protocol.
• This protocol uses neural stem cells (NSCs) and MNPs as an intermediate to generate functionally mature sMNs.
Keywords: hiPSCGraphical overview
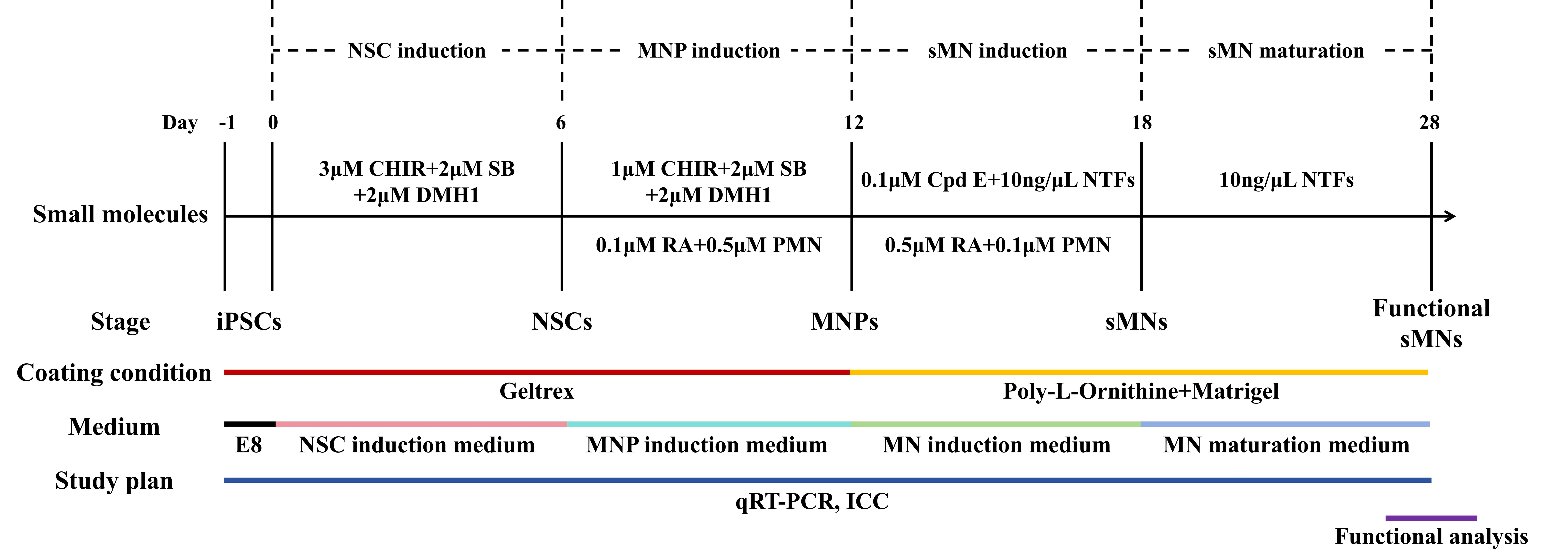
Schematic diagram of the spinal motor neuron (MN) differentiation protocol from iPSCs. CHIR, chir-99021; SB, SB-431542; RA, retinoic acid; PMN, purmorphamine; Cpd E, compound E; NTFs, neurotrophic factor.
Background
Spinal motor neurons (sMNs) are responsible for transmitting commands from the central nervous system (CNS) to muscles to control motor activity. sMNs are degenerated in motor neuron diseases (MNDs) such as amyotrophic lateral sclerosis (ALS), spinal muscular atrophy, and spinal and bulbar muscular atrophy. These MNDs are fatal diseases, and no effective treatment is available so far. This is thought to be caused, in part, by the lack of suitable human disease models [1,2]. The development of a robust protocol for the derivation of functional sMNs is, therefore, crucial for understanding disease mechanisms and developing novel therapies for MNDs.
As the signaling pathways and transcription factors contributing to sMN development have been unveiled [3–6], several protocols for sMN differentiation from pluripotent stem cells have been developed by mimicking in vivo neurogenesis using extrinsic signaling molecules [7,8]. Different doses, timing, and delivery procedures of signaling molecules among different protocols led to substantial variations in the duration (1–2 months) and efficiency (30%–90%) of MN differentiation [7–9]. In addition, the derived sMNs are often immature, and a lengthy maturation period is required before the detection of electrophysiological activity [10,11]. As most MNDs, including ALS, are late-onset diseases, these deficiencies limit the value of the derived sMNs in revealing disease characteristics.
By strategically advancing the temporal window for administering Compound E, a Notch signaling inhibitor, we have established a more efficient monolayer-based differentiation protocol compared to the previous method [12]. This precise timing intervention further enables the detection of extensive network activities in hiPSC–sMNs using a multielectrode array (MEA) as early as day 28 of differentiation. This novel MN differentiation protocol facilitates the production of mature sMNs for MND research, high-throughput drug screening, and potential cell transplantation therapy.
Materials and reagents
Biological materials
1. Human induced pluripotent stem cells (hiPSCs) (generated in-house [13])
Reagents
A. For cell culture
1. Essential 8TM Flex medium kit (E8 Flex medium) (Gibco, catalog number: A2858301)
2. GeltrexTM (Gibco, catalog number: A1413302)
3. KnockOutTM DMEM (Gibco, catalog number: 10829018)
4. Dulbecco’s phosphate-buffered saline, no calcium, no magnesium (DPBS w/o Ca2+/Mg2+) (Gibco, catalog number: 14190144)
5. Gentle cell dissociation reagent (Stem Cell Technologies, catalog number: 100-0485)
6. StemProTM AccutaseTM cell dissociation reagent (Thermo Fisher Scientific, catalog number: A1110501)
7. Knockout serum supplement (Gibco, catalog number: 10828028)
8. ROCK inhibitor Y-27632 (STEMCELL Technologies, catalog number: 72304)
9. DMEM/F12 with HEPES (Lonza, catalog number: LZBE12-719F)
10. Neurobasal medium (Gibco, catalog number: 21103049)
11. Penicillin/streptomycin (Gibco, catalog number: 15140122)
12. GlutaMAXTM supplement (Gibco, catalog number: 35050061)
13. N2 supplement (Gibco, catalog number: 17502048)
14. B27 supplement (Gibco, catalog number: 17504044)
15. L-ascorbic acid (Sigma, catalog number: A4544)
16. CHIR-99021 (MedChemExpress, catalog number: HY-10182)
17. SB-431542 (MedChemExpress, catalog number: HY-10431)
18. DMH-1 (MedChemExpress, catalog number: HY-12273)
19. Retinoic acid (MedChemExpress, catalog number: HY-14649)
20. Purmorphamine (MedChemExpress, catalog number: HY-15108)
21. Compound E (MedChemExpress, catalog number: HY-14176)
22. BDNF (PeproTech, catalog number: 450-02)
23. CNTF (PeproTech, catalog number: 450-13)
24. IGF-1 (PeproTech, catalog number: 100-11)
25. Poly-L-Ornithine (POL) (Sigma, catalog number: P4957)
26. Dulbecco’s phosphate-buffered saline (DPBS) (Sigma, catalog number: D8662)
27. Matrigel (Corning, catalog number: 354277)
28. Ethanol (Lennox, catalog number: 32221)
29. Dimethylsulfoxide (DMSO) (Sigma, catalog number: D2650)
30. Trypsin/EDTA (Gibco, catalog number: 25200056)
B. For immunocytochemistry (ICC)
1. Paraformaldehyde (PFA) (Santa Cruz, catalog number: 30525-89-4)
2. Bovine serum albumin (BSA) (Sigma, catalog number: A2153)
3. Phosphate-buffered saline (PBS) tablet (Fisher Scientific, catalog number: 12821680)
4. Triton X-100 (Sigma, catalog number: T8787)
5. Anti-NESTIN (Abcam, catalog number: ab22035)
6. Anti-PAX6 (Cell Signalling Technology, catalog number: 60433S, 1:500 dilution)
7. Anti-SOX2 (Cell Signalling Technology, catalog number: 3579, 1:500 dilution)
8. Anti-OCT4 (Cell Signalling Technology, catalog number: 2840, 1:500 dilution)
9. Anti-OLIG2 (Merck Millipore, catalog number: AB9610, 1:500 dilution)
10. Anti-NKX6.1 (DSHB, catalog number: F55A10-s, 1:50 dilution)
11. Anti-ISL1 (Sigma, catalog number: HPA057416, 1:500 dilution)
12. Anti-MNX1 (DSHB, catalog number: 81.5C10-s, 1:20 dilution)
13. Anti-CHAT (Merck Millipore, catalog number: AB144P, 1:100 dilution)
14. Anti-MAP2 (Proteintech, catalog number: 17490-1-AP, 1:500 dilution)
15. Anti-mouse IgG (H+L), F(ab’)2 fragment (Alexa Fluor® 488 conjugate) (Cell Signalling Technology, catalog number: 4408s, 1:1,000 dilution)
16. Anti-rabbit IgG (H+L), F(ab’)2 fragment (Alexa Fluor® 555 conjugate) (Cell Signalling Technology, catalog number: 4413s, 1:1,000 dilution)
17. Donkey anti-goat IgG (H+L) cross-adsorbed secondary antibody, Alexa Fluor 488 (Millipore, catalog number: A-11055, 1:1,000 dilution)
18. Donkey anti-rabbit IgG (H+L) cross-adsorbed secondary antibody, Alexa Fluor 647 (Millipore, catalog number: A-31573, 1:1,000 dilution)
19. Hoechst 33342 (Invitrogen, catalog number: H3569, 1:2,000 dilution)
Note: All reagents should be stored following the manufacturer's instructions.
Solutions
1. Complete E8 Flex medium for iPSC culture (see Recipes)
2. Geltrex solution for coating (see Recipes)
3. Basal medium for MN differentiation (see Recipes)
4. NSC induction medium (see Recipes)
5. MNP induction medium (see Recipes)
6. MN induction medium (see Recipes)
7. MN maturation medium (see Recipes)
8. Matrigel solution for coating (see Recipes)
9. Stock solutions of small molecules/growth factors for MN differentiation:
a. 100 mM L-ascorbic acid (see Recipes)
b. 10 mM CHIR99021 (see Recipes)
c. 10 mM DMH1 (see Recipes)
d. 10 mM SB431542 (see Recipes)
e. 10 mM retinoic acid (see Recipes)
f. 10 mM Purmorphamine (see Recipes)
g. Compound E (see Recipes)
h. 100 μg/mL BDNF (see Recipes)
i. 100 μg/mL CNTF (see Recipes)
j. 100 μg/mL IGF-1 (see Recipes)
10. 0.1% BSA in PBS (see Recipes)
11. 1× PBS solution (see Recipes)
12. 70% and 90% ethanol (see Recipes)
13. 1% BSA + 0.3% Triton X-100 in PBS (see Recipes)
Recipes
1. Complete E8 Flex medium for hiPSCs
a. Thaw the frozen E8 Flex supplement at 2–8 °C overnight. Protect the supplement from light, as it is light-sensitive.
b. Mix the thawed supplement by gently inverting the vial several times, then aseptically transfer all contents to the bottle of E8 Flex basal medium. Swirl the bottle to mix.
c. Complete E8 Flex medium can be stored at 2–8 °C for up to 2 weeks. Alternatively, aliquot 45 mL of complete E8 Flex medium into 50 mL Falcon tubes and store at -20 °C for up to 6 months. Avoid repeated freeze-thaw cycles.
d. Before use, warm the complete E8 Flex medium at room temperature and protect it from light.
Critical: Do not warm the medium in a 37 °C water bath.
2. Geltrex solution for coating
a) Geltrex stock solution (1:1 dilution)
i. Thaw the frozen Geltrex overnight at 2–8 °C.
ii. Mix 5 mL of Geltrex with 5 mL of pre-chilled KnockOutTM DMEM/F12.
iii. Aliquot Geltrex stock solution (1:1 dilution) into a 1 mL cryogenic tube and store at -80 °C for several months.
b) Geltrex working solution (1:100 dilution)
i. Thaw 1 mL of frozen Geltrex stock solution at 2–8 °C overnight.
ii. Dilute 1 mL of Geltrex stock solution into 49 mL of pre-chilled KnockOutTM DMEM/F12 to get a 1:100 diluted Geltrex working solution.
iii. The working solution can be stored at 2–8 °C for up to 2 weeks.
3. Basal medium for MN differentiation
| Reagent | Final concentration | Volume |
| DMEM/F12 with HEPES | 49% | 245 mL |
| Neurobasal medium (0.5× volume) | 49% | 245 mL |
| 1× Penicillin/streptomycin | 1% | 5 mL |
| 1× GlutaMAXTM supplement | 1% | 5 mL |
| Filter the above components through a 0.22 μm sterile filter | ||
| 0.5× N2 supplement | 2.5 mL | |
| 0.5× B27 supplement | 5 mL | |
The basal medium can be stored at 2–8 °C for up to 4 weeks. Alternatively, aliquot 45 mL of basal medium into 50 mL Falcon tubes and store at -20 °C for up to 6 months.
Note: Protect the medium from light, as N2 and B27 are light-sensitive. Avoid repeated freeze-thaw cycles.
4. NSC induction medium
| Reagent | Final concentration | Volume |
|---|---|---|
| Basal medium | ~100% | accordingly |
| L-ascorbic acid | 0.1 mM | see Recipe 9a |
| CHIR99021 | 3 μM | see Recipe 9b |
| DMH1 | 2 μM | see Recipe 9c |
| SB431542 | 2 μM | see Recipe 9d |
Before use, warm the basal medium at room temperature and protect it from light. Do not warm the medium in a 37 °C water bath. To prepare the NSC induction medium, first aliquot the basal medium according to the volume required. Add small molecules to the medium in the amounts specified in the table.
5. MNP induction medium
| Reagent | Final concentration | Volume |
|---|---|---|
| Basal medium | ~100% | accordingly |
| L-ascorbic acid | 0.1 mM | see Recipe 9a |
| Retinoic acid | 0.1 μM | see Recipe 9e |
| Purmorphamine | 0.5 μM | see Recipe 9f |
| CHIR99021 | 1 μM | see Recipe 9b |
| DMH1 | 2 μM | see Recipe 9c |
| SB431542 | 2 μM | see Recipe 9d |
Before use, warm the basal medium at room temperature and protect it from light. Do not warm the medium in a 37 °C water bath. To prepare the MNP induction medium, first aliquot the basal medium according to the volume required. Add small molecules to the medium in the amounts specified in the table.
6. MN induction medium
| Reagent | Final concentration | Volume |
|---|---|---|
| Basal medium | ~100% | accordingly |
| L-ascorbic acid | 0.1 mM | see Recipe 9a |
| Retinoic acid | 0.5 μM | see Recipe 9e |
| Purmorphamine | 0.1 μM | see Recipe 9f |
| Compound E | 0.1 μM | see Recipe 9g |
| BDNF | 10 ng/μL | see Recipe 9h |
| CNTF | 10 ng/μL | see Recipe 9i |
| IGF-1 | 10 ng/μL | see Recipe 9j |
Before use, warm the basal medium at room temperature and protect it from light. Do not warm the medium in a 37 °C water bath. To prepare the MN induction medium, first aliquot the basal medium according to the volume required. Add small molecules to the medium in the amounts specified in the table.
7. MN maturation medium
| Reagent | Final concentration | Volume |
|---|---|---|
| Basal medium | ~100% | accordingly |
| L-ascorbic acid | 0.1 mM | see Recipe 9a |
| BDNF | 10 ng/μL | see Recipe 9h |
| CNTF | 10 ng/μL | see Recipe 9i |
| IGF-1 | 10 ng/μL | see Recipe 9j |
Before use, warm the basal medium at room temperature and protect it from light. Do not warm the medium in a 37 °C water bath. To prepare MN maturation medium, first aliquot the basal medium according to the volume required. Add neurotrophic factors (NTFs) to the medium in the amounts specified in the table.
8. Matrigel solution for coating
a) Matrigel stock solution (1:1 dilution)
i. Thaw the frozen Matrigel at 2–8 °C overnight.
ii. Mix 5 mL of Matrigel with 5 mL of pre-chilled KnockOutTM DMEM/F12.
iii. Aliquot Matrigel stock solution (1:1 dilution) into a 1 mL cryogenic tube and store at -80 °C for up to one year.
b) Matrigel working solution (1:50 dilution)
i. Thaw 1 mL of frozen Matrigel stock solution at 2–8 °C overnight.
ii. Dilute 1 mL of Matrigel stock solution in 24 mL of pre-chilled KnockOutTM DMEM/F12 to obtain a 1:50 diluted Matrigel working solution.
iii. The working solution can be stored at 2–8 °C for up to 2 weeks.
Note: Avoid repeated freeze-thaw cycles of the working Matrigel solution.
9. Stock solutions of small molecules/growth factors for MN differentiation
Notes:
1. Information regarding the stability of small molecules in solution is rarely reported. The recommended storage time for each small molecule after preparation should follow the supplier’s guidelines.
2. Protect all small molecules from light.
a. 100 mM L-ascorbic acid
Dissolve 88.06 mg of L-ascorbic acid in 5 mL of sterile MilliQ water. Filter the solution through a 0.22 μm sterile filter. Aliquot 50 μL per tube and store at -80 °C for up to two years.
b. 10 mM CHIR99021
Dissolve 5 mg of CHIR99021 in 1.0745 mL of DMSO. Aliquot 10 μL per tube and store at -80 °C for up to one year.
c. 10 mM DMH1
Dissolve 10 mg of DMH1 in 2.6285 mL of DMSO. Aliquot 10 μL per tube and store at -80 °C for two years.
d. 10 mM SB431542
Dissolve 10 mg of SB431542 in 2.6012 mL of DMSO. Aliquot 10 μL per tube and store at -80 °C for two years.
e. 10 mM retinoic acid
Dissolve 100 mg of retinoic acid in 3.3285 mL of DMSO to prepare a 100 mM stock solution. Further dilute to 10 mM as needed. Aliquot 10 μL per tube and store at -80 °C for 6 months.
f. 10 mM Purmorphamine
Dissolve 5 mg of Purmorphamine in 960.393 μL of DMSO. Aliquot 10 μL per tube and store at -80 °C for up to two years.
g. Compound E
Dissolve 1 mg of Compound E in 203.874 μL of DMSO. Aliquot 10 μL per tube and store at -80 °C for up to 6 months.
h. 100 μg/mL BDNF
Dissolve 100 μg of BDNF in 1 mL of sterile 0.1% BSA in PBS. Aliquot 20 μL per tube and store at -80 °C for up to one year.
i. 100 μg/mL CNTF
Dissolve 100 μg of CNTF in 1 mL of sterile 0.1% BSA in PBS. Aliquot 20 μL per tube and store at -80 °C for up to one year.
j. 100 μg/mL IGF-1
Dissolve 100 μg IGF-1 in 1 mL of sterile 0.1% BSA in PBS. Aliquot 20 μL per tube and store at -80 °C for up to one year.
10. 0.1% BSA in PBS
Dissolve 0.1 g of BSA in 100 mL of sterile DPBS solution. Filter the solution through a 0.22 μm sterile filter. Aliquot 5 mL per tube and store at -20 °C for up to one year.
11. 1× PBS solution
Dissolve one PBS tablet in 100 mL of distilled water using a magnetic stirrer and stir bar. Store at room temperature for up to one month.
12. 70% and 90% ethanol
To prepare 500 mL of 70% ethanol, combine 350 mL of 100% ethanol with 150 mL of distilled water. To prepare 500 mL of 90% ethanol, combine 450 mL of 100% ethanol with 50 mL of distilled water.
13. 1% BSA + 0.3% Triton X-100 in PBS
Dissolve 0.1 g of BSA in 10 mL of 1× PBS solution by vortexing. Once fully dissolved, add 30 μL of Triton X-100 to the solution and vortex thoroughly to mix. Store at 4 °C for up to one month.
Laboratory supplies
1. 6-well tissue culture plate (Sarstedt, catalog number: 83.3920.300)
2. Centrifuge tubes, 15 and 50 mL (Sarstedt, catalog numbers: 62.554.502 and 62.547.254)
3. Microcentrifuge tube, 1.5 mL (Sarstedt, catalog number: 72.690.001)
4. Cell scraper (Sarstedt, catalog number: 83.1831)
5. μ-slide 8 well (ibidi Gmbh, catalog number: 80826)
6. 0.22 μm disposable sterile filters (Lennox, catalog number: A16534K)
7. 1.5 mL microcentrifuge tube (Sarstedt, catalog number: 72.690.001)
8. Filter tip 100–1,000 μL (Cruinn, catalog number: S1122-1830)
9. Filter tip 0.1–10 μL (Cruinn, catalog number: S1121-3810)
Equipment
1. CO2 incubator (Thermo Fisher Scientific, model: 371)
2. Stereo microscope (Thermo Fisher, model: EVOS XL Core)
3. Inverted light microscope (Zeiss, model: Axiovert 40 CFL)
4. Confocal microscope FV1000 (Olympus)
5. Maestro Original (Axion BioSystems)
6. CytoView MEA 48 plate (Axion BioSystems)
7. Water bath (Fisher Scientific, model: FSGPD28)
8. Operetta Imaging system with Harmony image analysis software (Perkin Elmer)
Software and datasets
1. AxIS Navigator, CiPA Analysis Tool: https://www.axionbiosystems.com/products/mea/mea-software
2. Fiji ImageJ-win64
Procedure
Note: The following steps are described based on a 6-well plate.
A. hiPSC preparation
1. hiPSC restoration
a. Prepare Geltrex-coated plates by adding 1 mL of Geltrex working solution per well, and incubate at 37 °C, 5% CO2 for at least 1 h.
b. Prewarm E8 Flex medium at room temperature until no longer cool to the touch.
Critical: Do not warm the medium in a 37 °C water bath.
c. Take out a cryovial of hiPSCs from liquid nitrogen and quickly thaw in a 37 °C water bath until only a sliver of ice remains.
d. Clean the vial surface with 70% ethanol before transferring it to a flow cabinet.
e. Transfer the cells to a 15 mL centrifuge tube using a 1 mL filter tip.
f. Slowly dilute the cell suspension by adding 9 mL of KnockOutTM DMEM dropwise.
g. Secure the lid and invert several times to mix gently.
h. Centrifuge at 200× g for 3 min and then carefully aspirate the supernatant without disturbing the pellet.
i. Resuspend the cell pellet in 2 mL of E8 Flex medium + 10 μM of Y-27632.
Note: Y-27632 is used to increase iPSC survival after thawing.
j. Transfer the cell suspension into one well of a Geltrex-coated 6-well plate.
Note: Avoid pipetting to prevent the dissociation of iPSC clusters into single cells, as this will reduce the recovery rate.
k. Gently move the plate in quick back-and-forth and side-to-side motions to distribute cells evenly.
l. Place the plate in a 37 °C, 5% CO2 incubator. Replace the medium with fresh E8 Flex (without Y-27632) within 24 h.
Note: Prolonged Y-27632 exposure may increase spontaneous differentiation of hiPSCs.
m. Culture cells for 4–5 days until 70%–80% confluency is reached. Renew medium every two days.
2. iPSC passage
a. Prepare a Geltrex-coated 6-well plate and prewarm E8 Flex medium as described in the previous section.
b. Discard the spent medium and gently rinse the cells with 1 mL of DPBS w/o Ca2+/Mg2+.
c. Add 1 mL of gentle cell dissociation reagent and incubate at room temperature for 2 min. Then, discard the solution.
d. Allow the plate to stand at room temperature for another 2–3 min. Monitor under a microscope until small gaps appear in iPSC clones.
e. Firmly hold the plate with one hand and tap the side of the plate with the other hand to detach the cells (Figure 1).
Note: iPSCs are more sensitive to the dissociation reagent than differentiation cells. The tapping method helps with iPSC purity.
f. Add E8 Flex medium to the well and split the cells into 4–6 new wells based on confluency.
g. Return the plate to the 37 °C, 5% CO2 incubator and change the medium with fresh E8 Flex medium the next day.
h. Continue culturing for 4–5 days, changing the medium every other day.

Figure 1. Example of the plate-tapping method to detach induced pluripotent stem cell (iPSC) clones after dissociation with gentle cell dissociation reagent
B. MN differentiation
1. NSC generation (timing: 7 days)
Day -1:
a. Begin NSC generation when hiPSCs are on day 3–4 after passaging, as cells are at a highly proliferative state.
Critical: The quality of the hiPSCs (with minimal or no differentiated colonies) is critical for efficient neural induction. Remove any fully or partially differentiated colonies before dissociation.
b. Prepare a Geltrex-coated 6-well plate and prewarm E8 Flex medium as previously described.
c. Aspirate the medium and wash cells with 1 mL of DPBS w/o Ca2+/Mg2+.
d. Add 1 mL of Accutase per well of a 6-well plate and incubate at 37 °C for 5–8 min.
e. Detach cells using a cell scraper and transfer the suspension to a sterile 15 mL centrifuge tube.
f. Gently pipette the cells up and down with a 1 mL filter tip to obtain a single-cell suspension.
g. Add 9 mL of prewarmed KnockOutTM DMEM + 10 μM Y-27632 to the tube.
h. Secure the lid and gently invert the tube to mix thoroughly.
i. Take out 20 μL of cell suspension for cell counting.
j. Centrifuge the remaining suspension at 200× g for 3 min.
Note: Perform cell counting during the centrifuge. Typical hiPSC yield is around 1–2 × 106 per well of a 6-well plate.
k. Carefully discard the supernatant without disturbing the cell pellet.
l. Resuspend the pellet in E8 Flex medium + 10 μM Y-27632 to a density of 1 × 106 cells per mL.
m. Plate 3 × 105 cells per well in a Geltrex-coated 6-well plate with 2 mL of E8 Flex medium + 10 μM Y-27632.
Note: To characterize the cells on day 6 of differentiation, 3 × 104 cells are seeded per well in a μ-slide 8-well chamber for ICC staining.
n. Move the plate in quick back-and-forth and side-to-side motions to distribute cells evenly.
o. Gently place the plate in a 37 °C, 5% CO2 incubator.
Days 0–6:
p. On day 0 of differentiation, replace the E8 Flex medium with 2 mL of NSC induction medium. Representative morphology of hiPSCs is shown in Figure 2.
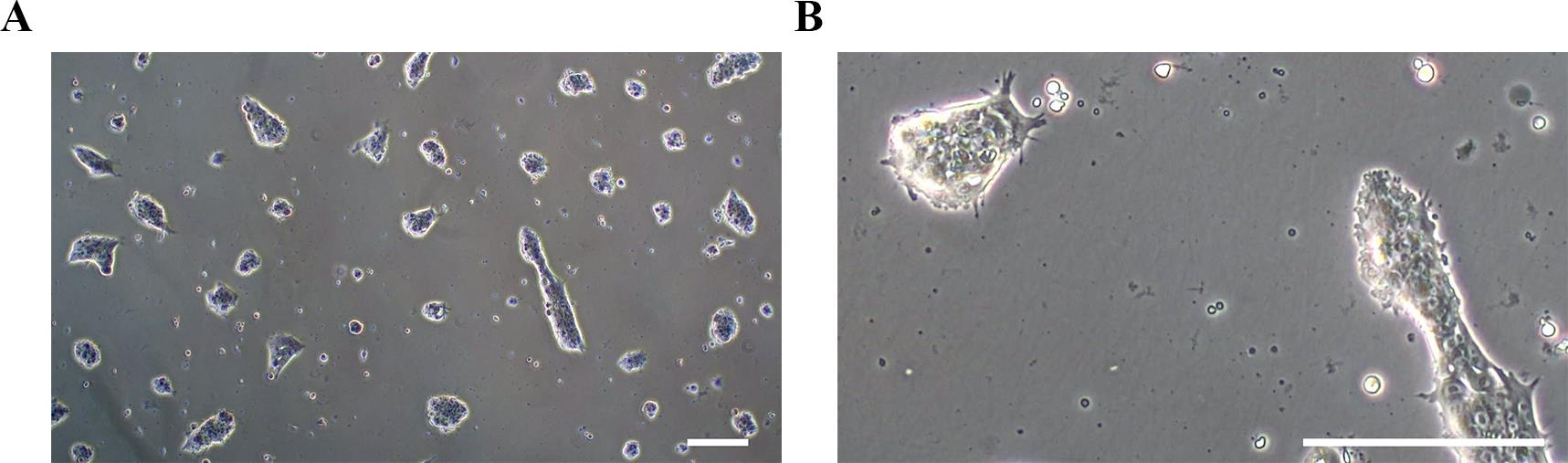
Figure 2. Representative morphologies of human induced pluripotent stem cells (hiPSCs) on day 0 of differentiation. (A) At lower magnification (4×) and (B) at higher magnification (20×). Scale bars, 100 μm.
q. Continue culturing the cells for 6 days, replenishing the medium every other day. Representative morphology and ICC staining for high-quality NSCs are shown in Figures 3 and 4A.
Note: As cell growth progresses, the confluency of the culture increases gradually. To meet the increased nutritional needs of the cells in each well, the volume of induction medium could be increased to 3 mL.
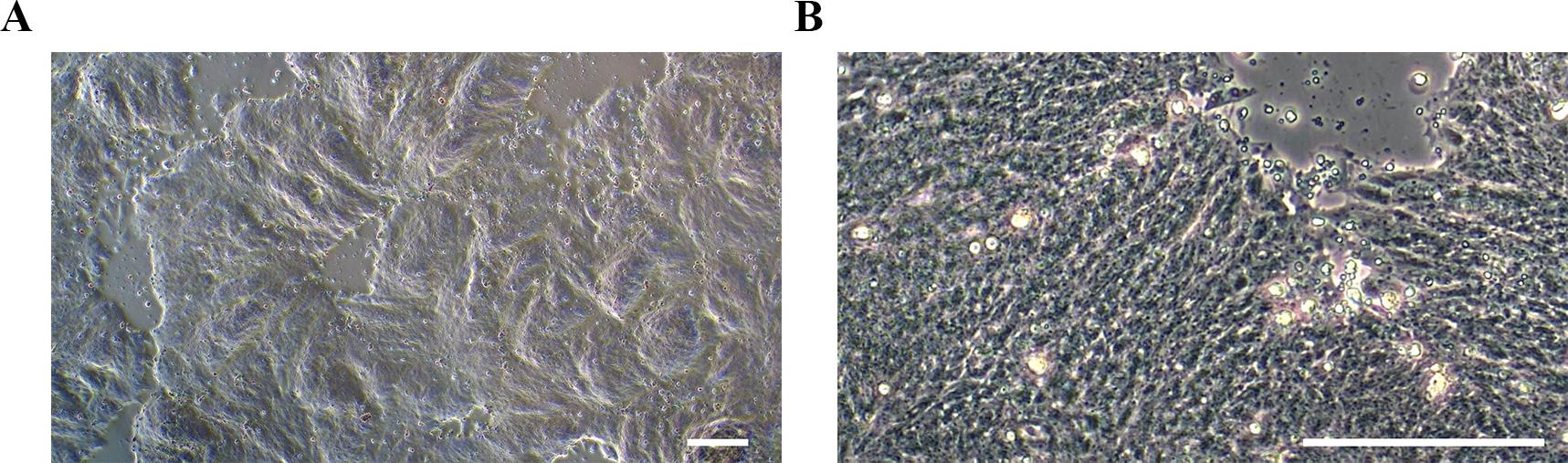
Figure 3. Representative morphology of neural stem cells (NSCs) on day 6 of differentiation. (A) At lower magnification (4×) and (B) at higher magnification (20×). Scale bars, 100 μm.
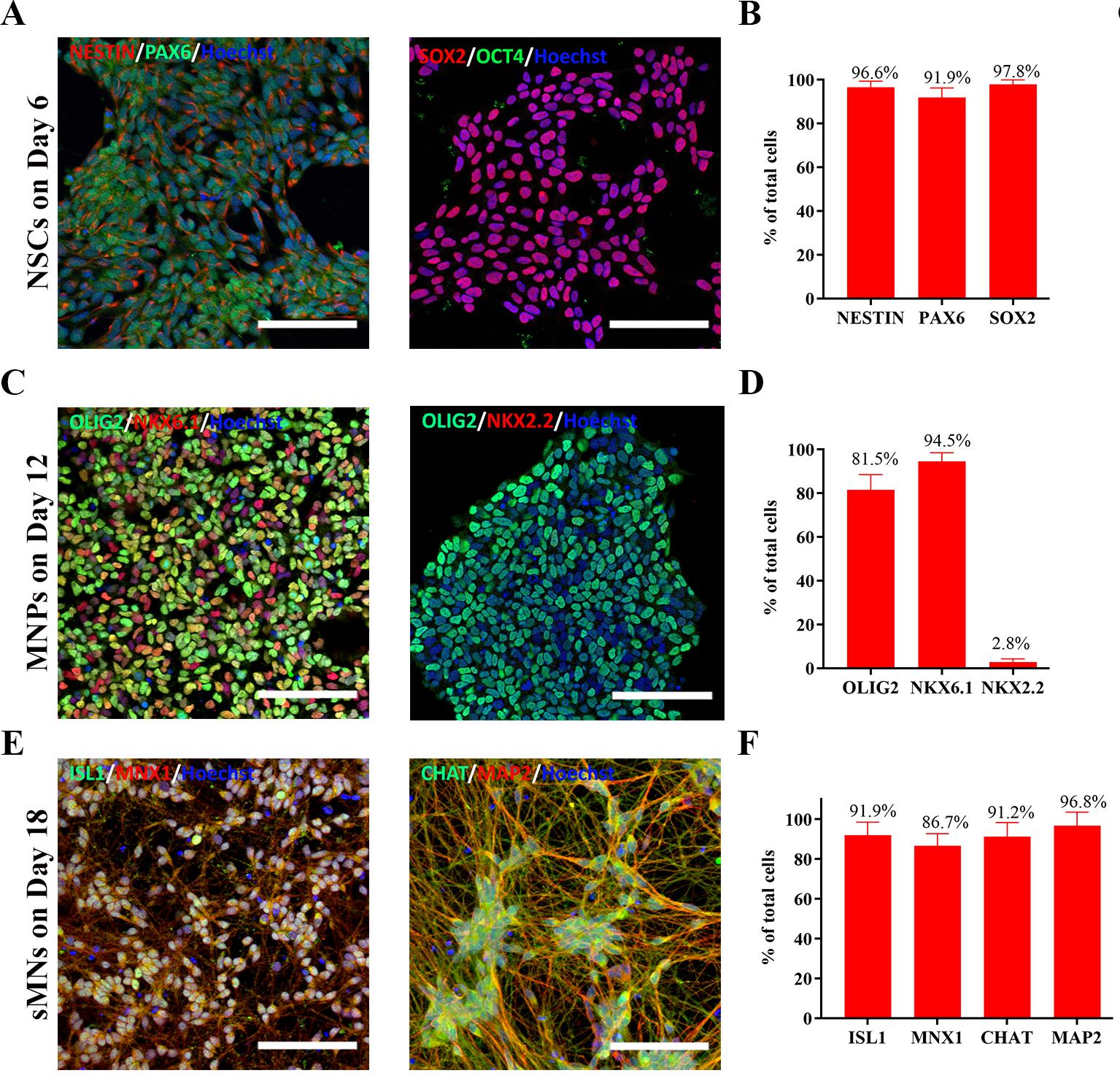
Figure 4. Characterization of cells at each differentiated stage. (A) Representative images of NESTIN+/PAX6+ and SOX2+/OCT4- neural stem cells (NSCs) on day 6 of differentiation. (B) Proportion of NESTIN+, PAX6+, SOX2+, and OCT4- NSCs on day 6 of differentiation. N = 7 cell lines, n = 3 replicates. (C) Representative images of OLIG2+/NKX6.1+ and OLIG2+/NKX2.2- MNPs on day 12 of differentiation. (D) Proportion of OLIG2+, NKX6.1+, and NKX2.2- MNPs on day 12 of differentiation. N = 2 cell lines, n = 3 replicates. (E) Representative images of MNX1+/ISLET1+ and MAP2+/CHAT+ spinal motor neurons (sMNs) on day 18 of differentiation. (F) Proportion of MNX1+, ISLET1+, MAP2+, and CHAT+ sMNs on day 18 of differentiation. N = 7 cell lines, n = 3 replicates. Scale bars: 100 μm. The quantification procedure is described in the Data analysis section. Data are presented by mean ± SEM.
2. MNP generation (timing: 6 days)
Days 6–12:
a. On day 6 of differentiation, discard the medium and wash the cells with 1 mL of DPBS w/o Ca2+/Mg2+.
b. Dissociate the cells with Accutase as previously described.
c. Split the cell suspension at a 1:3 ratio into a Geltrex-coated 6-well plate using MNP induction medium + 10 μM Y-27632.
Note: The cell number at this step is approximately 2–3 × 106 cells per well of a 6-well plate. To characterize cells on day 12 of differentiation, 5 × 104 cells are seeded per well in a μ-slide 8-well chamber for ICC staining.
Critical: Add 10 μM Y-27632 to the medium for the first 1 or 2 days after passaging to increase cell survival.
d. Move the plates in quick back-and-forth and side-to-side motions to distribute cells evenly.
e. Gently place the plate in a 37 °C, 5% CO2 incubator.
f. Continue culturing cells for 6 days, renewing the MNP induction medium (without Y-27632) every other day. Representative morphology and ICC staining of high-quality MNPs are shown in Figures 4B and 5.
Note: As cell growth progresses and confluency increases, the volume of the induction medium can be increased to 3 mL to meet the increased nutritional needs of the cells in each well.
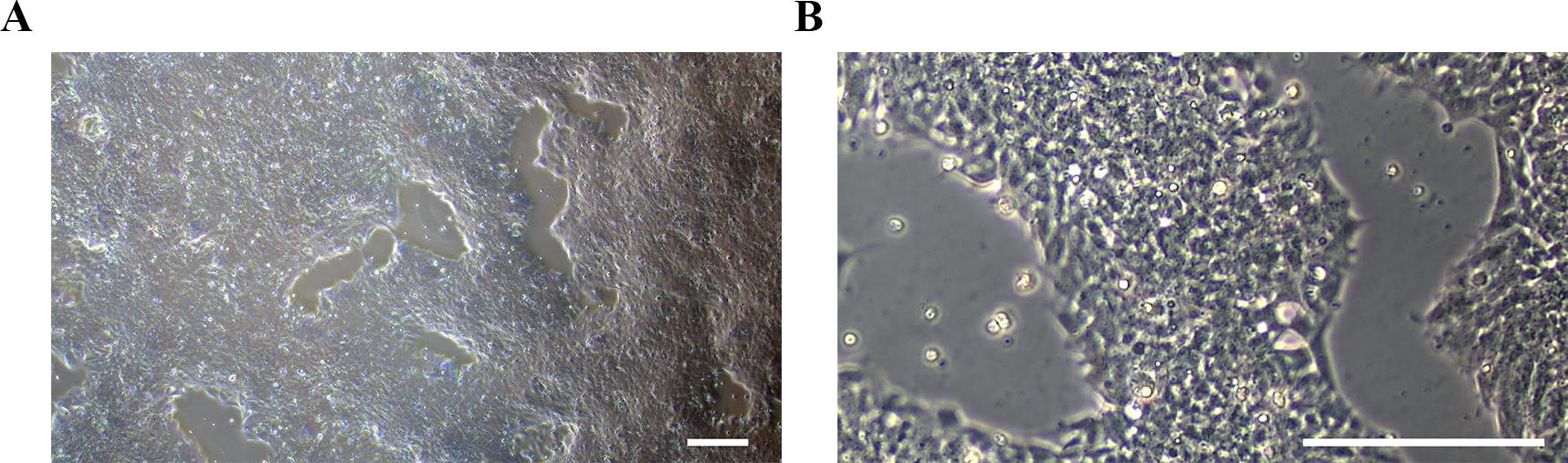
Figure 5. Representative morphology of MNPs on day 12 of differentiation. (A) At lower magnification (4×) and (B) at higher magnification (20×). Scale bars, 100 μm.
3. Spinal MN generation (timing: 6 days)
Days 12–18:
a. Prepare the Poly-L-ornithine/Matrigel (POM)-coated plate
i. Dilute 100 μg/mL of POL in sterile DPBS to a final concentration of 20 μg/mL.
ii. Add 1 mL/well of 20 μg/mL POL to a 6-well plate and incubate at 4 °C overnight.
iii. Wash the POL-coated wells twice with sterile DPBS and avoid drying the coated plate.
iv. Add 1 mL/well of Matrigel working solution (1:50 dilution) to POL-coated wells and incubate at 37 °C for at least 1 h.
v. Remove the Matrigel solution before use and do not let the coated plate dry out.
b) MNP passage
i. On day 12 of differentiation, discard the medium and wash cells with 1 mL of DPBS w/o Ca2+/Mg2+.
ii. Dissociate cells with Accutase as previously described.
iii. Resuspend cells in MN induction medium to a density of 1 × 106 cells per mL.
iv. Plate 1 × 105 cells per well in a POL/Matrigel (POM)-precoated 24-well plate using MN induction medium.
Critical: Add 10 μM Y-27632 to the medium for the first 2 days after passaging to increase cell survival.
Notes:
1. Cell seeding density is 5 × 104 cells/cm2. The seeding number should be adjusted proportionally based on the culture plate size.
2. To characterize the cells on day 18 and day 28 of differentiation, 5 × 104 cells are seeded on μ-slide 8-well chambers for ICC staining.
v. Move the plates in quick back-and-forth and side-to-side motions to distribute the cells evenly.
vi. Gently place the plate in a 37 °C, 5% CO2 incubator.
vii. Continue culturing cells for 6 days and perform a half-medium change every other day. Representative morphology and sampled ICC staining for high-quality sMNs are shown in Figures 4C and 6.
Note: Given that the cells on this stage have formed extensive neurite networks and established intercellular connections, harsh medium changes may cause cell detachment. To minimize disruption, a half-medium change is strongly recommended. The term “half-medium change” refers to the process of removing half of the old medium and replacing it with an equal volume of fresh medium.
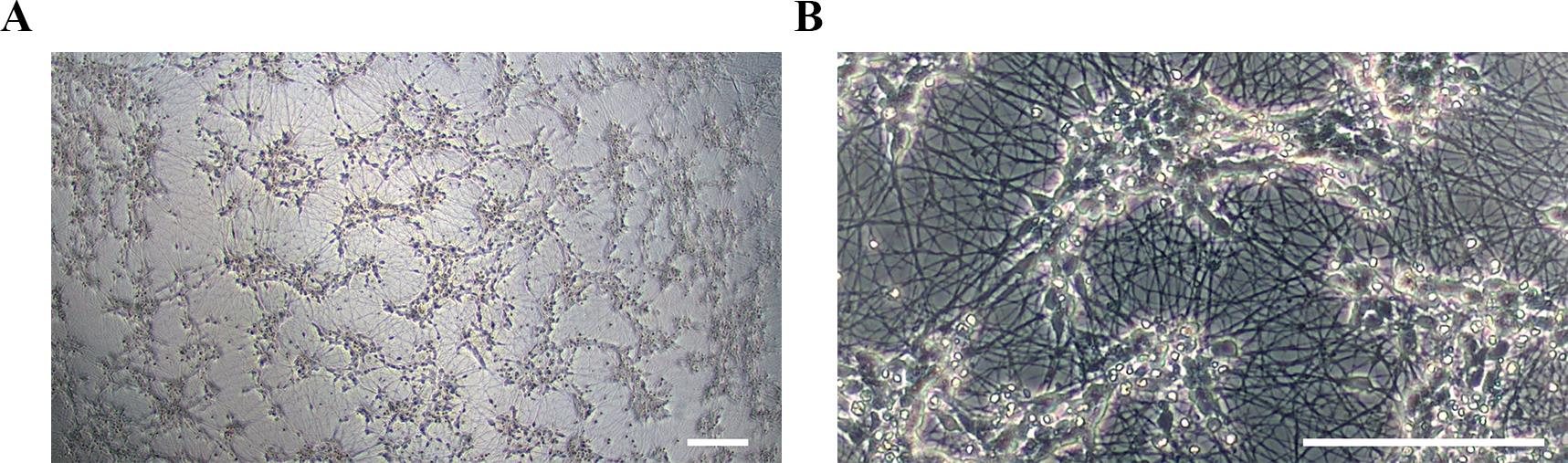
Figure 6. Representative morphology for spinal motor neurons (sMNs) on day 18 of differentiation. (A) At lower magnification (4×) and (B) at higher magnification (20×). Scale bars, 100 μm.
4. MN maturation (timing: 10 days)
Days 18–28:
a. On day 18 of differentiation, replace the medium with MN maturation medium.
Note: Small molecules are withdrawn at this stage, and only three growth factors are retained to support in vitro maturation of iPSC-derived sMNs.
b. Continue to culture the cells for 10 days without passaging. Gently perform a half-medium change every other day.
C. hiPSC-derived sMN characterization
1. Immunostaining of cells at each differentiated stage
Note: 3 × 104 hiPSCs on day 0, 5 × 104 hiPSC-derived NSCs on day 6, and 5 × 104 hiPSC-derived MNPs on day 12 of differentiation are seeded per well in a μ-slide 8-well chamber. The cells are cultured for 6 days using their stage-specific medium. Immunostaining is then performed on days 6, 12, and 18 to characterize cell identity.
a. Wash cells twice with 300 μL/well of 1× PBS.
b. Fix cells with 300 μL/well of 4% PFA for 15 min at room temperature.
c. Wash cells three times with 300 μL/well of 1× PBS.
d. Permeabilize and block cells with 300 μL/well of 1% BSA + 0.3% Triton X-100 in PBS for 30 min at room temperature.
e. Incubate cells with 200 μL/well of primary antibodies (diluted as specified in the Reagents list) at 4 °C overnight.
f. Wash cells three times with 300 μL/well of 1× PBS.
g. Incubate cells with secondary antibodies and Hoechst 33342 (diluted as recommended in the Reagents list) for 1 h at room temperature.
h. Wash cells three times with 300 μL/well of 1× PBS.
i. Retain 300–400 μL/well of 1× PBS to prevent drying.
j. Capture images using the confocal microscope. Representative immunostaining results for each differentiation stage are shown in Figure 4.
2. Functional analysis by multielectrode array (MEA)
a. Prepare a POM-coated CytoView 48-well MEA plate.
i. Apply 8 μL/well of 20 μg/mL POL solution to cover the recording electrodes of the CytoView 48-well MEA plate.
ii. Incubate the POL-coated plate at 37 °C, 5% CO2 incubator for at least 1 h.
iii. Carefully aspirate the POL solution without touching the recording electrodes. Avoid letting the plate dry out.
iv. Immediately rinse twice with 500 μL/well of sterile DPBS.
v. Apply 8 μL/well of Matrigel (1:50 dilution) to POL-precoated wells.
vi. Incubate the plate at 37 °C, 5% CO2 incubator for at least 1 h.
vii. Aspirate the Matrigel solution immediately before plating cells. Avoid letting the plate dry out.
b. Seed MNPs on the MEA plate on day 12 of differentiation.
i. Place 5 × 104 cells in an 8 μL drop of MN induction medium onto the central area (with 16 recording electrodes) of POM-precoated CytoView 48-well MEA plate.
ii. Incubate the plate for 2 h at 37 °C and 5% CO2 to allow cell attachment.
Note: Add approximately 8 mL of sterile water to the reservoir of the MEA plate to maintain humidity during incubation.
iii. Verify cell attachment using an inverted microscope, then gently add 300 μL/well of MN induction medium. Representative images of cells after replating on the MEA plate are shown in Figure 7.
Note: Add 10 μM Y-27632 to the medium for the first two days to enhance cell survival.
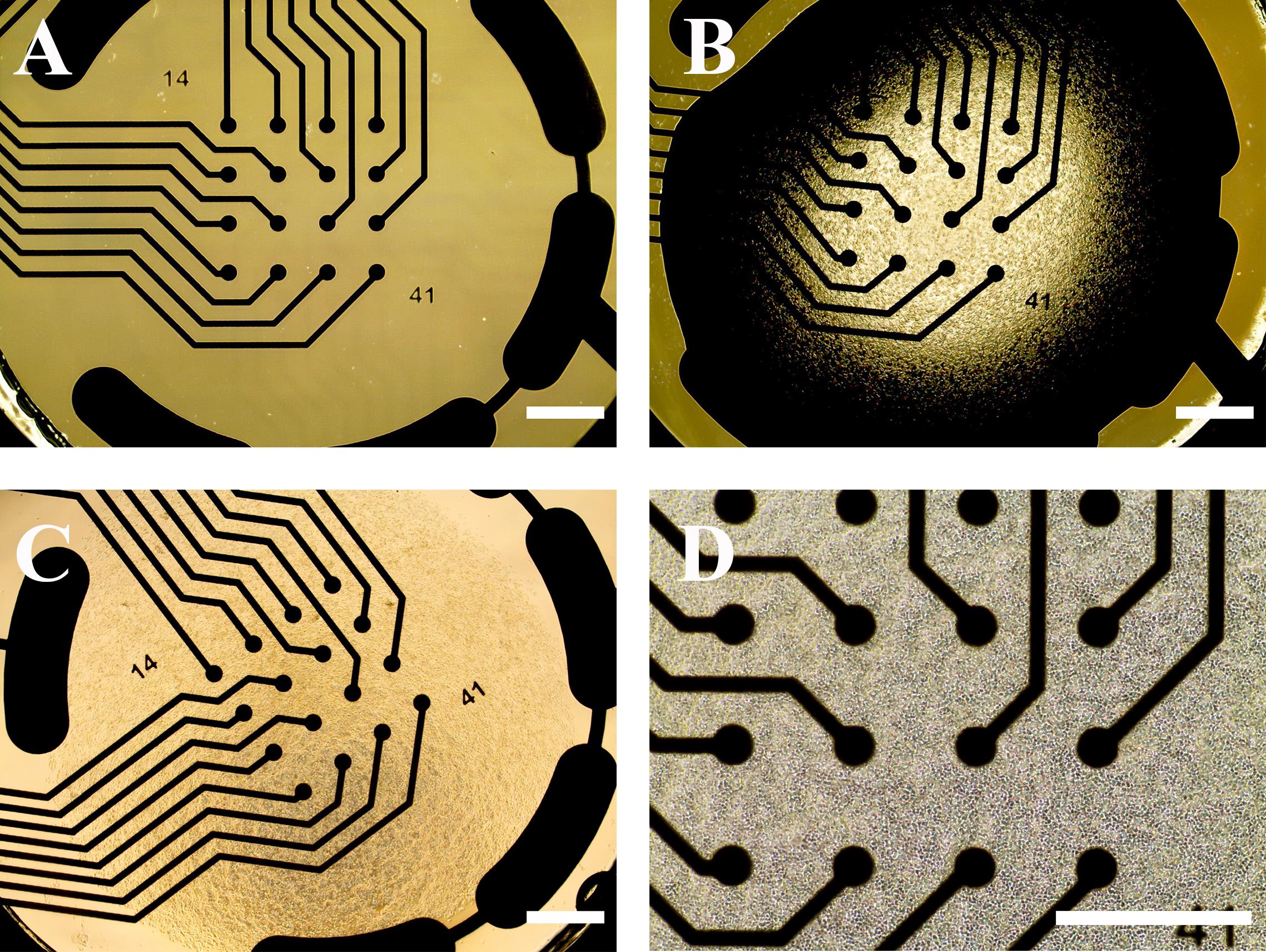
Figure 7. Representative images of the multielectrode array (MEA) plate and human induced pluripotent stem cell (hiPSC)-derived spinal motor neuron (sMN) plating. (A) An empty well of a 48-well MEA plate before coating. (B) The recording area (with electrodes) coated with 8 μL of Matrigel following initial POL coating. (C) 5 × 104 iPSC-derived MNPs plated on MEA in 8 μL of the medium. (D) Representative image of MNPs attached to the MEA plate after 2 h of incubation. Scale bars, 100 μm.
iv. Perform half-medium change every other day using MN induction medium for the first 6 days; thereafter, replace the medium with MN maturation differentiation for the subsequent 10 days.
Note: Avoid touching the electrodes when changing medium.
3. MEA data recording
a. Record field potentials every other day starting from day 18 of differentiation (6 days after replating), prior to medium change, using AxIS Navigator software (Version 2.5.2). Recommended acquisition settings are shown in Figure 8.
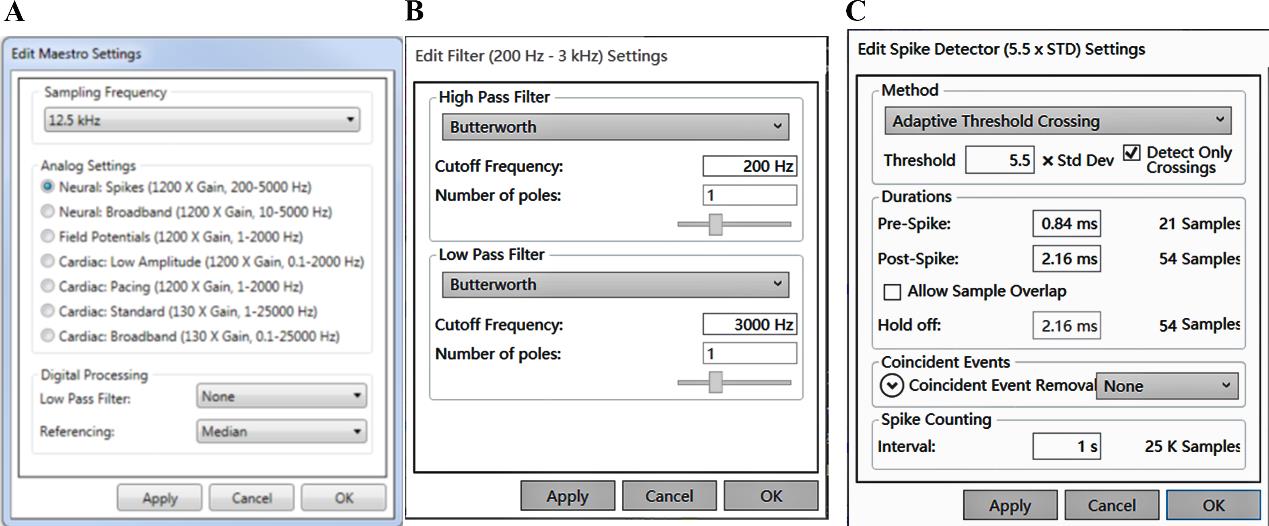
Figure 8. AxIS software setting for multielectrode array (MEA) recording. (A) Sampling frequency is set to 12.5 Hz and Analog Settings to Neural Spikes. (B) The digital filter is set to 200 Hz for the High Pass Filter and 3,000 Hz for the Low Pass Filter. (C) The threshold for detecting spikes is set to 5.5 times the standard deviation for each electrode with 1 s binning.
b. The exported spike files (.spk) are batch-processed with Neural Metric Tool (Axion Biosystems). The software settings are presented in Figure 9.
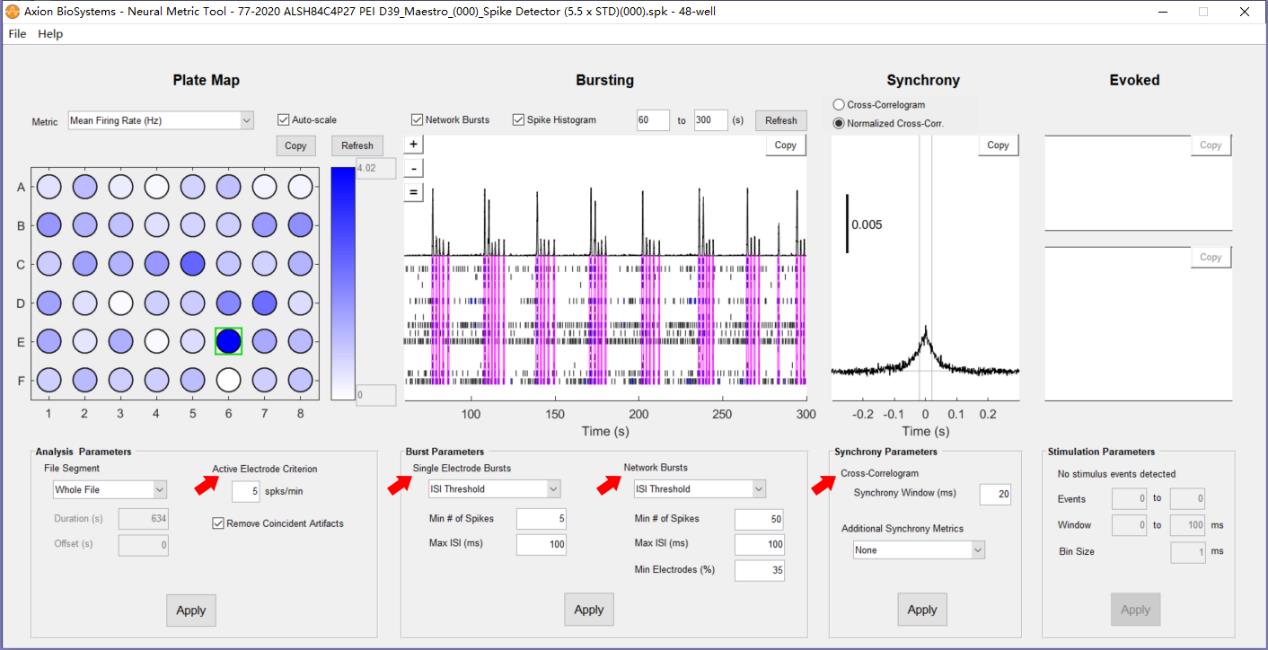
Figure 9. Neural metric settings. Red arrows indicate the settings for defining Active Electrode, Burst, Network Burst, and Synchrony.
c. Representative network activities of hiPSC–sMNs are exported from the Bursting panel, which are shown in Figure 10. A detailed comparison of MEA metrics (e.g., firing rate, burst duration, synchrony) is beyond the scope of this protocol but can be found in our previous publication [14].
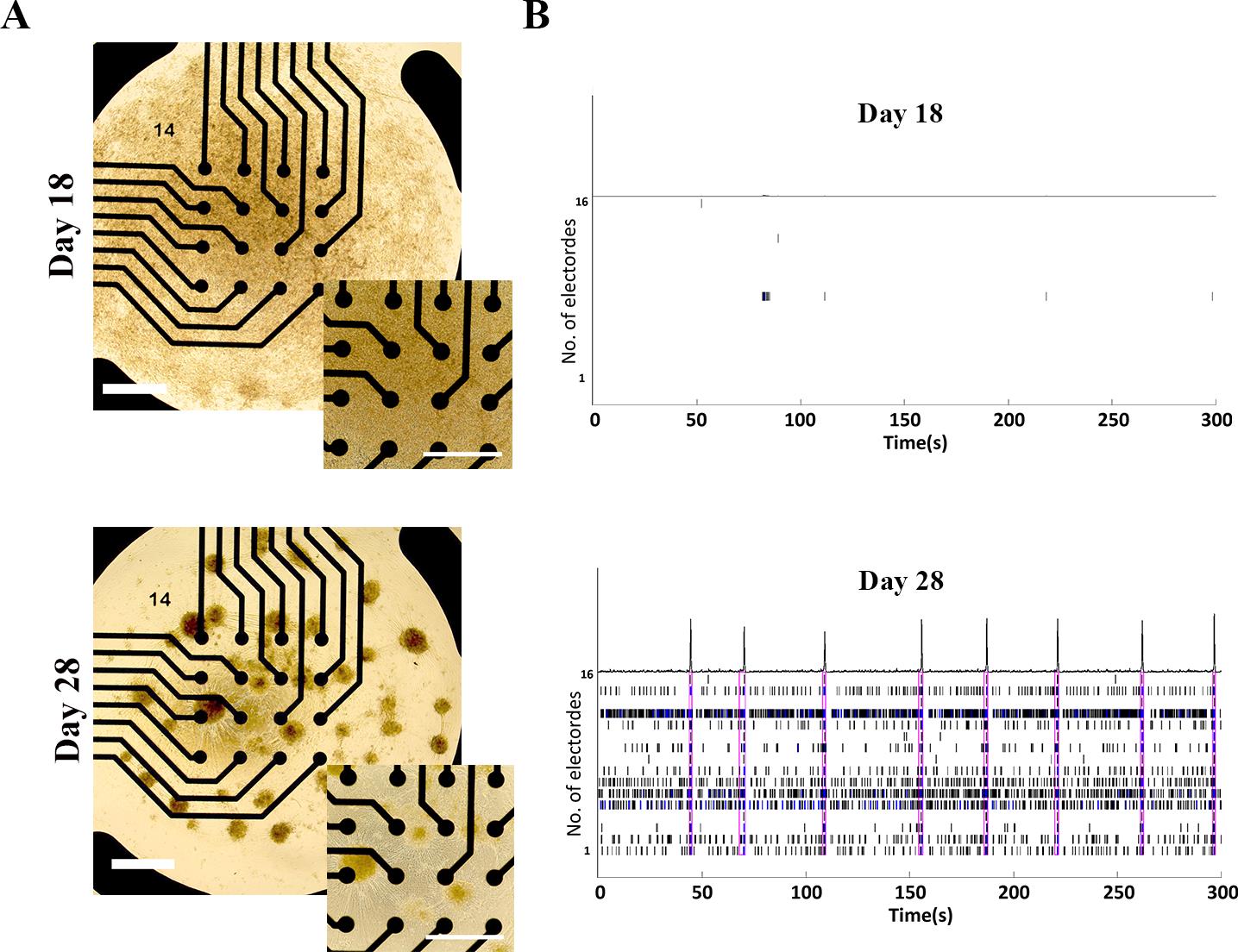
Figure 10. Functional network formation of human induced pluripotent stem cell (hiPSC)-derived spinal motor neurons (sMNs) on days 18 and 28 of differentiation. (A) Representative brightfield images of iPSC-derived day 18 (top) and day 28 (bottom) sMNs on a Poly-L-ornithine/Matrigel (POM)-coated 48-well multielectrode array (MEA) plate with 16 microelectrodes per well. Scale bars, 100 μm. (B) Representative MEA recording demonstrating typical network firing patterns of iPSC-derived sMNs on day 18 (top) and day 28 (bottom) of differentiation.
4. MEA clean up and sterilization for reuse
a. Rinse MEA plates three times with 500 μL of sterile DPBS w/o Ca2+ and Mg2+ to remove residual medium and cellular debris.
b. Add 50 μL of 0.05% Trypsin/EDTA to the center of each well of the CytoView 48-well MEA plate and incubate the plate at 37 °C for 30 min to digest residual proteins.
c. Wash each well three times with 500 μL of sterile deionized water to remove debris.
d. Examine the plate under a microscope to confirm complete removal of debris. Cleaning steps can be repeated if necessary.
e. Fill all wells and reservoirs of the MEA plate with 70% ethanol and incubate for 10 min at room temperature in the biological safety cabinet for sterilization.
f. Discard 70% ethanol and rinse once with 90% ethanol.
g. Discard 90% ethanol and rinse three times with sterile deionized water.
h. Remove residual water by gently shaking the plate upside-down.
i. Air-dry the plate with the lid open under UV light in a biological safety cabinet for additional sterilization.
j. MEA plates can be reused after proper cleaning and sterilization.
Data analysis
Quantification of ICC staining using the Operetta High Content Imaging System with Harmony software (PerkinElmer): For each sample, 15 fields are randomly selected for imaging by the Operetta High Content Imaging System. Image analysis is conducted with Harmony software. The steps for Harmony Image analysis are described below:
1. Nuclei identification: Nuclei are identified using Hoechst/DAPI channel with the Find Nuclei button. Settings for diameter and shape are adjusted to ensure only intact, whole nuclei are quantified.
2. Cytoplasm identification: Cytoplasmic regions are detected using the appropriate channels (e.g., 488, 555, or 647 nm) with the Find Cytoplasm button.
4. Intensity quantification: Following the identification of cellular compartments, staining intensities are quantified for both the nuclei and cytoplasm using the Calculate Intensity Properties: Nuclei/Cytoplasm button.
All image analysis settings are kept consistent across all replicates within a given experiment.
Validation of protocol
Using this protocol, we generated enriched CHAT+ cervical spinal MNs (sMNs) (88%–97% purity) from three healthy hiPSC lines and four sporadic ALS hiPSC lines within 18 days and functionally mature sMNs within 28 days, as validated by whole-cell patch clamp, MEA assay, and calcium imaging. This simple and reproducible protocol enabled the detection of hyperexcitability phenotypes in iPSC-derived sMNs from patients with sporadic ALS. These findings have been published in the following research articles:
• Yang et al. [9]. A novel protocol to derive cervical motor neurons from induced pluripotent stem cells for amyotrophic lateral sclerosis. Stem Cell Reports. (Figure 1A–D, Figure 2B–D, Figure 3F–D, Figure 4B, D, Figure 5A–E, and Figure 6A–D).
• Yang et al. [14]. Polyethyleneimine facilitates the growth and electrophysiological characterization of iPSC-derived motor neurons. Scientific Reports (Figure 3B–C and Figure 4A–D, F, H).
General notes and troubleshooting
General notes
1. Since the E8 Flex medium for iPSC culture does not contain antibiotics, aseptic techniques must be maintained. To prevent mycoplasma contamination, perform mycoplasma testing upon each new iPSC recovery and conduct routine mycoplasma testing every two months for all cultures.
2. The quality of hiPSCs significantly impacts differentiation efficiency. It is recommended to culture at least two but no more than five passages after iPSC recovery from liquid nitrogen to ensure a stable and undifferentiated state.
3. Typically, hiPSCs at 70%–80% confluency 3–4 days after passaging are suitable for differentiation. Avoid using hiPSC clones with dense centers, as these often contain differentiated cells inside. If observed, re-passage the hiPSCs to obtain uniformly sized hiPSC clusters and ensure even distribution after replating.
4. Due to inherent variability in iPSC differentiation, closely monitor cell morphology throughout the process. Quality checks at the NSCs and/or MNPs stages are recommended before proceeding to terminal differentiation.
Troubleshooting
Problem 1: Nonspecific differentiation observed on days 6 and 12 (Figure 11).
Possible cause: Poor hiPSC quality or instability of the hiPSC line used.
Solution: Restart differentiation using a high-quality hiPSC or a stable hiPSC line without spontaneous differentiation. If limited to a subset of cells, thoroughly remove differentiated cells before MN differentiation.
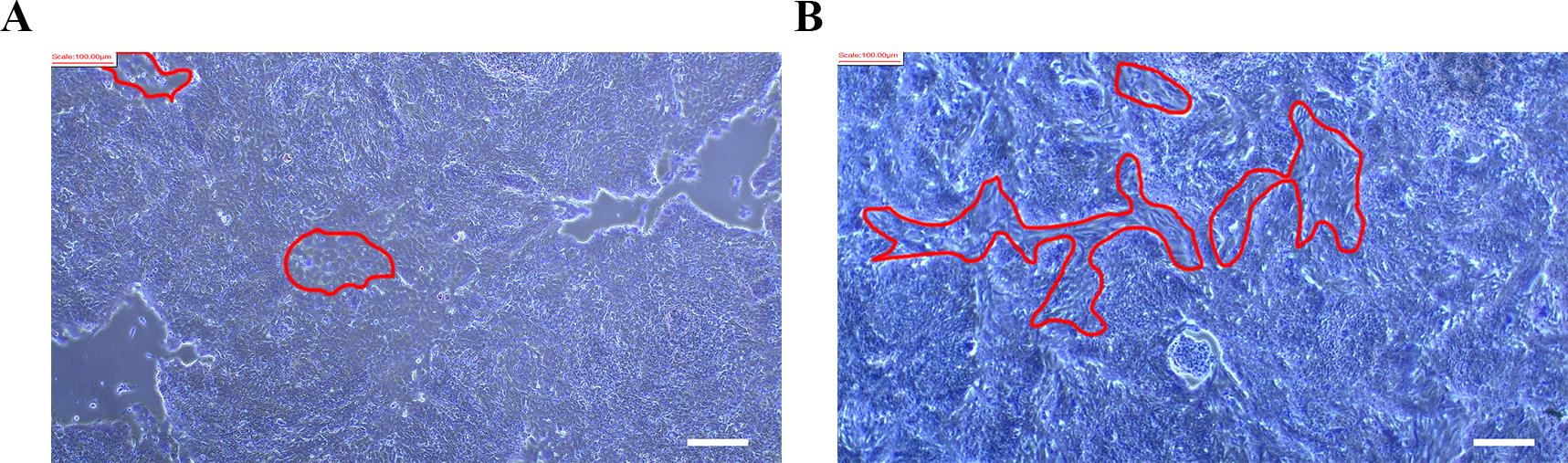
Figure 11. Nonspecific differentiation on day 6 (A) and day 12 (B). Red circles indicate nonspecific differentiation cells. Scale bars, 100 µm.
Problem 2: Detachment of hiPSC–sMNs from the culture plates after day 18 of differentiation.
Possible cause: As hiPSC–sMNs mature, cell attachment becomes weaker due to extensive neurite outgrowth and network formation. Aggressive medium changes, mechanical disturbance (e.g., violent shaking), or prolonged exposure to room temperature can promote detachment.
Solution: Handle plates gently, perform half-medium change instead of full replacements, and limit exposure to room temperature to less than 15 min.
Problem 3: It has been observed that hiPSC–sMNs tend to form neural aggregates during in vitro maturation (Figure 10A), which may be attributed to the degradation of peptide- or protein-based coating substrates such as Poly-L-ornithine or Matrigel. To alleviate this phenomenon, 0.1% polyethyleneimine in ddH2O can be used as an alternative coating material for the generation and maturation of hiPSC–sMNs. Although cells cultured on PEI-coated plates exhibit delayed maturation compared to those on POM-coated substrates, they demonstrate improved distribution uniformity [14]. The detailed coating method is described in the published reference [14].
Acknowledgments
1) Specific contributions of each author: Conceptualization: S.S., T.O., and M.Y.; experimental investigation and data analysis: M.Y., and Y.C.; original draft: Y.C., and Y.F.; review & editing: S.S., and M.Y.
2) Funding sources that supported the work: This research was supported by the SFI Investigator award (13/IA/1787), Centre Grant Number 16/RC/3948 and 21/RC/10294_P2 (which was co-funded under the European Regional Development Fund and by FutureNeuro industry partners), Galway University Foundation, the Doctoral Research Initiation Fund Project of Hebei Normal University (L2024B36), the Hebei Natural Science Foundation (C2024205026), and Provincial Medical Outstanding Talents Project funded by the Provincial Government in 2022 (Leader): Research on the Pathogenesis, Diagnosis and Treatment of childhood Migraine.
This protocol was used in [9,14].
Competing interests
The authors declare that no competing interests exist.
Ethical considerations
All procedures and protocols have been reviewed and approved by the Galway Clinical Research Ethics Committee. Title: "Making Stem Cells from Somatic Tissues (iPS Study)," Approval Number: C.A.750, Date of Approval: 12 March 2020. Written informed consent was obtained from all subjects and/or their legal guardian(s). All methods were performed in accordance with the relevant guidelines and regulations.
References
- Hardiman, O., Al-Chalabi, A., Chio, A., Corr, E. M., Logroscino, G., Robberecht, W., Shaw, P. J., Simmons, Z., van den Berg, L. H. (2017). Amyotrophic lateral sclerosis. Nat Rev Dis Primers. 3: 17071. https://doi.org/10.1038/nrdp.2017.71
- Goutman, S. A., Hardiman, O., Al-Chalabi, A., Chió, A., Savelieff, M. G., Kiernan, M. C. and Feldman, E. L. (2022). Emerging insights into the complex genetics and pathophysiology of amyotrophic lateral sclerosis. Lancet Neurol. 21(5): 465–479. https://doi.org/10.1016/s1474-4422(21)00414-2
- Cave, C. and Sockanathan, S. (2018). Transcription factor mechanisms guiding motor neuron differentiation and diversification. Curr Opin Neurobiol. 53: 1–7. https://doi.org/10.1016/j.conb.2018.04.012
- Jessell, T. M. (2000). Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 1(1): 20–29. https://doi.org/10.1038/35049541
- Muñoz-Sanjuán, I. and Brivanlou, A. H. (2002). Neural induction, the default model and embryonic stem cells. Nat Rev Neurosci. 3(4): 271–280. https://doi.org/10.1038/nrn786
- Shirasaki, R. and Pfaff, S. L. (2002). Transcriptional Codes and the Control of Neuronal Identity. Annu Rev Neurosci. 25(1): 251–281. https://doi.org/10.1146/annurev.neuro.25.112701.142916
- Sances, S., Bruijn, L. I., Chandran, S., Eggan, K., Ho, R., Klim, J. R., Livesey, M. R., Lowry, E., Macklis, J. D., Rushton, D., et al. (2016). Modeling ALS with motor neurons derived from human induced pluripotent stem cells. Nat Neurosci. 19(4): 542–553. https://doi.org/10.1038/nn.4273
- Giacomelli, E., Vahsen, B. F., Calder, E. L., Xu, Y., Scaber, J., Gray, E., Dafinca, R., Talbot, K. and Studer, L. (2022). Human stem cell models of neurodegeneration: From basic science of amyotrophic lateral sclerosis to clinical translation. Cell Stem Cell. 29(1): 11–35. https://doi.org/10.1016/j.stem.2021.12.008
- Yang, M., Liu, M., Sánchez, Y. F., Avazzadeh, S., Quinlan, L. R., Liu, G., Lu, Y., Yang, G., O'Brien, T., Henshall, D. C., et al. (2023). A novel protocol to derive cervical motor neurons from induced pluripotent stem cells for amyotrophic lateral sclerosis. Stem Cell Rep. 18(9): 1870–1883. https://doi.org/10.1016/j.stemcr.2023.07.004
- Bianchi, F., Malboubi, M., Li, Y., George, J. H., Jerusalem, A., Szele, F., Thompson, M. S. and Ye, H. (2018). Rapid and efficient differentiation of functional motor neurons from human iPSC for neural injury modelling. Stem Cell Res. 32: 126–134. https://doi.org/10.1016/j.scr.2018.09.006
- Qu, Q., Li, D., Louis, K. R., Li, X., Yang, H., Sun, Q., Crandall, S. R., Tsang, S., Zhou, J., Cox, C. L., et al. (2014). High-efficiency motor neuron differentiation from human pluripotent stem cells and the function of Islet-1. Nat Commun. 5(1): e1038/ncomms4449. https://doi.org/10.1038/ncomms4449
- Du, Z. W., Chen, H., Liu, H., Lu, J., Qian, K., Huang, C. L., Zhong, X., Fan, F. and Zhang, S. C. (2015). Generation and expansion of highly pure motor neuron progenitors from human pluripotent stem cells. Nat Commun. 6(1): e1038/ncomms7626. https://doi.org/10.1038/ncomms7626
- Yang, M., Liu, M., Ding, Y., Vajda, A., Ma, J., Cui, H., O'Brien, T., Henshall, D., Hardiman, O., Shen, S., et al. (2020). Generation of twelve induced pluripotent stem cell lines from two healthy controls and two patients with sporadic amyotrophic lateral sclerosis. Stem Cell Res. 44: 101752. https://doi.org/10.1016/j.scr.2020.101752
- Yang, M., You, D., Liu, G., Lu, Y., Yang, G., O’Brien, T., Henshall, D. C., Hardiman, O., Cai, L., Liu, M., et al. (2024). Polyethyleneimine facilitates the growth and electrophysiological characterization of iPSC-derived motor neurons. Sci Rep. 14(1): e1038/s41598–024–77710–1. https://doi.org/10.1038/s41598-024-77710-1
Article Information
Publication history
Received: Jul 21, 2025
Accepted: Sep 9, 2025
Available online: Sep 23, 2025
Published: Oct 20, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Chen, Y., Yang, F., O'Brien, T., Shen, S. and Yang, M. (2025). Rapid and Simplified Induction of Spinal Motor Neurons From Human Induced Pluripotent Stem Cells. Bio-protocol 15(20): e5478. DOI: 10.21769/BioProtoc.5478.
Category
Stem Cell > Pluripotent stem cell > Cell differentiation
Neuroscience > Nervous system disorders > Cellular mechanisms
Medicine
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link