- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
An Effective and Safe Maize Seed Chipping Protocol Using Clipping Pliers With Applications in Small-Scale Genotyping and Marker-Assisted Breeding
Published: Vol 15, Iss 3, Feb 5, 2025 DOI: 10.21769/BioProtoc.5200 Views: 1853
Reviewed by: Samik BhattacharyaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Optimized Protocol for DNA Extraction in Three Theobroma Species
Angie F. Riascos-España [...] Pedro A. Velasquez-Vasconez
May 5, 2025 2050 Views

CRISPR/Cas9-Induced Targeted Mutagenesis of the Moss Physcomitrium patens by Particle Bombardment-Mediated Transformation
So Takenaka [...] Setsuyuki Aoki
Sep 20, 2025 1009 Views

Identifying Causal Genes and Building Regulatory Networks in Crops Using the CisTrans-ECAS Method
Yutong Yan [...] Weibo Xie
Feb 5, 2026 101 Views
Abstract
In applications such as marker-assisted breeding and positional cloning, tissue sampling and plant tracking are vital steps in the genotyping pipeline. They enable the identification of desirable seedlings, saving time and reducing the cost, space, and handling required for growing adult plants, especially for greenhouses and winter nurseries. Small-scale marker-assisted selection laboratories rely heavily on leaf-based genotyping, which involves over-planting large, segregating populations followed by leaf sampling, genotyping, and backtracking to identify desired individuals, which is costly and laborious. Thus, there is a need to adopt seed-based genotyping to reduce costs and save time. Therefore, we developed a safe and cheap seed-chipping protocol using clipping pliers to chip seeds to genotype before planting. To identify a cost-effective and high-throughput DNA extraction method, we tested four extraction methods and assessed the quality of the seed DNA using PCR. For three of the methods, seed-based DNA was of comparable quality to DNA extracted from leaf punches. We also compared seed- and leaf-derived DNA from the same individuals in a segregating population to test for genotyping miscalls that could arise due to the presence of maternally derived pericarp in the seed samples. Out of 43 potential instances, we found zero miscalled samples and, therefore, no evidence supporting consequential pericarp inclusion. Germination rates of chipped and unchipped seeds were the same for the inbreds tested, B73 and Mo17. However, chipped seeds grew slower until ~14 days after sowing. Overall, seed sampling using clipping pliers provides a simple, reliable, and high-throughput method to identify specific genotypes before planting.
Key features
• Provides a quick, safe, and cheap sampling technique for maize kernels that may also be suitable for other plants with relatively large seeds.
• Includes procedures and materials to track and organize samples within and across batches involving tens to thousands of seeds.
• Seeds can be sampled and genotyped relatively quickly for planting; in one day, 384 seeds can be sampled, processed for DNA, and genotyped by PCR.
Keywords: MaizeBackground
Over the last decades, marker-assisted selection (MAS) has become integral to most plant breeding systems because quantitative trait loci (QTL) and molecular markers linked to desired traits have enhanced the selection efficiency of desirable plants and shortened breeding cycles [1]. Unlike commercial breeding companies with highly automated and streamlined MAS and trait-discovery programs, public-sector breeding programs and small-scale genotyping laboratories rely on laborious and time-consuming tissue sampling and DNA extraction procedures [2]. Compared to the proprietary seed-based MAS programs utilized by industry scientists, public-sector genotyping systems that rely heavily on leaf-based DNA genotyping may be particularly resource- and time-demanding [1] as the marker genotyping process must be coupled to the sowing and germinating of seeds, caring for seedlings, tracking of live plants to static samples, and the immediate interpretation of data in order to identify, retain, and rear the desired, growing plants for their intended purpose. Thus, to improve the efficiency of MAS and related approaches, there is a need to adopt seed-based genotyping as a separate, pre-planting process, which reduces overall labor costs and saves space and time through selection of desirable genotypes during the off-season [1,2]. During trait discovery, seed-based DNA genotyping ensures that seeds with desirable traits are selected and planted, thus saving time and costs associated with greenhouse use and winter fields. In addition to tissue sampling, optimizing rapid and cost-effective DNA extraction methods to screen large populations will be essential for a successful MAS and genotyping pipeline.
Seed-based DNA genotyping methods have been implemented in small-scale maize breeding programs and genotyping laboratories but are still ineffective based on safety and scalability [2,3]. One seed-based DNA genotyping method is based on soaking maize kernels in water before endosperm chipping [2]. The caveats of this procedure include susceptibility to fungal contamination and low seed germination if kernels are not dried and stored well. Another common method uses a razor blade for seed chipping [3], which presents injury hazards and is time-consuming, thus making it impractical to quickly chip a large population. In this study, we used clipping pliers, a safe alternative to previous chipping methods that is scalable to high-throughput applications. This seed-chipping method did not significantly impact germination rates for maize B73 and Mo17 seeds, although seedlings from chipped seeds had reduced vigor that was evident for 14 days after planting, after which they appeared comparable to their non-chipped sibling seedlings. In addition, we tested four DNA extraction methods, based on each of cetyltrimethylammonium bromide (CTAB) [4], sodium dodecyl sulfate (SDS) [5], urea [6], and guanidine hydrochloride [7], and optimized a cost- and time-efficient DNA extraction protocol for maize endosperm samples.
Materials and reagents
Biological materials
1. Maize kernels (seeds)
Reagents
1. GoTaq GreenMaster Mix 2× (Promega, catalog number: M7123)
2. Dimethyl sulfoxide (DMSO) (Promega, CAS 67-68-5)
3. Betaine (Promega, CAS 107-43-7)
Laboratory supplies
1. 2.2 mL 96-well sample collection plates (n+1 plates for n sets of 96 tissue samples; see step A1) (VWR, catalog number 43001-0020)
2. 1.2 mL sampling tubes (96 for one set of 96 tissue samples, if using SDS or filter plate DNA extraction) (VWR, catalog number 83009-678)
3. 2.0 mL safe-lock microcentrifuge tubes (96 for one set of 96 tissue samples, if using CTAB DNA extraction) (Eppendorf, catalog number 022363352)
4. Labeling tape
Equipment
1. Forceps (VWR, catalog number 82027-446)
2. Pet nail clipping pliers (Resco, catalog number: PF0728); see General Note 1
3. Mini ice cube tray, used for seed storage, 135 cubes per tray 0.5 inch size (e.g., Source 1 or Source 2).
Software
1. Tissue Sample Plate Mapper software; available as a Google sheet plug-in (Google Workspace
Marketplace, plug-in number: 115639436496)
2. Excel or other spreadsheets may be useful for additional notetaking
3. Primer3 software (https://bioinfo.ut.ee/primer3-0.4.0/)
4. MaizeGDB (http://www.maizegdb.org) for primer blast analysis
Procedure
A. Labeling seed trays and collection plates
1. During seed chipping, two 96-well sample collection plates are needed. One plate (the tube holder; Figure 1A) holds one sampling tube at a time to aid tissue collection during chipping.
2. The second 96-well sample collection plate (for tissue storage; Figure 1A) stores up to 96 accumulated sampling tubes containing chipped tissue. Optionally, prepare the tissue storage plate by printing a plate map using the tissue-sample plate mapper software, overlaying the map on a 96-well sample collection plate and securing it with clear tape (see General Note 4). Label this plate with a unique ID using tape and a marker.
3. One seed storage tray is used to hold chipped maize seeds until analysis and decision-making are completed. Lab tape may be used to block off superfluous sections so the tray holds 96 seeds in an 8 × 12 format (Figure 1A). Label this tray with the same unique ID to pair it with a tissue storage plate.
B. Maize seed chipping
See Video 1 for a demonstration of the complete chipping procedure.
1. On a large laboratory bench, place the tube holder plate, labeled tissue storage plate, and seed storage tray with the tube holder plate closest to you and the seed storage tray farthest from you (Figure 1A).
2. Place one sampling tube in one of the tube holder plate’s bottommost corner wells. To help contain debris produced in the chipping process, unused wells in the tube holder plate may be covered with masking tape (optional). The corner location for the sampling tube facilitates positioning the clipping pliers right above the sampling tube (Figure 2H, 2K).
3. Using the dominant hand, hold the clipping pliers' blade side up, with the thumb on the top, stationary handle, and the other fingers holding the lower handle with the spring attached that moves the blade.
4. With your other hand’s thumb and index finger, pick up and hold one maize seed by the tip cap with the embryo side facing toward the thumb (Figure 2H–2I).
5. While holding the chipper orifice directly and slightly above the sampling tube in the tube holder plate, hold the seed approximately perpendicular to the pliers, place its crown (Figure 2A) into the chipper hole, and gently chip the seed repeatedly to shave off fine tissue particles (Figures 2H–2J). For example, three to five passes of the chipping blade are usually required with B73 and Mo17 kernels.
Critical: Chip and shave the tissue finely (Figures 2B, 2D) because large chip particles (Figure 2F) grind poorly and barely yield extracted DNA (see General Note 2).
Critical: It is imperative to avoid damaging the embryo (Figure 2A) while chipping the endosperm. If necessary, secondary chips may be collected, e.g., from small to or rounded kernels like from the W22 inbred, by slightly tilting the seed at a 45° angle to the first plane of chipping. Then, chip and shave fine endosperm particles as described above for the crown, but instead chip from the side opposite the embryo (Figure 2K–2M). A combination of such primary and secondary chips also increases the proportion of endosperm tissue in the sample relative to maternal pericarp tissue.
Critical: Processing too much tissue may reduce DNA yield in the extraction step. Using the extraction methods tested, approximately 15–30 mg of tissue optimally yields approximately 5 μg of DNA at concentrations appropriate for use as PCR template.
6. If the SDS, filter plate, or urea method is used for DNA extraction (section C), then remove the sample tube with chipped tissue from the tube holder plate and place it into the tissue storage plate; then, place the chipped seed into the appropriate well in the seed tray. For tracking purposes, match the chipped seed’s position in the seed tray grid with the position of the sampling tube on the tissue storage plate grid. If CTAB is used for DNA extraction (section C), then transfer the chipped tissue into a labeled 2 mL microcentrifuge tube instead.
7. Clean the clipping pliers by wiping with a dry tissue and/or blowing off any chip residue. Application of compressed air while flexing the pliers’ handle may be used to ensure no chip dust carries over in or on the tool; more thorough cleaning of the chipping pliers may be performed periodically using 70% ethanol and compressed air.
8. Repeat steps B2–B7 with a new sampling tube and kernel until 96 or the desired number of samples have been collected.
9. When sampling is completed, seed storage trays may be stacked and placed in the sealed plastic tubs they are shipped in (see Equipment) and set aside for temporary storage. For a turnaround to sowing within six months, storage at room temperature and humidity had no apparent effects on germination frequency.
10. Proceed to DNA extraction.
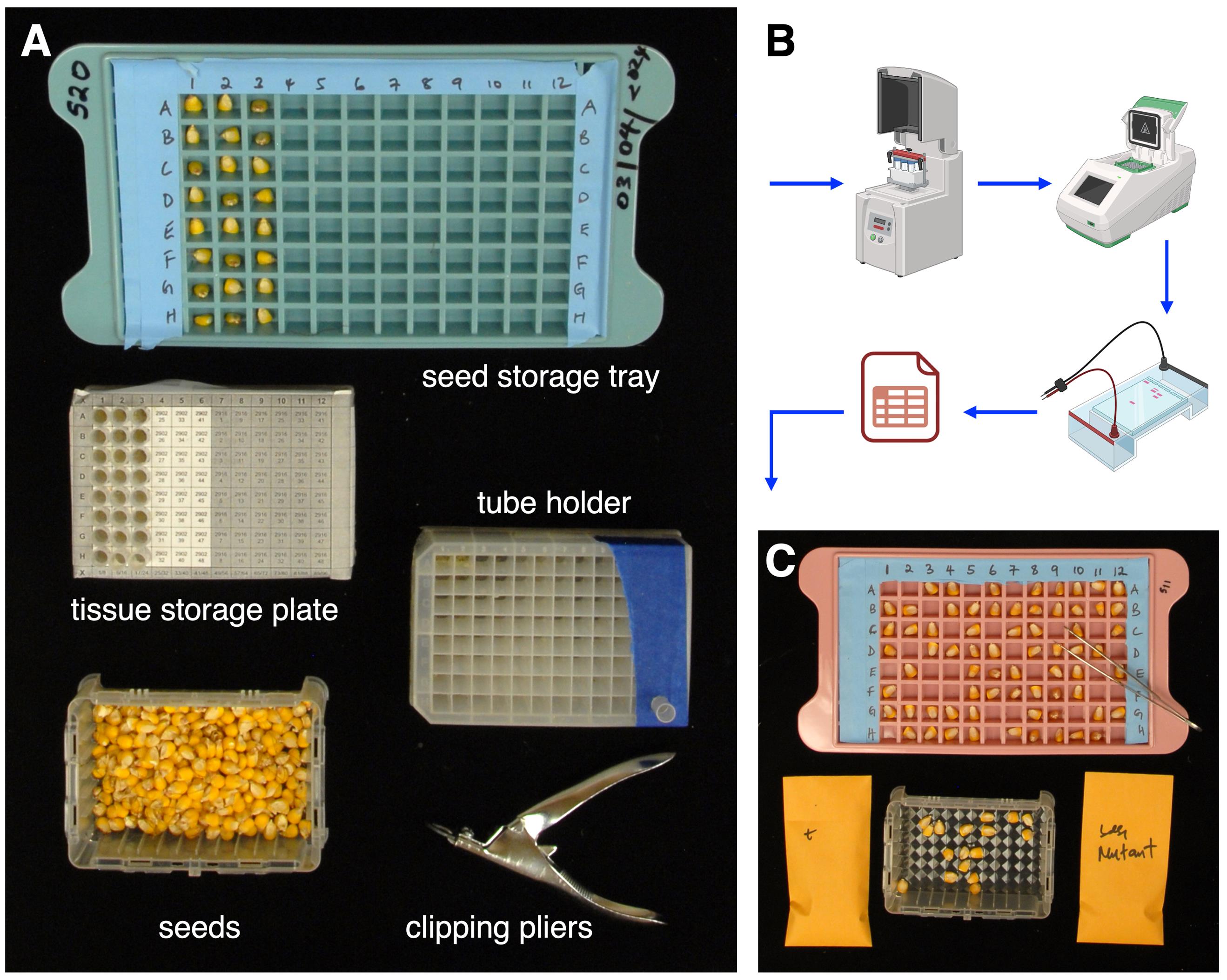
Figure 1. Maize seed chipping setup and genotyping. A. Sampling includes clipping pliers, seeds, a 96-well plate acting as a tube holder plate, a 96-well tissue storage plate overlaid with a sampling map to accumulate chipped samples in tubes, and a seed storage tray. B. Once chipping is completed, chipped tissue samples are processed for DNA extraction, which is genotyped using PCR and an agarose gel and data analysis. C. With data tracking, seeds with the genotype of interest are selected for downstream applications.
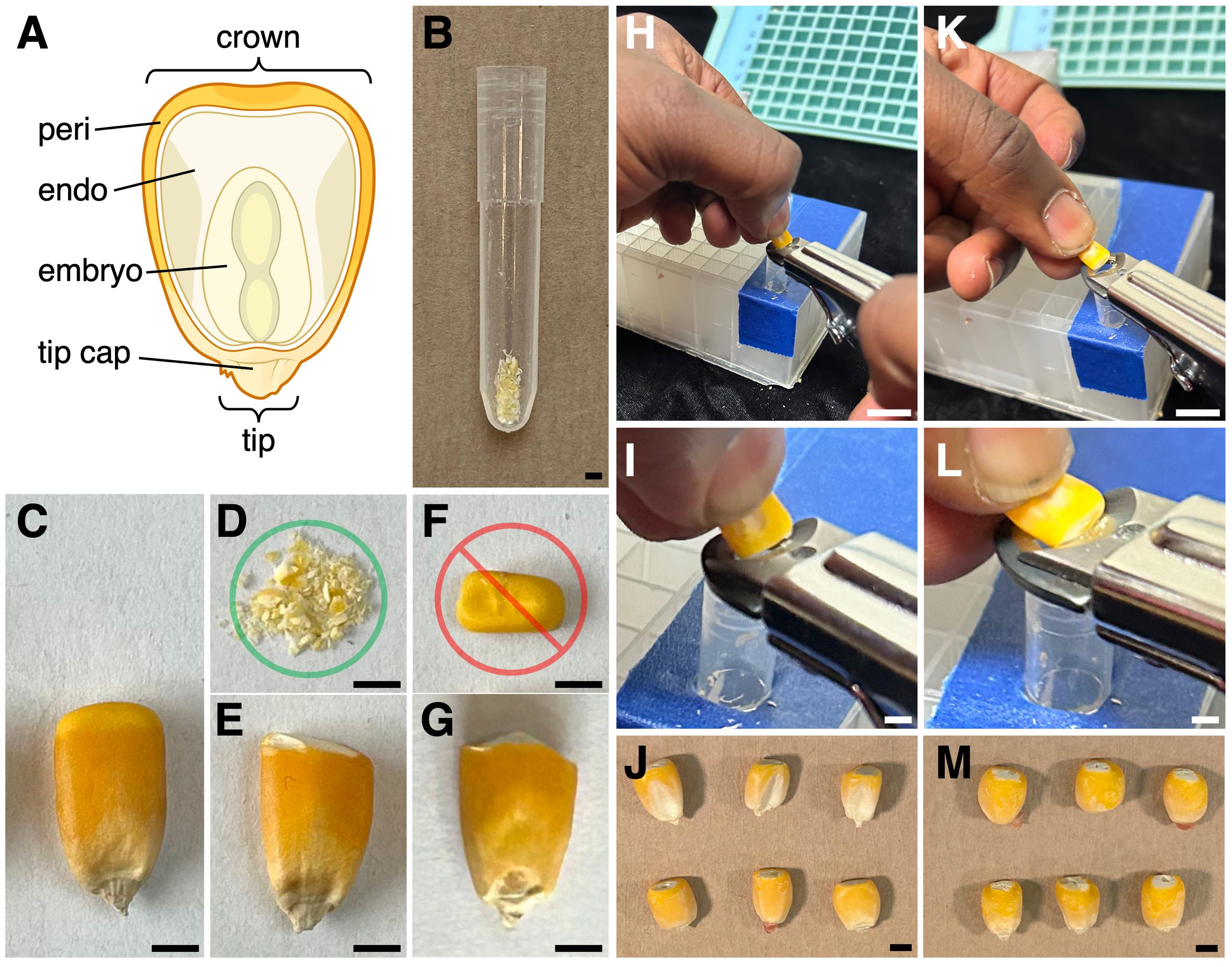
Figure 2. Maize kernels and the seed chipping process. A. Anatomy of a maize kernel in longitudinal section; peri: pericarp; endo: endosperm. B. An appropriate volume of seed chips in a collection tube. C. Intact, unchipped kernel. D, E. Properly chipped kernel shavings (D, green circle) removed from the crown of the kernel (E). F, G. Improperly chopped, large chunk(s) (F, red circle) removed from the crown (G). H–J. Seed chipping of the crown is done with the embryo facing you and the seed nearly perpendicular to the clipping pliers. K–M. Secondary chips, e.g., from round-shaped kernels, produced by holding the seed with the embryo facing upward and nearly parallel to the chipping pliers’ blade to shave chips from the back of the seed, i.e., at a 45° angle to the first chipping surface. Scale bars = 10 mm for panels H and K, scale bars = 2 mm for the remaining panels.
C. DNA extraction options
1. Guanidine-HCl filter plate extraction protocol (see Supplemental Protocol 1)
2. SDS-based DNA extraction protocol (see Supplemental Protocol 2)
3. Urea-based DNA extraction protocol (see Supplemental Protocol 3)
4. Modified CTAB DNA extraction protocol with steel-bead (https://bio-protocol.org/en/bpdetail?id=2906&type=0; [4])
D. PCR and genotyping
1. Design high-quality genotyping markers using the maize reference genome (http://www.maizegdb.org) and Primer3 (https://bioinfo.ut.ee/primer3-0.4.0/).
2. Set up a 13 μL PCR reaction composed of 2× GoTaq GreenMaster Mix (6 μL), DMSO (0.5 μL), forward and reverse primer mix (0.5 μL of 5 μM each), water (2.5 μL), and DNA (3 μL). For amplifying GC-rich regions, substitute the water in the PCR reaction with betaine (2.5 μL of 5 M).
3. Run standard PCR cycling parameters according to the primers used and product size. For instance, for Figure 4, the primers used were BZ316 (5'-ATCTGCATCCTGCGACGCAAC-3') and BZ317 (5'-GTCGGCGGTCTTTCTCGAGTC-3') and PCR conditions were 94 °C, 3 min (1 cycle); 94 °C, 30 s, 60 °C, 30 s, 72 °C, 1 min (34 cycles); 72 °C, 5 min (1 cycle); hold at 10 °C for forever. This reaction produced amplicon sizes of 555 bp (B73) and 416 bp (Mo17).
4. To genotype, run the PCR-amplified products with, e.g., 100 bp ladder marker on a 2% agarose gel in TAE buffer at 120 V for 20 min or longer depending on the product size.
5. Record the genotypes. If using the tissue-sample plate mapper software, then genotype calls may be entered in the ordered Sample List tab generated by the software.
6. After data analysis, use forceps to carefully pick chipped seeds with the genotype(s) of interest from the seed storage trays.
Data analysis
Seed chipping with nail clipping pliers provides safer, scalable, and better throughput than razor blade–based methods. Thirty to forty minutes are required to chip 96 maize seeds, depending on seed size and shape. Smaller or round seeds are challenging to hold and need more time to chip than larger ones. To examine the effects of seed chipping on germination, we planted chipped and unchipped seeds from the same two seed packets in three replications of 32 kernels in the greenhouse. One packet contained inbred B73 and the other contained inbred Mo17. Seeds were sown in prewet Metro-Mix 830 or similar in a standard maize greenhouse (16/8 h day/night photoperiod, ~27/18.5 °C, respectively) and covered with a 7-inch humidity dome to maintain moisture until emergence. Seedlings were watered sparingly, as needed. We scored germination per each kernel based on whether or not a coleoptile emerged above the soil line; we also measured vigor per each replicate as an aggregate (chipped or unchipped) based on any differences overall in growth rate or in appearance by a response to any unintended stresses that may have been present in their shared environmental conditions. We noticed that chipping did not impact the germination rate, but seedlings from chipped seeds initially grew more slowly (Figure 3; see General Note 3) while other observed responses occurred uniformly across both treatments, e.g., some yellowing occurred in all seedlings in the B73 experiment (Figure 3B).

Figure 3. Effect of seed chipping on germination and seedling growth across B73 and Mo17 maize inbreds. A. Germination frequency (y-axis, showing from 90% to 100%) of unchipped (blue) and chipped (red) seeds across two maize inbreds, Mo17 and B73 (x-axis). Each circle indicates the percentage germination from one replicate of 32 sown kernels. Each line indicates the mean for three replicates. B. Post-germination seedlings at 14 days to compare the growth of unchipped (left) and chipped seeds (right) for B73. C. Post-germination seedlings at 14 days to compare the growth of unchipped (left) and chipped seeds (right) for Mo17. Scale bars = 10 cm.
We compared the quality of DNA extracted from seeds to DNA extracted from leaves using several different DNA extraction methods. The results indicated that the quality of the DNA extracted from seeds and leaves was comparable using three of the four methods tested (Figure 4A). The exception was the urea-based method, where DNA extracted from seed chips performed poorly (Figure 4A). Regarding usability and high throughput, the extraction methods are user-friendly and high-throughput, except for CTAB. For instance, with CTAB extraction, we process a maximum of 48 samples at a time, whereas with other extraction methods, 384 samples can be processed at a time once chipping is complete. The guanidine-HCl with filter plate method produced the best-quality DNA but can include significant expenses for reagents and the filter plate. Thus, we conclude that for a seed chip sample type, the SDS extraction method optimizes the combination of usability, high throughput, and cost-effectiveness (e.g., figure 4B).
In maize seed-based genotyping, maternal pericarp tissue has led to incorrect genotype results. Gao et al. [2] reported that among the homozygotes within an F2 population, false heterozygous genotyping errors occurred at a rate of 3.8% due to maternal pericarp DNA when chipping was performed after soaking maize kernels in water. While pericarp “contamination” could be reduced or eliminated by carefully peeling pericarps away prior to chipping endosperm, that approach is labor intensive. To minimize pericarp contribution, we included secondary chips that enriched chip samples for endosperm, diluting the proportion of pericarp sampled. To quantify the significance of pericarp “contamination,” we compared genotype results in an F2 population from B73 × Mo17 by first seed-chipping 96 F2 kernels and then germinating and leaf-sampling their corresponding 96 F2 seedlings. DNA was prepared using the SDS method for both seed chip and leaf tissue and PCR was performed as above (Section D). For 88 of the 96 individuals, sufficient PCR product was detected to call genotypes for both the chip- and leaf-based methods. Among the 88 individuals, we focused on the homozygotes because in F2 seed-chip DNA, either homozygous genotype could theoretically be miscalled as heterozygous due to PCR amplification of DNA extracted from the heterozygous, maternal pericarp tissue. 47 individuals were homozygous, as called by the leaf-based method. Among their corresponding chip-DNA genotypes, 43 had the homozygous call, 4 could not be called due to no PCR product, and none were miscalled. Thus, we observed zero disparities between the seed and leaf DNA groups, and therefore found no evidence supporting pericarp contamination among the 43 homozygotes (e.g., see Figure 4B–4C).
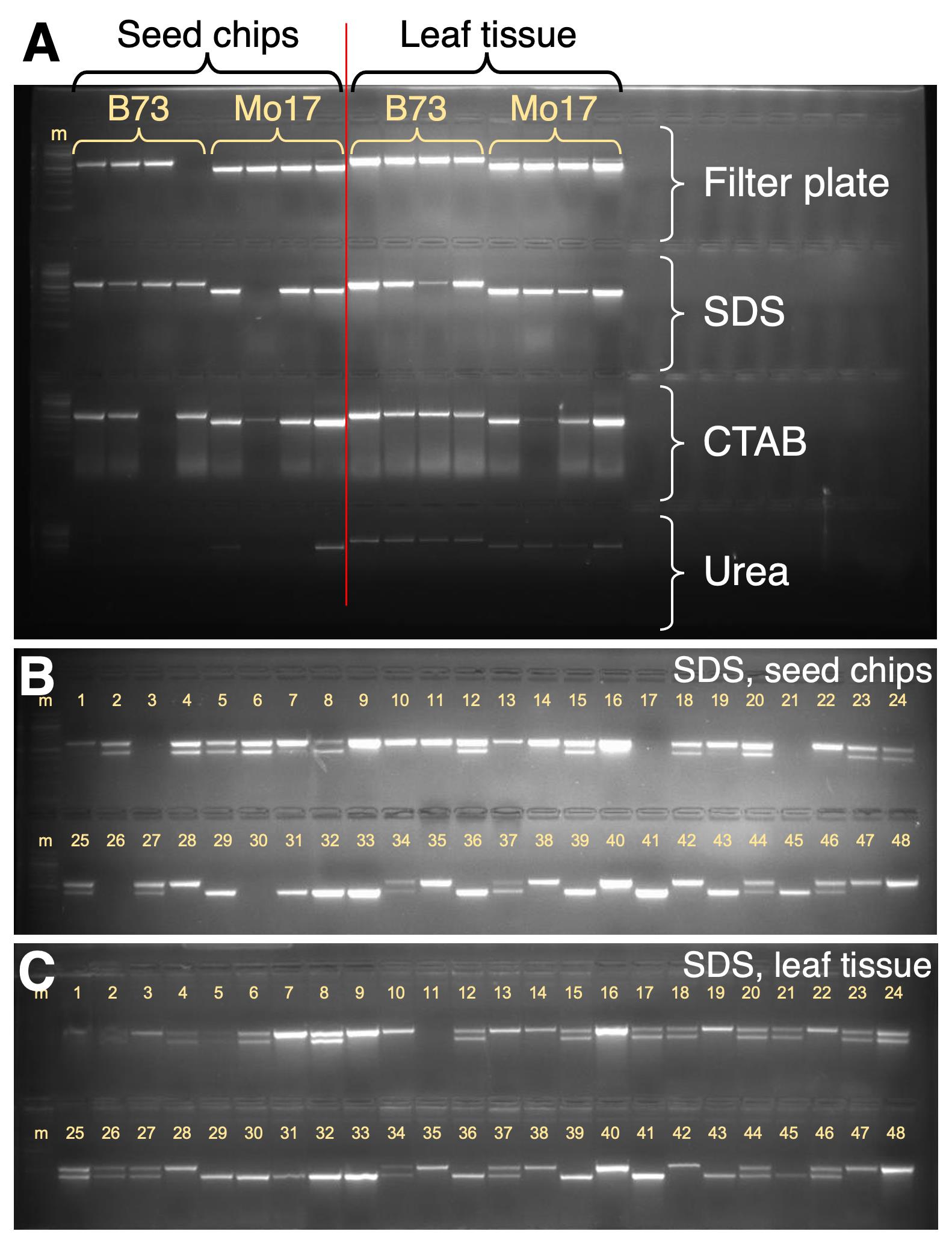
Figure 4. PCR-based genotyping using DNA extracted from chipped seed endosperm and comparison with DNA from leaves. A. Comparison of seed-based and leaf-based DNA across four DNA extraction methods. B–C. Testing for pericarp-based genotyping miscalls by comparing corresponding chipped (B) seed- and (C) leaf-based DNA from the same 96 individuals; representative data from 48 individuals (1–48, yellow numbers) are shown. Homozygotes show upper band alone (e.g., individual 7) or lower band alone (e.g., individual 29); heterozygotes show both bands (e.g., individual 6). M: marker DNA; New England Biolabs 100 bp DNA ladder; intense bands are 500 bp and 1,000 bp.
Validation of protocol
This protocol was used in the authors’ research over the last year to chip and genotype over 1,300 kernels and then selectively germinate the desired class(es). We used SDS-based DNA extraction and genotyped at six different loci for which we had previously developed PCR assays using DNA extracted from leaves. All six assays worked well with DNA extracted from chipped endosperm and the results depicted in Figure 4B are highly representative; genotype calls were returned for upward of 90% of the chipped kernels. Chipped kernels generally germinated at the same frequency as unchipped ones.
General notes and troubleshooting
General notes
1. Plastic and metal nail clipping pliers were tested (Figure 5); we recommend the metal pliers because they are durable and the blade can be removed for sharpening or replacement.
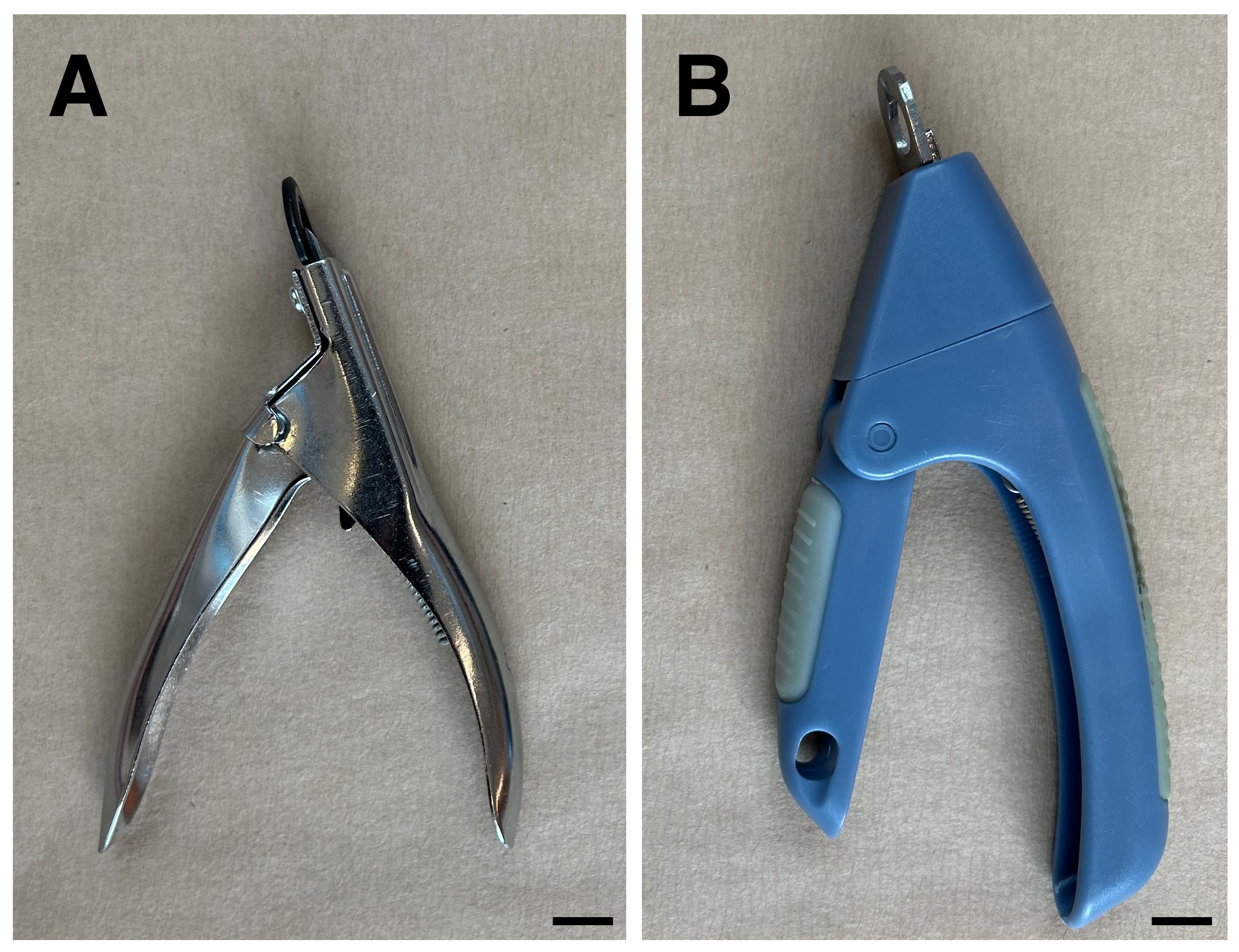
Figure 5. Nail clipping pliers tested. Clippers for seed chipping were either metal with an exchangeable blade (A, preferred) or plastic with a fixed blade (B).
2. Sufficient chipped seed tissue (15–30 mg) should be collected for adequate DNA for one to several genotyping reactions. Within that constraint, keep the chipped tissue pieces as small as possible because larger particles are more challenging to grind in the DNA extraction step. During the chipping process, chip more endosperm to dilute and minimize the amount of pericarp sampled.
3. Seedlings from chipped seeds grew slower, perhaps due to reduced mobilization of nutrients from the reduced volume of aleurone and endosperm. Thus, we also recommend not chipping more endosperm than required for adequate DNA yield (see note 2). Moreover, chipped seeds are susceptible to mold infection, especially if germinated in paper towels. Thus, treating or disinfecting the seed before planting may be warranted.
4. Additional details on installing and using the plate mapping software as a Google sheet plug-in are available through the Vollbrecht lab website (https://faculty.sites.iastate.edu/vollbrec/tissue-sample-plate-mapper). When printing plate maps using the software, navigate to the Plate labels for printing tab and execute the print command with these settings: Print current sheet; Paper size, letter; Landscape orientation; Scale, normal (100%); Margins, normal; Formatting, show gridlines and show notes; Page order, over then down; Alignment horizontal, center; Alignment vertical, center; Headers and Footers selections according to your preferences.
5. Trays with chipped seeds may be stored at room temperature for up to six months. Cool (to prevent damage from pests such as mice or moths) and low-humidity conditions (e.g., 8 °C, ~25% relative humidity) would be optimal for maize, especially for long-term storage [8]. A refrigerator or cold room that is not humidity-controlled would result in high humidity levels (~ 100% relative humidity) and would be detrimental to viability over time; it is less desirable than storing at room-temperature conditions. We have not investigated how chipping affects overall seed storage longevity.
Supplementary information
The following supporting information can be downloaded here
- Supplemental Protocol 1
- Supplemental Protocol 2
- Supplemental Protocol 3
Acknowledgments
The tissue-sample plate mapper software was developed by Takao Shibamoto and Kokulapalan Wimalanathan with support from the National Science Foundation NSF-IOS grant 1238202 to E. Vollbrecht. Some figures were generated with BioRender.com. This research was also supported by funding from the Iowa State University Crop Bioengineering Center.
Competing interests
The authors declare no competing interests.
References
- Xu, Y. and Crouch, J. H. (2008). Marker‐Assisted Selection in Plant Breeding: From Publications to Practice. Crop Sci. 48(2): 391–407. https://doi.org/10.2135/cropsci2007.04.0191
- Gao, S., Martinez, C., Skinner, D. J., Krivanek, A. F., Crouch, J. H. and Xu, Y. (2008). Development of a seed DNA-based genotyping system for marker-assisted selection in maize. Mol Breed. 22(3): 477–494. https://doi.org/10.1007/s11032-008-9192-4
- Mills, A., Allsman, L., Leon, S. and Rasmussen, C. (2020). Using Seed Chipping to Genotype Maize Kernels. Bio Protoc. 10(6): e3553. https://doi.org/10.21769/bioprotoc.3553
- Yi, S., Jin, W., Yuan, Y. and Fang, Y. (2018). An Optimized CTAB Method for Genomic DNA Extraction from Freshly-picked Pinnae of Fern, Adiantum capillus-veneris L. Bio Protoc. 8(13): e2906. https://doi.org/10.21769/bioprotoc.2906
- Edwards, K., Johnstone, C. and Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19(6): 1349–1349. https://doi.org/10.1093/nar/19.6.1349
- Leach, K. A., McSteen, P. C. and Braun, D. M. (2016). Genomic DNA Isolation from Maize (Zea mays) Leaves Using a Simple, High‐Throughput Protocol. Curr Protoc plant Biol. 1(1): 15–27. https://doi.org/10.1002/cppb.20000
- Gao, H., Smith, J., Yang, M., Jones, S., Djukanovic, V., Nicholson, M. G., West, A., Bidney, D., Falco, S. C., Jantz, D., et al. (2010). Heritable targeted mutagenesis in maize using a designed endonuclease. Plant J. 61(1): 176–187. https://doi.org/10.1111/j.1365-313x.2009.04041.x
- Guzzon, F., Gianella, M., Velazquez Juarez, J. A., Sanchez Cano, C. and Costich, D. E. (2021). Seed longevity of maize conserved under germplasm bank conditions for up to 60 years. Ann Bot. 127(6): 775–785. https://doi.org/10.1093/aob/mcab009
Article Information
Publication history
Received: Apr 4, 2024
Accepted: Dec 5, 2024
Available online: Jan 9, 2025
Published: Feb 5, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Zebosi, B., Ssengo, J., Geadelmann, L. F., Unger-Wallace, E. and Vollbrecht, E. (2025). An Effective and Safe Maize Seed Chipping Protocol Using Clipping Pliers With Applications in Small-Scale Genotyping and Marker-Assisted Breeding. Bio-protocol 15(3): e5200. DOI: 10.21769/BioProtoc.5200.
Category
Plant Science > Plant molecular biology > Genetic analysis
Molecular Biology > DNA > DNA extraction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









