- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
From Llama to Nanobody: A Streamlined Workflow for the Generation of Functionalised VHHs
Published: Vol 14, Iss 6, Mar 20, 2024 DOI: 10.21769/BioProtoc.4962 Views: 6288
Reviewed by: Luis Alberto Sánchez VargasJaveena HussainDogan Can KirmanThirupugal Govindarajan

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation of Thylakoid Membranes from the Cyanobacterium Synechocystis sp. PCC 6803 and Analysis of Their Photosynthetic Pigment-protein Complexes by Clear Native-PAGE
Josef Komenda [...] Tomas Zakar
Jan 5, 2019 8633 Views

Purification of Native Dentilisin Complex from Treponema denticola by Preparative Continuous Polyacrylamide Gel Electrophoresis and Functional Analysis by Gelatin Zymography
Pachiyappan Kamarajan [...] Yvonne L. Kapila
Apr 5, 2024 2110 Views

Thermus thermophilus CRISPR Cas6 Heterologous Expression and Purification
Junwei Wei [...] Yingjun Li
Jul 20, 2025 2193 Views
Abstract
Nanobodies are recombinant antigen-specific single domain antibodies (VHHs) derived from the heavy chain–only subset of camelid immunoglobulins. Their small molecular size, facile expression, high affinity, and stability have combined to make them unique targeting reagents with numerous applications in the biomedical sciences. From our work in producing nanobodies to over sixty different proteins, we present a standardised workflow for nanobody discovery from llama immunisation, library building, panning, and small-scale expression for prioritisation of binding clones. In addition, we introduce our suites of mammalian and bacterial vectors, which can be used to functionalise selected nanobodies for various applications such as in imaging and purification.
Key features
• Standardise the process of building nanobody libraries and finding nanobody binders so that it can be repeated in any lab with reasonable equipment.
• Introduce two suites of vectors to functionalise nanobodies for production in either bacterial or mammalian cells.
Graphical overview
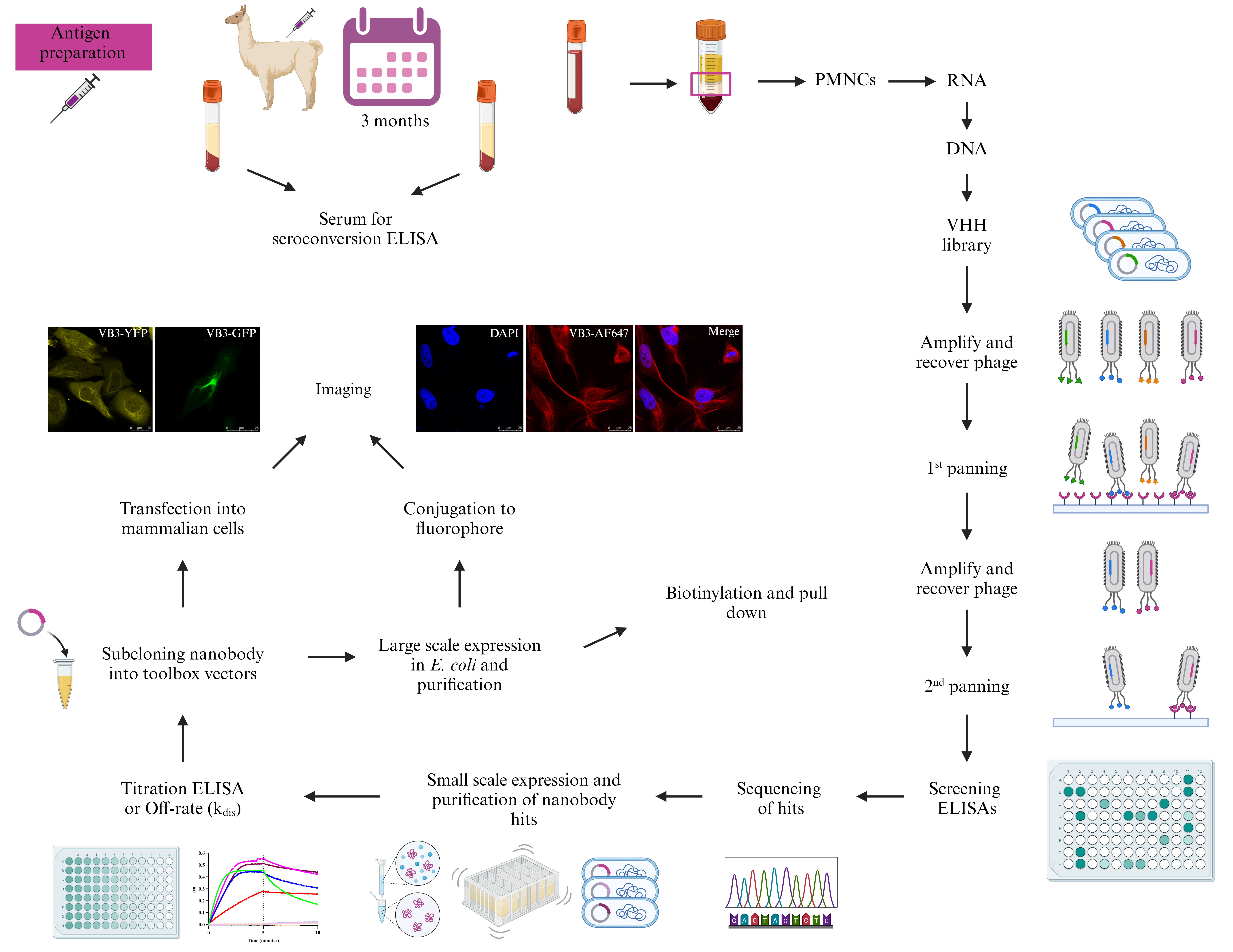
Background
The fact that camelids produce a unique heavy-chain antibody was discovered serendipitously some thirty years ago by researchers at the Vrije Universiteit Brussel [1]. Pioneering work from the Belgian group showed that the variable heavy chain domain of these antibodies, termed VHH, could be produced as a single domain binding protein, referred to as a nanobody. Subsequently, nanobodies have been generated to a wide variety of antigens for applications in cell and structural biology, including as crystallization chaperones for high-value membrane and unstructured proteins and as probes for super-resolution microscopy [2]. Typically, nanobodies are generated by screening phage display libraries of VHH domains cloned from the peripheral blood cells of immunised llamas and alpacas with the target immunogen [3]. Since 2019, our group have generated nanobodies to 75 different antigens. The antigens varied from complexes, membranes, and soluble proteins. Some of the nanobodies that we have identified have been applied as structural chaperones [4], diagnostics [5], and anti-viral therapeutics for SARS-CoV-2 [6] and for in vitro and in vivo cell biology [7]. Although several protocols for producing nanobodies have been published previously [8], we have identified areas for streamlining the process, e.g., incorporating ligation-independent cloning to facilitate the construction of VHH domain libraries and small-scale expression screening to identify high-producing clones.
Furthermore, by using generic VHH cloning primers, we have designed a suite of expression vectors that enable the functionalisation of any cloned VHH with a variety of carboxy terminal tags, e.g., Flag, Avi-tag®, or SNAP-tag® for subsequent site-specific labelling. Overall, our aim is to make this technology readily accessible to any research group with an appropriately equipped laboratory.
Materials and reagents
Biological materials
Post-immune heparinised llama blood
Pre- and post-immune llama sera
Electrocompetent TG1 (Agilent, catalog number: 200123)
CM13K trypsin-sensitive helper phages (Antibody Design Laboratories, catalog number: PH050L)
StellarTM competent cells (Takara Bio, catalog number: 636766)
Escherichia coli (Migula) Castellani and Chalmers, strain WK6 (ATCC, catalog number: 47078)
BL21(DE3)-R3-pRARE2-BirA E. coli cells for in vivo biotinylation, SGC (Structural Genomics Consortium)
HeLa cells (ATCC, catalog number: CCL-2)
Reagents
Gerbu adjuvant F (Biotechnik GmbH, catalog number: 3030). Store at 4 °C
NeutrAvidinTM biotin-binding protein (Invitrogen, catalog number: A2666). Store at -20 °C
10× PBS (Fisher BioReagents, catalog number: BP399-20), for general use. Store at room temperature (RT)
Skim milk powder (Oxoid, catalog number: LP0031). Store at RT
Tween-20 (Sigma-Aldrich, catalog number: P1379). Store at RT
BSA (Sigma-Aldrich, catalog number: A2153). Store at 4 °C
Goat anti-llama IgG (H+L) HRP (Invitrogen, catalog number: A16060). Store at -20 °C in 5 µL aliquots
KPL ABTS peroxidase solution A (SeraCare, catalog number: 5120-0034). Store at 4 °C
KPL peroxidase substrate solution B (SeraCare, catalog number: 5120-0037). Store at 4 °C
ChemgeneTM (Chemgene&Trade, catalog number: XTM308). Store at RT
Caution: This chemical is corrosive.
Ethanol (EtOH) (Fisher BioReagents, catalog number: E/0650DF/17). Store at RT in a flammable liquid storage cupboard
Caution: This chemical is flammable.
Virkon® (Rely+onTM, catalog number: 12358667). Store at RT
Caution: This chemical is corrosive.
PBS, pH 7.4 for polymorphonuclear cell (PMNC) isolation (Gibco, catalog number: 10010015). Store at 4 °C
Histopaque®-1077 (Sigma-Aldrich, catalog number: 10771). Store at 4 °C
RNaseZapTM RNase decontamination solution (Invitrogen, catalog number: AM9782). Store at RT
Trypan blue solution, 0.4% (Sigma-Aldrich, catalog number: T8154-20ML). Store at RT
Caution: This chemical is potentially carcinogenic.
TRIzolTM reagent (Invitrogen, catalog number:15596026). Store at 4 °C
Caution: This chemical is corrosive and toxic.
Chloroform (Fluorchem, catalog number: D007F). Store at RT in a flammable liquid storage cupboard
Caution: This chemical is flammable.
2-propanol (Sigma-Aldrich, catalog number: I9516). Store at RT in a flammable liquid storage cupboard
Caution: This chemical is flammable.
Nuclease-free water (not DEPC treated) (Invitrogen, catalog number: AM9932). Store at RT
SuperScriptTM IV one-step RT-PCR system (Invitrogen, catalog number: 12594025). Store at -20 °C
CALL_001 primer 5′ GTCCTGGCTGCTCTTCTACAAGG 3′. Ordered primer from IDT. Store at RT prior to reconstitution in water, then store at -20 °C
CALL_002 primer 5′ GGTACGTGCTGTTGAACTGTTCC 3′. Ordered primer from IDT. Store at RT prior to reconstitution in water, then store at -20 °C
6× DNA gel loading buffer (New England Biolabs, catalog number: B7024S). Store at 4 °C
10× TBE buffer (Thermo Scientific, catalog number: B52). Store at RT
Agarose (Fisher BioReagents, catalog number: BP1356-500). Store at RT
1,000× SYBRTM safe DNA gel stain (Invitrogen, catalog number: S33102). Store at RT
GeneRuler 1 kb DNA ladder (Thermo Scientific, catalog number: SM0311). Store at 4 °C
Nucleospin Gel and PCR clean-up kit (Macherey-Nagel, catalog number: 740609.50). Store at RT
Purelink PCR purification kit (Invitrogen, catalog number: K310002). Store at RT
HyperLadderTM 1 kb DNA ladder (Meridian Bioscience®, Bioline, catalog number: BIO-33053). Store at 4 °C
Phusion flash high fidelity PCR master mix (Invitrogen, catalog number: F548L). Store at -20 °C
VHHFor2 primer 5′ TACTCGCGGCCCAGCCGGCCATGGCCCAGGTGCAGCTGCAGGAGTCT GGRGGA 3′. Ordered primer from IDT. Store at RT prior to reconstitution in water, then store at -20 °C
VHHRev primer 5′ GTGATGGTGTTGGCCTCCTGAGGAGACGGTGACCTGG 3′. Ordered primer from IDT. Store at RT prior to reconstitution in water, then store at -20 °C
pADL23c vector (Antibody Design Laboratories, catalog number: PD0111). Store at -20 °C
SfiI (New England Biolabs, catalog number: R0123S). Store at -20 °C
10× rCutSmartTM buffer (New England Biolabs, catalog number: B6004S). Store at 4 °C
ClonExpress II one step cloning kit, which contains 5× CEII buffer and Exnase II (Vazymbiotech, catalog number: C112-02). Store at -20 °C
Recovery medium (Sigma-Aldrich, catalog number: S1797). Store at 4 °C
LB medium mix (Formedium, catalog number: LBL0103). Store at RT
Bacto tryptone (Melford, catalog number: T60065-2000.0). Store at RT
Yeast extract (Melford, catalog number: Y20020-1000.0). Store at RT
Bacto agar (Formedium, catalog number: AGR10). Store at RT
Ampicillin (Formedium, catalog number: AMP100). Store at 4 °C
Caution: Ampicillin is a sensitiser.
25 mM dNTP (Thermo Scientific, catalog number: R1122). Store at -20 °C in 30 µL aliquots
Taq polymerase with standard Taq reaction buffer (New England Biolabs, catalog number: M0273X). Store at -20 °C
PhD_seq_Fwd primer 5′ GCTTCCGGCTCGTATGTTG 3′. Store at -20 °C
PhD_seq_Rev primer 5′ GTCGTCTTTCCAGACGTTAG 3′. Store at -20 °C
Glycerol (Sigma-Aldrich, catalog number: G5516-1L). Store at RT
PEG 6000 (Sigma-Aldrich, catalog number: 81260). Store at RT
NaCl (Sigma-Aldrich, catalog number: S9888). Store at RT
Kanamycin (Sigma-Aldrich, catalog number: K1377). Store at 4 °C
Caution: Kanamycin is a sensitiser and may damage fertility.
StartingBlockTM (PBS) blocking buffer (Invitrogen, catalog number: 37538). Store at 4 °C
DynabeadsTM M-280 streptavidin (Invitrogen, catalog number: 11205D). Store at 4 °C
Tris base (Melford, catalog number: T60040-100.0). Store at RT
CaCl2·2H2O (Sigma-Aldrich, catalog number: C3306). Store at RT
Trypsin (Sigma-Aldrich, catalog number: T1426). Store at -20 °C
Anti-M13 HRP (Sino Biological, catalog number: 11973-MM05T-H). Store at -20 °C in 5 µL aliquots
Highprep PCR magnetic beads (Magbio, catalog number: AC60050). Store at 4 °C
Glucose (Sigma-Aldrich, catalog number: G8270). Store at RT
MgCl2·6H2O (Sigma-Aldrich, catalog number: M2670). Store at RT
IPTG (NeoBiotech, catalog number: NB-45-00030). Store at -20 °C
Polymyxin B sulfate (Gibco, catalog number: 21850029). Store at RT
Caution: Polymyxin B sulfate is toxic.
2× Laemmli buffer (Sigma-Aldrich, catalog number: S3401). Store at 4 °C
20× NuPAGETM MES SDS running buffer (Invitrogen, catalog number: NP000202). Store at RT
Mark12TM unstained standard (Invitrogen, catalog number: LC5677). Store at 4 °C
InstantBlue® Coomassie protein stain (Abcam, catalog number: ab119211). Store at 4 °C
Ni-NTA spin columns (QIAGEN, catalog number: 31014). Store at 4 °C
Imidazole (Sigma-Aldrich, catalog number: I202-500g). Store at RT
Caution: Imidazole is toxic, corrosive, and an irritant and may damage fertility.
ZebaTM spin desalting columns, 7K MWCO, 0.5 mL (Thermo Scientific, catalog number: 89882). Store at 4 °C
Rabbit anti-camelid VHH HRP (GenScript, catalog number: A01861-200). Store at -20 °C in 5 µL aliquots
pOPINVHH_his vector (Addgene, catalog number: 210405). Store at -20 °C
pOPINVHH_cys_his vector (Addgene, catalog number: 210403). Store at -20 °C
pOPINVHH_BAP_his vector (Addgene, catalog number: 210402). Store at -20 °C
pOPINVHH_sort_his vector (Addgene, catalog number: 210406). Store at -20 °C
pOPINVHH_flag_his vector (Addgene, catalog number: 210404). Store at -20 °C
pOPINVHH_myc_his vector (Addgene, catalog number: 210526). Store at -20 °C
pOPINVHH_snap_his vector (Addgene, catalog number: 210407). Store at -20 °C
pOPINE-3C-eGFP vector (Addgene, catalog number: 41125). Store at -20 °C
pOPINE-3C-eYFP vector (Addgene, catalog number: 214028). Store at -20 °C
pOPINE-3C-mCherry vector (Addgene, catalog number: 214060). Store at -20 °C
Common Fwd primer 5′ GCGGCCCAGCCGGCCATGGCCCAGGTGCAGCTGGTGGAG 3′. Ordered primer from IDT. Store at RT prior to reconstitution in water, then store at -20 °C
His FLAG Rev primer 5′ GTGATGGTGGCCTGAGGAGACGGTGACCTGGGTC 3′. Ordered primer from IDT. Store at RT prior to reconstitution in water, then store at -20 °C
Cys Rev primer 5′ ATGGTGACAGCCTGAGGAGACGGTGACCTGGGTC 3′. Ordered primer from IDT. Store at RT prior to reconstitution in water, then store at -20 °C
BAP Rev primer 5′ ATCATTCAAGCCTGAGGAGACGGTGACCTGGGTC 3′. Ordered primer from IDT. Store at RT prior to reconstitution in water, then store at -20 °C.
Sort Rev primer 5′ CGGCAGGCCGCCTGAGGAGACGGTGACCTGGGTC 3′. Ordered primer from IDT. Store at RT prior to reconstitution in water, then store at -20 °C
Myc Rev primer 5′ GTGATGGTGGTGGCCTGAGGAGACGGTGACCTGGGTC 3′. Ordered primer from IDT. Store at RT prior to reconstitution in water, then store at -20 °C
SNAP Rev primer 5′ GTCCTTGTCGCCTGAGGAGACGGTGACCTGGGTC 3′. Ordered primer from IDT. Store at RT prior to reconstitution in water, then store at -20 °C
pOPINE common Fwd primer 5′ AGGAGATATACCATGCAGGTGCAGCTGGTGGAG 3′. Ordered primer from IDT. Store at RT prior to reconstitution in water, then store at -20 °C
pOPINE common Rev primer 5′ CAGAACTTCCAGTTTAGGGGAGACGGTGACCTGGGTC 3′. Ordered primer from IDT. Store at RT prior to reconstitution in water, then store at -20 °C
Bsu36I (New England Biolabs, catalog number: R0524S). Store at -20 °C
NcoI (New England Biolabs, catalog number: R01093S). Store at -20 °C
X-gal ready to use (Thermo Scientific, catalog number: R0941). Store at 4 °C
QIAprep spin miniprep kit (QIAGEN, catalog number: 27104). Store at RT
DNA sequence of anti-GFP nanobody [9] after codon optimisation for expression in E. coli. CAG GTC CAA TTA GTG GAG TCC GGT GGG GCA CTT GTC CAG CCT GGA GGT TCA CTT CGC TTG TCT TGC GCA GCG TCT GGA TTC CCG GTG AAC CGC TAT AGT ATG CGT TGG TAC CGT CAA GCT CCG GGG AAA GAA CGT GAA TGG GTA GCA GGG ATG TCT TCC GCC GGT GAC CGC TCT TCA TAC GAG GAC TCG GTC AAG GGG CGC TTC ACA ATC TCT CGT GAT GAT GCC CGT AAC ACC GTT TAC TTG CAA ATG AAC AGC CTG AAA CCG GAA GAC ACT GCG GTG TAT TAC TGC AAT GTT AAT GTA GGG TTT GAA TAC TGG GGT CAA GGT ACA CAA GTT ACA GTT TCG TCA. Ordered gBlock from IDT. Store at RT prior to reconstitution in TE buffer, incubate at 50 °C for 20 min, then store at -20 °C
Chloramphenicol (Sigma-Aldrich, catalog number: C1919). Store at 4 °C
Caution: Chloramphenicol is toxic, can cause eye damage, and is a carcinogen.
Spectinomycin dihydrochloride pentahydrate (Sigma-Aldrich, catalog number: S4014). Store at 4 °C
Caution: Spectinomycin is an irritant.
Carbenicillin disodium salt (Sigma-Aldrich, catalog number: C3416). Store at 4 °C
Caution: Carbenicillin is a sensitiser.
Biotin (Sigma-Aldrich, catalog number: B4639). Store at 4 °C
Streptavidin, Alexa FluorTM 488 conjugate (Invitrogen, catalog number: S11223). Store at -20 °C
BenchMarkTM fluorescent protein standard (Invitrogen, catalog number: LC5928). Store at -20 °C
DNA sequence of anti-vimentin (VB3) nanobody [10] after codon optimisation for expression in E. coli: CAG GTC CAA CTT GTA GAG TCA GGA GGT GGA AGC GTG CAA GCT GGG GAC TCT CTG CGC CTG TCT TGT GCT TCG AGC GGA AAT ACC TTC TCG ATC AAA GTC ATG GGA TGG TAC CGC CAG GCA CCT GGA AAG CAA CGT GAA TTA GTC GCG GTT TCA ACC AAT AGC GGG GCC TCT GTT AAT TAT GCC AAC TCT GTG AAG GGA CGC TTT ACC ATT TCT ATT GAT TCA GTA AAA AAA ACA ACC TAC TTA CAG ATG AAT TCC TTG AAG CCA GAA GAT ACA GCC GTC TAC TTT TGC AAT GCA TAT GAT GGG CGT TAT GAG GAC TAT TAC GGT CAG GGG ACC CAA GTG ACA GTA TCA TCA. Ordered gBlock from IDT. Store at RT prior to reconstitution in TE buffer, incubate at 50 °C for 20 min, then store at -20 °C
TCEP, hydrochloride (Merck, Millipore®, catalog number: 580567-5GM). Store at RT
Alexa FluorTM 647 C2-maleimide (Invitrogen, catalog number: A20347). Store at -20 °C
ZebaTM dye and biotin removal columns (Thermo Scientific, catalog number: A44296). Store at 4 °C
PageRulerTM prestained protein ladder (Thermo Scientific, catalog number: 26616). Store at 4 °C
DMEM, high glucose, HEPES, no phenol red (Gibco, catalog number: 21063-029). Store at 4 °C
Fetal bovine serum (FBS) (Biowest, catalog number: S00NB1001Y). Store at -20 °C
100× GlutaMAXTM (Gibco, catalog number: 35050-038). Store at -20 °C
100× penicillin-streptomycin (Gibco, catalog number: 15140-122). Store at -20 °C
4% paraformaldehyde in PBS (Thermo Scientific chemicals, catalog number: J61899.AK). Store at 4 °C
Caution: Paraformaldehyde is an eye irritant, skin sensitiser, and carcinogen.
TritonTM X-100 (Sigma-Aldrich, catalog number: T8787-250mL). Store at RT
Fluoroshield with DAPI (Abcam, catalog number: ab104139). Store at RT
Nail varnish. Store at RT
Type F immersion liquid (Leica, catalog number: 11513859). Store at RT
Fugene transfection reagent (Promega, catalog number: E2311). Store at 4 °C
Solutions
0.5 mg/mL neutrAvidin biotin-binding protein (see Recipes)
10 µg/mL neutrAvidin for coating ELISA plate (see Recipes)
1× PBS (see Recipes)
Blocking solution (see Recipes)
Washing buffer (PBST) (see Recipes)
0.1% (w/v) BSA-PBS (see Recipes)
5% (v/v) Chemgene (see Recipes)
70% (v/v) ethanol (EtOH) (see Recipes)
2% (w/v) Virkon (see Recipes)
75% (v/v) EtOH (see Recipes)
1× TBE buffer (see Recipes)
0.7% (w/v) agarose gel containing 1× SYBRTM Safe DNA gel stain (see Recipes)
1% (w/v) agarose gel containing 1× SYBRTM Safe DNA gel stain (see Recipes)
2× YT medium (see Recipes)
LB medium (see Recipes)
100 mg/mL ampicillin (see Recipes)
1% (w/v) agar LB plates containing 100 µg/mL ampicillin (see Recipes)
2% (w/v) agar LB plates containing 100 µg/mL ampicillin (see Recipes)
50% glycerol (see Recipes)
2× YT medium containing 25% glycerol (see Recipes)
1% (w/v) agar LB plates without antibiotic (see Recipes)
2× YT containing 100 µg/mL ampicillin (see Recipes)
50 mg/mL kanamycin (see Recipes)
2× YT containing 25 µg/mL kanamycin (see Recipes)
PEG/NaCl precipitation solution (see Recipes)
2× YT containing 100 µg/mL ampicillin and 25 µg/mL kanamycin (see Recipes)
TBSC (see Recipes)
1 mg/mL trypsin (see Recipes)
250 µg/mL trypsin (see Recipes)
Terrific broth (see Recipes)
20% (w/v) glucose (see Recipes)
1 M MgCl2·6H2O (see Recipes)
Terrific broth containing 100 µg/mL ampicillin, 0.1% glucose, 2 mM MgCl2·6H2O (see Recipes)
1 M IPTG (see Recipes)
1 mg/mL polymyxin B sulfate in PBS (see Recipes)
1× NuPAGETM MES SDS running buffer (see Recipes)
Equilibration buffer (see Recipes)
Elution buffer (see Recipes)
Octet dilution buffer (see Recipes)
1% (w/v) agar LB plate containing 100 µg/mL ampicillin, 2 mM IPTG, 40 µg/mL X-gal (see Recipes)
34 µg mL chloramphenicol (see Recipes)
50 µg/mL spectinomycin (see Recipes)
50 µg/mL carbenicillin (see Recipes)
1% (w/v) agar LB plate containing 34 µg/mL chloramphenicol, 50 µg/mL spectinomycin, 50 µg/mL carbenicillin (see Recipes)
0.2 M Biotin (see Recipes)
Terrific broth containing 50 µg/mL spectinomycin and 50 µg/mL carbenicillin (see Recipes)
1 mg/mL Streptavidin Alexa FluorTM 488 conjugate (see Recipes)
50 µg/mL Streptavidin Alexa FluorTM 488 conjugate (see Recipes)
500 mM TCEP, pH 8.0 (see Recipes)
1 mM TCEP, pH 8.0 (see Recipes)
10 mg/mL Alexa FluorTM 647 C2-maleimide (see Recipes)
DMEM medium without phenol red, 10% (v/v) FBS, 1× GlutaMAXTM, 1× Penicillin-streptomycin (see Recipes)
Confocal blocking buffer (see Recipes)
Confocal dilution buffer (see Recipes)
Recipes
0.5 mg/mL neutrAvidin biotin-binding protein
Prepare 200 µL aliquots and store at -20 °C. Stable for at least one year.
Reagent Final concentration Quantity or Volume neutrAvidinTM 0.5 mg 5 mg ddH2O n/a 10 mL Total n/a 10 mL 10 µg/mL neutrAvidin for coating ELISA plate
Prepare just before use.
Reagent Final concentration Quantity or Volume neutrAvidinTM (Recipe 1) 10 µg/mL 200 µL PBS (1×) 1× 9.8 mL Total n/a 10 mL 1× PBS
Prepare both sterile (by autoclaving) and non-sterile PBS. Store at RT. Stable for at least one month.
Reagent Final concentration Quantity or Volume PBS (10×) 1× 100 mL ddH2O n/a 900 mL Total n/a 1,000 mL Blocking solution
Prepare on day of use.
Reagent Final concentration Quantity or Volume Milk 2% (w/v) 2 g PBS (Recipe 3) (or buffer that is compatible with the protein) n/a 100 mL Total n/a 100 mL Washing buffer (PBST)
Store at RT. Stable for at least one month.
Reagent Final concentration Quantity or Volume PBS (Recipe 3) 1× 1 000 mL Tween-20 0.05% (v/v) 500 µL Total n/a 1,000 mL 0.1% (w/v) BSA-PBS
Prepare on day of use.
Reagent Final concentration Quantity or Volume BSA 0.1 % (w/v) 50 mg PBS (Recipe 3) 1× 50 mL Total n/a 50 mL 5% (v/v) Chemgene
Store at RT.
Reagent Final concentration Quantity or Volume Chemgene (100%) 5% (v/v) 50 mL H2O n/a 950 mL Total n/a 1,000 mL 70% (v/v) EtOH
Store at RT.
Reagent Final concentration Quantity or Volume Ethanol (absolute) 70% (v/v) 70 mL H2O n/a 30 mL Total n/a 100 mL 2% (w/v) Virkon
Prepare weekly and store at RT.
Reagent Final concentration Quantity or Volume Virkon powder 2% (w/v) 100 g H2O n/a 5,000 mL Total n/a 5,000 mL 75% (v/v) EtOH
Store at RT.
Reagent Final concentration Quantity or Volume Ethanol (absolute) 75% (v/v) 75 mL H2O n/a 25 mL Total n/a 100 mL 1× TBE buffer
Store at RT. Stable for at least one month.
Reagent Final concentration Quantity or Volume TBE (10×) 1× 100 mL H2O n/a 900 mL Total n/a 1,000 mL 0.7% (w/v) agarose gel containing 1× SYBRTM Safe DNA gel stain
Heat in a microwave until all the agarose has dissolved and allow to cool to 55 °C before the addition of the gel stain. Pour into the DNA casting apparatus and allow to solidify. Prepare on day of use.
Reagent Final concentration Quantity or Volume Agarose 0.7 % (w/v) 0.7 g TBE (Recipe 11) 1× 100 mL Total n/a 100 mL SYBRTM Safe DNA gel stain (1,000×) 1× 10 µL 1% (w/v) agarose gel containing 1× SYBRTM Safe DNA gel stain
Heat in a microwave until all the agarose has dissolved and allow to cool to 55 °C before the addition of the gel stain. Pour into the DNA casting apparatus and allow to solidify. Prepare on day of use.
Reagent Final concentration Quantity or Volume Agarose 1% (w/v) 1 g TBE (Recipe 11) 1× 100 mL Total n/a 100 mL SYBRTM Safe DNA gel stain (1,000×) 1× 10 µL 2× YT medium
Autoclave and store at RT. Stable for at least one month.
Reagent Final concentration Quantity or Volume Bacto tryptone 16 g/L 4 g NaCl 5 g/L 1.25 g Yeast extract 10 g/L 2.5 g ddH2O n/a 250 mL Total n/a 250 mL LB medium
Autoclave and store at RT. Stable for at least one month.
Reagent Final concentration Quantity or Volume LB medium mix 25 g/L 6.25 g ddH2O n/a 250 mL Total n/a 250 mL 100 mg/mL ampicillin
Filter sterilise using a 0.22 µm filter. Prepare 500 µL aliquots and store at -20 °C. Stable for at least one year.
Caution: Ampicillin is a sensitiser.
Reagent Final concentration Quantity or Volume Ampicillin 100 mg/mL 2 g ddH2O n/a 20 mL Total n/a 20 mL 1% (w/v) agar LB plates containing 100 µg/mL ampicillin
Autoclave LB medium and bacto agar mix and allow to cool to 55 °C before the addition of antibiotic. Invert to mix and pour into 8.5 cm Petri dishes in a safety cabinet. Allow to cool at RT. Prepare on day of use.
Reagent Final concentration Quantity or Volume LB medium mix 25 g/L 6.25 g Bacto agar 1% (w/v) 2.5 g ddH2O n/a 250 mL Total n/a 250 mL Ampicillin (Recipe 16) 100 µg/mL 250 µL 2% (w/v) agar LB plates with 100 µg/mL ampicillin
Autoclave LB medium and bacto agar mix and allow to cool to 55 °C before the addition of antibiotic. Invert to mix and pour into 8.5 cm Petri dishes in a safety cabinet. Allow to cool at RT. Prepare on day of use.
Reagent Final concentration Quantity or Volume LB medium mix 25 g/L 6.25 g Bacto agar 2% (w/v) 5 g ddH2O n/a 250 mL Total n/a 250 mL Ampicillin (Recipe 16) 100 µg/mL 250 µL 50% (v/v) glycerol
Autoclave and store at RT. Stable for at least one year.
Reagent Final concentration Quantity or Volume Glycerol 50% (v/v) 100 mL ddH2O n/a 100 mL Total n/a 200 mL 2× YT medium containing 25% (v/v) glycerol
Prepare just before use in a safety cabinet.
Reagent Final concentration Quantity or Volume 2× YT medium n/a 20 mL Glycerol (Recipe 19) 25% (w/v) 20 mL Total n/a 40 mL 1% (w/v) agar LB plates without antibiotic
Autoclave and allow to cool to 55 °C. Invert to mix and pour into 8.5 cm Petri dishes in a safety cabinet. Allow to cool and set at RT. Store at 4 °C for two weeks.
Reagent Final concentration Quantity or Volume LB medium mix 25 g/L 6.25 g Bacto agar 1% (w/v) 2.5 g ddH2O n/a 250 mL Total n/a 250 mL 2× YT containing 100 µg/mL ampicillin
Prepare just before use in a safety cabinet.
Reagent Final concentration Quantity or Volume 2× YT n/a 100 mL Ampicillin (Recipe 16) 100 µg/mL 100 µL Total n/a 100 mL 50 mg/mL kanamycin
Filter sterilise using a 0.22 µm filter. Prepare 100 µL aliquots and store at -20 °C. Stable for at least one year.
Reagent Final concentration Quantity or Volume Kanamycin 50 mg/mL 0.5 g ddH2O n/a 10 mL Total n/a 10 mL 2× YT containing 25 µg/mL kanamycin
Prepare just before use in a safety cabinet.
Reagent Final concentration Quantity or Volume 2× YT n/a 100 mL Kanamycin (Recipe 23) 25 µg/mL 50 µL Total n/a 100 mL PEG/NaCl precipitation solution
Autoclave and store at RT. Stable for at least one year.
Reagent Final concentration Quantity or Volume PEG 6000 20% (w/v) 200 g NaCl 2.5 M 146.1 g ddH2O n/a Make up to 1,000 mL Total n/a 1,000 mL 2× YT containing 100 µg/mL ampicillin and 25 µg/mL kanamycin
Prepare just before use in a safety cabinet.
Reagent Final concentration Quantity or Volume 2× YT n/a 100 mL Ampicillin (Recipe 16) 100 µg/mL 100 µL Kanamycin (Recipe 23) 25 µg/mL 50 µL Total n/a 100 mL TBSC
Autoclave and store at RT. Stable for at least one year.
Reagent Final concentration Quantity or Volume Tris base 10 mM 0.788 g NaCl 137 mM 4 g CaCl2·2H2O 1 mM 73.5 mg ddH2O n/a 500 mL Total n/a 500 mL 1 mg/mL trypsin
Prepare 130 µL aliquots and store at -20 °C. Stable for at least one year.
Reagent Final concentration Quantity or Volume Trypsin 1 mg/mL 50 mg ddH2O n/a 50 mL Total n/a 50 mL 250 µg/mL trypsin
Prepare just before use.
Reagent Final concentration Quantity or Volume Trypsin (Recipe 28) 250 µg/mL 125 µL TBSC (Recipe 27) n/a 375 µL Total n/a 500 µL Terrific broth
Autoclave and store at RT. Stable for at least one month.
Reagent Final concentration Quantity or Volume Bacto tryptone 12 g/L 3 g Yeast extract 24 g/L 6 g Glycerol 0.4% (v/v) 1 mL KH2PO4 0.17 M 0.5775 g K2HPO4 0.72 M 3.135 g ddH2O n/a 250 mL Total n/a 250 mL 20% (w/v) glucose
Filter sterilise using a 0.22 µm filter. Prepare 500 µL aliquots and store at -20 °C. Stable for at least one year.
Reagent Final concentration Quantity or Volume Glucose 20% (w/v) 20 g ddH2O n/a Make up to 100 mL Total n/a 100 mL 1 M MgCl2·6H2O
Filter sterilise using a 0.22 µm filter. Prepare 500 µL aliquots and store at -20 °C. Stable for at least one year.
Reagent Final concentration Quantity or Volume MgCl2·6H2O 1 M 4.066 g ddH2O n/a 20 mL Total n/a 20 mL Terrific broth containing 100 µg/mL ampicillin, 0.1% glucose, 2 mM MgCl2·6H2O
Prepare just before use in a safety cabinet.
Reagent Final concentration Quantity or Volume Terrific broth (Recipe 30) n/a 99.2 mL Ampicillin (Recipe 16) 100 µg/mL 100 µL Glucose (Recipe 31) 0.1% (v/v) 500 µL MgCl2·6H2O (Recipe 32) 2 mM 200 µL Total n/a 100 mL 1 M IPTG
Filter sterilise using a 0.22 µm filter. Prepare 500 µL aliquots and store at -20 °C. Stable for at least one year.
Reagent Final concentration Quantity or Volume IPTG 1 M 4.766 g ddH2O n/a 20 mL Total n/a 20 mL 1 mg/mL polymyxin-B sulfate in PBS
Prepare 1 mL aliquots and store at -20 °C. Stable for at least one year.
Reagent Final concentration Quantity or Volume Polymyxin-B sulfate 1 mg/mL 2 mg PBS (Recipe 3) 1× 2 mL Total n/a 2 mL 1× NuPAGETM MES SDS running buffer
Store at RT. Stable for at least one month.
Reagent Final concentration Quantity or Volume MES (20×) 1× 100 mL H2O n/a 1,900 mL Total n/a 2,000 mL Equilibration buffer
Store at RT. Stable for at least one month.
Reagent Final concentration Quantity or Volume Imidazole 30 mM 0.51 g PBS (Recipe 3) 1× 250 mL Adjust pH to 7.4 Total n/a 250 mL Elution buffer
Store at RT. Stable for at least one month.
Reagent Final concentration Quantity or Volume Imidazole 300 mM 2.04 g PBS (Recipe 3) 1× 100 mL Adjust pH to 7.4 Total n/a 100 mL Octet dilution buffer
Store at RT. Stable for at least one month.
Reagent Final concentration Quantity or Volume BSA 0.1% (w/v) 50 mg PBS (Recipe 3) (or buffer that is compatible with the protein) 1× 50 mL Total n/a 50 mL 1% (w/v) agar LB plate containing 100 µg/mL ampicillin, 2 mM IPTG, 40 µg/mL X-gal
Autoclave and allow to cool to 55 °C before the addition of additives. Invert to mix and pour into 8.5 cm Petri dishes in a safety cabinet. Allow to cool and set at RT. Store at 4 °C for two weeks.
Reagent Final concentration Quantity or Volume LB medium mix 25 g/L 6.25 g Bacto agar 1% (w/v) 2.5 g ddH2O n/a Make up to 250 mL Total n/a 250 mL Ampicillin (Recipe 16) 100 µg/mL 250 µL IPTG (Recipe 34) 2 mM 500 µL X-gal (20 mg/mL) 40 µg/mL 500 µL 34 mg/mL chloramphenicol
Filter sterilise using a 0.22 µm filter. Prepare 100 µL aliquots and store at -20 °C. Stable for at least one year.
Reagent Final concentration Quantity or Volume Chloramphenicol 34 mg/mL 34 mg EtOH 100% 1 mL Total n/a 1 mL 50 mg/mL Spectinomycin
Filter sterilise using a 0.22 µm filter. Prepare 100 µL aliquots and store at -20 °C. Stable for at least one year.
Reagent Final concentration Quantity or Volume Spectinomycin dihydrochloride pentahydrate 50 mg/mL 50 mg ddH2O n/a 1 mL Total n/a 1 mL 50 mg/mL Carbenicillin
Filter sterilise using a 0.22 µm filter. Prepare 100 µL aliquots and store at -20 °C. Stable for at least one year.
Reagent Final concentration Quantity or Volume Carbenicillin disodium salt 50 mg/mL 50 mg ddH2O n/a 1 mL Total n/a 1 mL 1% (w/v) agar LB plate containing 34 µg/mL chloramphenicol, 50 µg/mL spectinomycin, 50 µg/mL carbenicillin
Autoclave LB medium and bacto agar mix and allow to cool to 55 °C before the addition of additives. Invert to mix and pour into 8.5 cm Petri dishes in a safety cabinet. Allow to cool and set at RT. Store at 4 °C for two weeks.
Reagent Final concentration Quantity or Volume LB medium mix 25 g/L 2.5 g Bacto agar 1% (w/v) 1 g ddH2O n/a Make up to 100 mL Total n/a 100 mL Chloramphenicol (Recipe 41) 34 µg/mL 100 µL Spectinomycin (Recipe 42) 50 µg/mL 100 µL Carbenicillin (Recipe 43) 50 µg/mL 100 µL 0.2 M biotin
Filter sterilise using a 0.22 µm filter. Prepare 100 µL aliquots and store at -20 °C. Stable for at least one year.
Reagent Final concentration Quantity or Volume Biotin 0.2 M 0.488 g NaOH (1 M) n/a Add to dissolve biotin ddH2O n/a Make up to 10 mL Total n/a 10 mL Terrific broth containing 50 µg/mL spectinomycin and 50 µg/mL carbenicillin
Prepare just before use in a safety cabinet.
Reagent Final concentration Quantity or Volume Terrific broth (Recipe 30) n/a 998 mL Spectinomycin (Recipe 42) 50 µg/mL 1 mL Carbenicillin (Recipe 43) 50 µg/mL 1 mL Total n/a 1 000 mL 1 mg/mL streptavidin Alexa FluorTM 488 conjugate
Protect from light, prepare 10 µL aliquots, and store at -20 °C. Stable for six months.
Reagent Final concentration Quantity or Volume Streptavidin Alexa FluorTM 488 conjugate 1 mg/mL 1 mg PBS (Recipe 3) 1× 1 mL Total n/a 1 mL 50 µg/mL streptavidin Alexa FluorTM 488 conjugate
Protect from light and prepare just before use.
Reagent Final concentration Quantity or Volume Streptavidin Alexa FluorTM 488 conjugate (1 mg/mL) 50 µg/mL 1 µL PBS (Recipe 3) 1× 19 µL Total n/a 20 µL 500 mM TCEP, pH 8.0
Store at 4 °C. Stable for at least one month.
Reagent Final concentration Quantity or Volume TCEP 500 mM 1.433 g ddH2O n/a 10 mL Adjust to pH 8.0 Total n/a 10 mL 1 mM TCEP, pH 8.0
Prepare just before use.
Reagent Final concentration Quantity or Volume TCEP (Recipe 49) 1 mM 2 µL ddH2O n/a 998 µL Total n/a 1 mL 10 mg/mL Alexa FluorTM 647 C2-Maleimide
Protect from light and store at -20 °C. Stable for at least one year.
Reagent Final concentration Quantity or Volume Alexa FluorTM 647 C2-Maleimide 10 mg/mL 1 mg DMSO 1× 500 µL Total n/a 500 µL DMEM medium without phenol red, 10% (v/v) FBS, 1× GlutaMAXTM, 1× penicillin-streptomycin
Store at 4 °C. Stable for at least one month.
Reagent Final concentration Quantity or Volume DMEM medium n/a 88 mL FBS 10% (v/v) 10 mL GlutaMAXTM (100×) 1× 1 mL Penicillin-streptomycin (100×) 1× 1 mL Total n/a 100 mL Confocal blocking buffer
Store at 4 °C. Use within one month.
Reagent Final concentration Quantity or Volume PBS (Recipe 3) 1× 10 mL BSA 5% (w/v) 0.5 g Triton X-100 0.25% (v/v) 25 µL Total n/a 10 mL Confocal dilution buffer
Store at 4 °C. Use within one month.
Reagent Final concentration Quantity or Volume PBS (Recipe 3) 1× 10 mL BSA 1% (w/v) 0.1 g Triton X-100 0.25% (v/v) 25 µL Total n/a 10 mL
Laboratory supplies
Vacuette® tube 4 mL CAT serum clot activator blood collection tubes (Greiner, Bio-one, catalog number: 454204)
Vacuette® tube 9 mL NH sodium heparin blood collection tubes (Greiner, Bio-one, catalog number: 455051)
Microcentrifuge tubes, 0.5 mL, 1.5 mL, 2 mL (Greiner, Bio-one, catalog numbers: 667201, 616201, 623201)
1.5 mL microcentrifuge tubes, RNase free (SARSTEDT, catalog number: 72.706.700)
PCR tubes and caps (VWR, catalog number: 20170-010)
50 mL conical centrifuge tubes (Greiner Bio-One, catalog number: 227261)
Serological pipettes, 2 mL, 5 mL, 10 mL, 50 mL (Greiner Bio-One, catalog numbers: 710180, 606180, 607180, 760180, 768160)
Screw cap microtubes (SARSTEDT, catalog number: 72.694.305)
MicrolanceTM 3 needles, 21G (BD, catalog number: 304432)
2.5 mL syringe (Greiner Bio-One, catalog number: SYR2)
8.5 cm Petri dishes (Greiner, Bio-one, catalog number: 633181)
Pipette tips, filtered, 20 µL, 200 µL, 1,000 µL (Mettler Toledo, Ranin, catalog numbers: 17005861, 17005863, 30389211)
Gene Pulser electroporation cuvettes, 0.1 cm gap (Bio-Rad, catalog number: 1652083)
Polystyrene semi micro cuvettes (Fisherbrand, catalog number: FB55147)
125 mL baffled flasks (VWR, catalog number: 214-0458)
250 mL baffled flasks (VWR, catalog number: 214-0460)
1 L baffled flasks (VWR, catalog number: 214-0464)
Micro quartz absorption cuvettes (Merck, Hellma®, catalog number: Z600210)
Square bioassay dish (Nunc, catalog number: 240845)
Reagent reservoirs, non-sterile (VWR, catalog number: 613-1176)
Reagent reservoirs, sterile (Axygen, catalog number: RES-V-25-S)
Deep-well 96-well 2 mL plate (Fisherbrand, catalog number: 11391555)
Deep-well 96-well 1 mL plate (Greiner, Bio-one, catalog number: 780215)
Deep-well 96-well 0.5 mL plate (Greiner, Bio-one, catalog number: 786261)
Cell culture adhesive seal (Azenta Life Sciences, catalog number: 4ti-0517)
Adhesive film for ELISA and general incubation (VWR, catalog number: 60941-062)
PCR foil seal (Azenta Life Sciences, catalog number: 4ti-0550)
Rigid semi-skirted 96-well PCR plate (Thermo Scientific, catalog number: AB-0990)
Framestar® 96-well skirted PCR plate (Azenta Life Sciences, catalog number: 4ti-0960)
96-well ELISA microplate (Greiner Bio-One, catalog number: 655061)
Octet SA biosensors (Sartorius, catalog number: 18-5019)
96-well black plate, polypropylene (Greiner Bio-One, catalog number: 655209)
96-well microtitre plate (Greiner Bio-One, catalog number: 65101)
250 mL PPCO centrifuge tube (Nalgene, catalog number: 3141-0250)
25 mL high speed PPCO centrifuge tube (Nalgene, catalog number: 3119-0010)
NuPage 4%–12% bis tris precast gel (Invitrogen, catalog number: NP0323BOX). A suitable alternative is homemade 12% running and 4% stacking SDS-PAGE gel
0.22 μm syringe filters (Agilent, catalog number: 5190-5116)
µ-Slide 8-well high polymer bottom chambered coverslip (ibidi, catalog number: 80807)
Microscope slides with ground edges (Fisherbrand, catalog number: 11572203)
Microscope coverslip, 12 mm diameter, no. 1.5 (Scientific Laboratory supplies, catalog number: MIC3334)
6-well cell culture dish (Greiner Bio-One, catalog number: 657160)
Dumont tweezer style 5 (Electron Microscopy Sciences, catalog number: 0203-5-PS)
Equipment
Pipetboy acu 2 pipette controller (Ingetra, catalog number: I155017)
Single-channel pipettes starter kit (Mettler Toledo, Ranin, catalog number: 30386738)
Multichannel pipettes 1–10 µL, 20–200 µL, 100–1,200 µL (Mettler Toledo, Ranin, catalog numbers: 17013802, 17013805, 17014496)
Microplate washer (Thermo Fisher, WellwashTM, catalog number: 5165000). If there is no well washer available, the wells can be washed manually using a multichannel pipette or using a wash bottle filled with PBST
CLARIOstar plus plate reader (BMG Labtech, catalog number: 430-501S-FL)
Class 2 microbiological safety cabinet (referred to as a safety cabinet and known as a laminar flow) (Contained Air solutions, catalog number: BioMAT 2). If no safety cabinet is available, working by a Bunsen burner is a suitable alternative. Working by an open flame with flammable EtOH should be done with caution
PCR workstation cabinet (Bigneat, catalog number: MW520-20)
NanoDropTM One/OneC Microvolume UV-Vis spectrophotometer (Thermo Scientific, catalog number: ND-ONE-W)
DynaMagTM 96-side magnet (Invitrogen, catalog number: 12331D)
PCR thermal cycler (Applied Biosystems, VeritiTM, catalog number: 4375305)
Eporator (Eppendorf, catalog number: 4309000027)
ChemiDocTM imaging system (Bio-Rad, catalog number: 12003154)
Cell density meter (Fisherbrand, catalog number: A0)
Multi Sub Electrophoresis System (Flowgen, catalog number: FMMS10)
SDS-PAGE equipment, mini gel tank (Invitogen, catalog number: A25977)
PowerPac basic power supply (Bio-Rad, catalog number: 1645050EDU)
JB Nova water bath (Grant, catalog number: JBN5)
Dry heating block (Grant, catalog number: QBD2)
Dual LED Blue/White Light Transilluminator (Thermo Fisher, catalog number: LB0100)
Safe ImagerTM viewing glasses (Thermo Fisher, catalog number: S37103)
HulaMixerTM sample mixer (Invitrogen, catalog number: 15920D). Agitation using this sample mixer is performed using the following settings: orbital = 5, rpm = 1
DynaMagTM-2 magnet (Invitrogen, catalog number: 12321D)
LSETM digital microplate shaker (Corning, catalog number: 4782-4). Agitation using the microplate shaker is performed at 500 rpm
Multifuge X4 Pro-MD (Thermo Scientific, catalog number: 75009500). In all centrifugation steps, maximum acceleration and deceleration rates are used unless otherwise specified
FrescoTM 21 microcentrifuge (Thermo Scientific, catalog number: 75002555)
Orbital shaking incubator (Shel Lab, catalog number: SI6/SI6R) (for 600 rpm agitation steps)
44R incubator shaker (New Brunswick, Innova®, catalog number: M1282-0006) (for 200 and 250 rpm agitation steps)
Octet® R8 (Sartorius, catalog number: 30-0518)
SP8 Lightning confocal microscope (Leica Microsystems Ltd.)
HeracellTM VIOS 160i CO2 Incubator (Thermo Scientific, catalog number: 51033559)
StuartTM SSM3 Gyratory Rocker (Cole-Parmer, catalog number: 51900-26)
Software and datasets
SnapGene (version 7.1.0, 2023, released 28 November 2023); requires a license
Octet® Analysis Studio Software (version 12.2.2.26); requires a license
LasX software for confocal microscope (version 5.2.1); requires a license
Procedure
Llama immunisation
Antibodies are raised in a llama by intramuscular immunisation with up to eight different proteins in parallel. The identity of the proteins depends on the intended application. We have raised nanobodies to integral membrane proteins (e.g., PEPT2 [4]), cell surface glycoproteins (e.g., GPC3 [7]) and viral antigens (e.g., SARS-CoV-2 spike protein [6]).
Store protein antigens at -80 °C in 3 × 200 µg aliquots, preferably 200 µL of 1 mg/mL per aliquot, in PBS or a buffer that is optimal for the protein (e.g., if in a detergent-solubilised membrane protein, then the required amount of appropriate detergent would be included in the buffer).
Before immunisation, collect 5 mL of blood in blood tubes without anticoagulant to prepare a sample of pre-immune serum. Preparation of the pre-immune serum sample is detailed in step C2.
Thaw an aliquot of each of the proteins that are to be included in the immunisation. Gently mix each protein with an equal volume of Gerbu adjuvant and inject subcutaneously (maximum 2 mL per site) in the neck base/shoulder of the llama.
The llama is immunised on day 0 followed by two boosts of antigen on days 28 and 56. Ten days following the final boost, 170 mL of blood from the jugular vein is collected into heparinised tubes (to prevent coagulation) for isolation of polymorphonuclear cells (PMNCs). In addition, collect 5 mL of blood in blood tubes without anticoagulant to prepare a sample of post-immune serum. Details for how to prepare the post-immune serum sample are described in step C2.
Seroconversion ELISA
A seroconversion ELISA is carried out to confirm if the immunisation of the llama was successful in generating a response to the injected antigen(s). A strong ELISA signal is strongly suggestive that binding nanobodies can be isolated from the created library by phage display.
Coat a 96-well ELISA microplate with 100 µL/well of 10 µg/mL neutrAvidinTM diluted in PBS and incubate overnight at 4 °C.
Using a plate washer, wash the plate five times with 300 µL/well of PBST.
Add 100 µL/well of 50 nM biotinylated target protein diluted in PBS and incubate for 1 h at RT on a microplate shaker. See General note 1.
Using a plate washer, wash the plate five times with 300 µL/well of PBST.
Add 250 µL/well of blocking solution and incubate for 1 h at RT on a microplate shaker.
Using a plate washer, wash the plate five times with 300 µL/well of PBST.
Add 110 µL of either pre- or post-immunisation serum diluted 1:10 in blocking solution to the first well of the 96-well plate. Prepare a serial dilution of the sera in the plate by adding 100 µL of blocking solution to the second well and add 10 µL of the solution from the first well. Repeat for a third well so that the sera are diluted 1 in 10, 100, and 1,000; include a no-serum control. Incubate for 1 h at RT on a microplate shaker.
Using a plate washer, wash the plate five times with 300 µL/well of PBST.
Add 100 µL/well of anti-llama-HRP diluted 1:2,500 in 0.1% BSA-PBS and incubate for 1 h at RT on a microplate shaker.
Using a plate washer, wash the plate five times with 300 µL/well of PBST.
Add 100 µL/well of ABTS substrate, prepared by mixing solution A and solution B in a 1:1 ratio. Protect from light and measure the absorbance at 405 nm after 15 min. See General note 2.
VHH library preparation
Below (Table 1) is a suggested timetable of experiments required for the preparation of a VHH library. Instances where the protocol can be interrupted/held are indicated by “pause point.”
Table 1. Experimental timetable for the preparation of a VHH library
Task 1 Task 2 Task 3 Day 1 Isolation of PMNCs
(step C1)
Isolation of immune serum
(step C2)
RNA extraction (steps C3a–g) Pause point Day 2 RNA extraction
(steps C3h–i)
Reverse transcription and PCR amplification of VHH1 (step C4) Production of VHH2 (step C5) Pause point Day 3 Digestion of pADL23c (step C6)
Pause point
Day 4 Test of library size
(steps C7a–g)
Day 5 Test of library size
(steps C7h–n)
Pause point
Day 6 Scaled library preparation (steps C8a–i) Day 7 Scaled library preparation (steps C8j)
Pause point
Isolation of PMNCs
The below instructions are based on receiving 170 mL of llama blood in 17 × 10 mL blood tubes without anticoagulant. Isolation of the PMNCs should be done on the day of the blood draw. All work is performed in a safety cabinet after spraying with 5% Chemgene followed by 70% EtOH.
Caution: Decontaminate all blood-contaminated consumables and liquid with 2% Virkon, followed by autoclaving.
Transfer blood from 2 × 10 mL blood tubes into 1 × 50 mL conical centrifuge tube using a 10 mL serological pipette.
Add 10 mL of PBS to each of the empty blood vials and transfer into the conical centrifuge tube with the 20 mL of blood (step C1a) to give a total volume of 40 mL of diluted blood per 50 mL conical centrifuge tube, or a total of 340 mL of diluted blood. Invert tubes gently to ensure a homogenous mixture.
In fresh 23 × 50 mL conical centrifuge tubes, add 15 mL of Histopaque®-1077 and then gently layer 15 mL of the diluted blood on top. See General note 3. Hold the tube at a 45° angle from the horizontal while adding the blood. See General note 4.
Critical: Avoid agitating the tubes at this point to maintain the density gradient that is forming.
Centrifuge the conical centrifuge tubes at 800× g for 20 min at 18 °C using an acceleration = 1 and deceleration = 0.
Critical: The acceleration and deceleration rates are very important, as they will create and maintain the desired gradient. The results of the density gradient and localisation of the PMNC layer is shown in Figure 1.
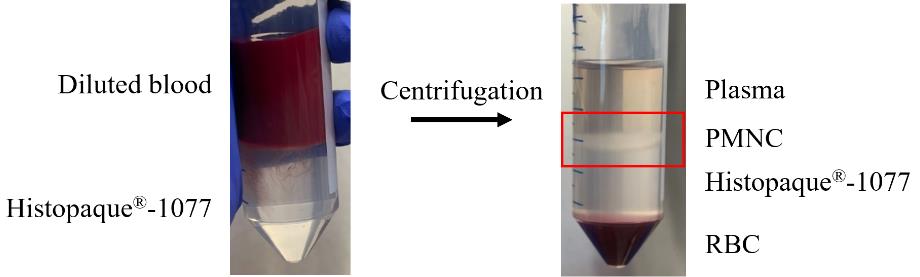
Figure 1. Isolation of polymorphonuclear cells (PMNCs) from llama blood using a density gradient. Diluted llama blood overlaid on Histopaque®-1007 and the resulting layer of PMNCs and red blood cells (RBCs) after centrifugation.From the 23 prepared blood–Histopaque®-1077 tubes, collect the PMNC layer at the plasma–Histopaque®-1077 interface (red square in Figure 1). This is achieved by using a 25 mL serological pipette and placing it just above the interface, drawing the liquid up whilst moving across the surface interface. Any Histopaque®-1077 or serum that is drawn up with the PMNCs will be washed away in the next steps.
Transfer 2× conical centrifuge tubes’ worth of isolated PMNCs into a single 50 mL conical centrifuge tube. Typically, 15–20 mL of PMNCs is retrieved from each conical centrifuge tube. If more is collected, ensure that 20 mL of isolated PMNCs is added to fresh 50 mL conical centrifuge tubes. There should be between 23 and 25 × 50 mL conical centrifuge tubes at this point.
To each conical centrifuge tube containing isolated PMNCs, add PBS to a total volume of 40 mL. One can just pour from the bottle of PBS, as long as it does not touch the conical centrifuge tube. Gently invert the conical centrifuge tubes several times and centrifuge at 250× g for 10 min at 18 °C.
Pour off the supernatant and gently resuspend the pellet from each conical centrifuge tube in 5 mL PBS using a new 5 mL serological pipette per pellet. Centrifuge the cells at 100× g for 5 min at 18 °C.
Note: At this point, start using RNaseZapTM to wipe the work surface, gloves, and any items that are brought into the safety cabinet.
Use a 5 mL serological pipette to remove the supernatant from each 50 mL conical centrifuge tube. Gently resuspend the pellets from three conical centrifuge tubes in a total of 1 mL of PBS into a single 50 mL conical centrifuge tube using a 2 mL serological pipette. Use the final amount of liquid in this serological pipette and dispense it into a 1.5 mL microcentrifuge tube. There should be six 50 mL conical centrifuge tubes and six 1.5 mL microcentrifuge tubes at this point.
Remove the six 1.5 mL microcentrifuge tubes from the safety cabinet. In the lid of each of the 1.5 mL microcentrifuge tubes, add 20 µL of cells and 20 µL of trypan blue and mix gently. Determine cell number and viability using an automated cell counter. Expect approximately 1×107 cells at 98%–100% viability per sample taken from each conical centrifuge tube. Thus, an overall yield of 6×107 cells should be expected. See General note 5.
Note: The following steps can be performed at a lab bench or in a PCR cabinet after being sprayed with RNaseZapTM and using RNase-free tips and 1.5 mL microcentrifuge tubes.
Transfer the cell suspension from each 50 mL conical centrifuge tube (step C1i) into their own 1.5 mL microcentrifuge tube. There should be six 1.5 mL microcentrifuge tubes in total. Pellet the cells at 100× g for 5 min at 4 °C. Discard the supernatant. The RNA can now be isolated from the pellets (step C3a).
Critical: Perform RNA extraction (step C3) immediately.
Isolation of pre- and post-immune serum
Isolation of serum must be done on the day of the blood draw. These steps can be done during the isolation of PMNCs at a lab bench.
Incubate the coagulated blood at RT for 30 min.
Transfer the serum and the blood clot into a 50 mL conical centrifuge tube. This is achieved by using a 2 mL serological pipette to stab holes in the blood clot and using the stripette to guide the clot and serum into a 50 mL conical centrifuge tube. Centrifuge the conical centrifuge tube at 1,000× g for 10 min at 4 °C.
Transfer the serum into a 15 mL conical centrifuge tube. A second centrifugation at 1,000× g for 10 min at 4 °C may be required to obtain clear serum. Prepare 500 µL aliquots of serum in screw cap microtubes, which are then stored at -20 °C.
RNA extraction
Note: All work should be done at a lab bench or in a PCR cabinet after spraying with RNaseZapTM. Use RNase-free consumables and spray gloves with RNaseZapTM. See General note 6.
To each cell pellet in each of the six 1.5 mL microcentrifuge tubes (step C1k), add 1 mL of cold TRIzolTM reagent. Using a 21 G needle and 2.5 mL syringe, homogenise the contents of each 1.5 mL microcentrifuge tube by aspirating and dispensing ten times through the needle.
Caution: Use caution when working with needles.
Incubate the resulting lysate on ice for 5 min to permit complete dissociation of the nucleoproteins complex and then add 0.2 mL of chloroform to each 1.5 mL microcentrifuge tube.
Vortex each microcentrifuge tube vigorously for approximately 10 s. A homogenous pink milky solution should occur. Incubate on ice for 3 min and then centrifuge at 12,000× g for 15 min at 4 °C. The mixture separates into a lower red phenol-chloroform layer, an interphase layer, and a colourless upper aqueous phase. If the upper layer is still cloudy or has not formed properly, vortex and centrifuge again.
Transfer approximately 500 µL of the upper aqueous phase containing the RNA to a new 1.5 mL microcentrifuge tube using a 200 µL pipette. Some of the upper aqueous layer can be left behind to ensure that none of the precipitate from the interphase is included. Add 0.5 mL of isopropanol, gently invert each tube several times, and incubate on ice for 10 min.
Centrifuge at 12,000× g for 10 min at 4 °C to pellet the RNA. Keep the lid of the 1.5 mL microcentrifuge tube facing outwards in the rotor to aid in the visualisation of the white gel-like pellet after centrifugation. Remove the approximately 900 µL of the isopropanol containing supernatant using a 200 µL pipette to ensure that the pellet is not disturbed or accidently aspirated with the waste.
Add 1 mL of 75% ethanol to the 1.5 mL microcentrifuge tube, vortex briefly, and centrifuge at 7,500× g for 5 min at 4 °C. The RNA pellet will look a little whiter and slightly bigger than what it did at the end of the previous step. Remove the approximately 900 µL of the supernatant using a 200 µL pipette to ensure that the pellet is not disturbed or accidently aspirated.
Add 1 mL of 75% EtOH and store the purified RNA at -20 °C. See General note 7.
Pause point.
Remove one microcentrifuge tube of RNA from the -20 °C and centrifuge at 7,500× g for 5 min at 4 °C. Discard the EtOH using a 200 µL and then a 10 µL pipette to remove all EtOH without accidently aspirating the pellet. Allow to dry at RT for 10 min.
Dissolve the RNA pellet in 30 µL of nuclease-free water and measure the A260 using the Nanodrop. Expect a concentration between 500 and 800 ng/µL per 1.5 mL tube with values of approximately 2.0 for both A260/280 and A260/230.
Critical: Use RNase-free tips and pipette from the PCR cabinet to measure the RNA concentration to avoid the introduction of RNases from the communal tips and pipette used at the Nanodrop. Once the RNA has been isolated, proceed to step C4 as soon as possible to produce cDNA, which is more stable than RNA.
Reverse transcription and PCR amplification of VHH1
Note: Work at a lab bench or in a PCR cabinet after spraying with RNaseZapTM. Use RNase-free consumables and spray gloves with RNaseZapTM. See General note 6.
Prepare the following reaction in a PCR tube (1 × 50 µL) (Table 2).
Table 2. PCR reaction composition
Reagent Final concentration Volume Nuclease-free water n/a 9.5 µL 10 µM CALL 001 primer 0.5 µM 2.5 µL 10 µM CALL 002 primer 0.5 µM 2.5 µL 2× PlatinumTM SuperFiTM RT-PCR Master Mix 1× 25 µL 1 µg Template RNA (step C3i) 200 ng 10 µL SuperScriptTM IV RT Mix 0.5 µL Total n/a 50 µL Briefly vortex and centrifuge the reaction tube and place on ice.
Set up the thermal cycler as below and perform the PCR (Table 3).
Table 3. PCR cycling conditions
Step Temperature (°C) Duration Number of cycles Reverse transcription 55 10 min 1 Reverse transcription inactivation/initial denaturation 98 2 min Denaturation 98 10 s 30 Annealing 66 10 s Extension 72 30 s Final extension 72 5 min 1 Hold 12 Infinite hold -
Note: There is no need to maintain RNase-free conditions from this point onwards. Work at a lab bench.Add 10 µL of 6× DNA gel loading buffer to the reaction tube and run alongside a lane of 1× GeneRuler 1 kb DNA ladder on a 0.7% agarose gel containing 1× SYBRTM safe DNA gel stain. Electrophorese in 1× TBE buffer at 80 V for 40 min.
Visualise the amplified 700 bp fragment (VHH-CH2) using a LED Blue transilluminator, excise from the gel using a scalpel knife, and gel extract using the Nucleospin Gel and PCR clean-up kit according to the manufacturer’s protocol. Elute in a final volume of 50 µL of NE buffer.
Caution: Use Safe ImagerTM viewing glasses when visualising the DNA during excision of the band from the agarose gel and use caution when using scalpel knives.
Using the Purelink PCR purification kit, add 4× volume of B2 buffer (200 µL) to the gel-extracted product and purify using one Purelink column from the Purelink PCR purification kit eluting in 25 µL E1 buffer. Measure the A280 and expect a concentration of approximately 40–50 ng/µL.
Mix 6× DNA gel loading buffer with 120 ng of VHH1 (step C4f) to a final concentration of 1× and run alongside a well of 1× HyperLadderTM 1 kb DNA ladder on a 1% agarose gel containing 1× SYBRTM safe DNA gel stain to determine the purity. Electrophorese in 1× TBE buffer at 80 V for 40 min. Expect a result as shown in Figure 2.
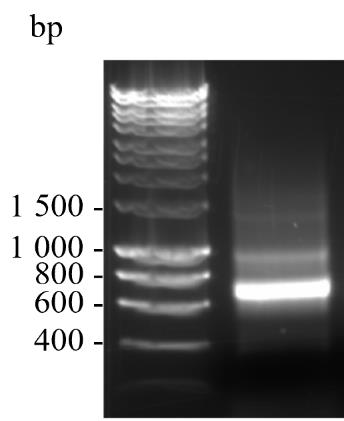
Figure 2. Sample of amplified VHH1 of approximately 700 bp visualised on a 1% agarose gel containing 1 × SYBRTM safe DNA gel stain. The sizes in base pairs of a DNA ladder run in parallel are shown.
Production of VHH2
Prepare the following PCR reaction in PCR tubes (6 × 50 µL reactions) (Table 4).
Table 4. PCR reaction composition
Reagent Final concentration Volume Nuclease-free water n/a 19 µL 10 µM VHH For2 primer 0.5 µM 2.5 µL 10 µM VHH Rev primer 0.5 µM 2.5 µL 2× Phusion flash PCR master mix 1× 25 µL 5 ng/µL VHH1 (step C4f) 1 ng 10 µL Total n/a 50 µL Briefly vortex and centrifuge the reaction tubes.
Set up the thermal cycler as below and perform the PCR (Table 5).
Table 5. PCR cycling conditions
Step Temperature (°C) Duration Number of cycles Denaturation 98 30 s 1 Annealing 98 10 s 30 Extension 55 30 s Final extension 72 20 s Hold 72 5 min 1 Denaturation 12 Infinite hold - Combine all six reactions into a single 2 mL microcentrifuge tube. Using the Purelink PCR purification kit, add 4× volume of B2 buffer (1.2 mL) to the amplified PCR product and purify between two Purelink columns, eluting each with 25 µL of E1 buffer. Combine both eluates into one 1.5 mL microcentrifuge tube, measure the A280, and expect a concentration between 200 and 300 ng/µL.
Mix 6× DNA gel loading buffer with 120 ng of VHH2 (step C5d) to a final concentration of 1× and run alongside a well of 1× HyperladderTM 1 kb DNA ladder on a 1% agarose gel containing 1× SYBRTM safe DNA gel stain to determine the purity. Electrophorese in 1× TBE buffer at 80 V for 40 min. Expect a result as shown in Figure 3.
Pause point.
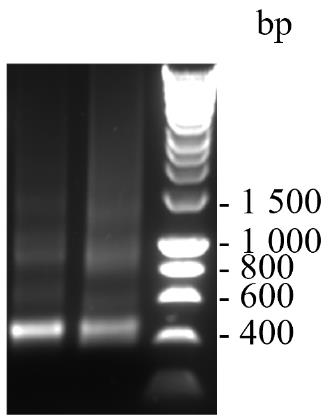
Figure 3. Sample of amplified VHH2 of approximately 400 bp visualised on a 1% agarose gel containing 1× SYBRTM safe DNA gel stain. The sizes in base pairs of a DNA ladder run in parallel are shown.
Digestion of pADL23c
Prepare the following restriction digestion reaction in PCR tubes (10 × 100 µL reactions) (Table 6).
Table 6. Restriction digestion reaction composition
Reagent Final concentration Volume Nuclease-free water n/a 83 µL rCutSmartTM buffer 1× 10 µL SfiI 5 units 2.5 µL 400 ng/µL vector DNA 1.8 µg 4.5 µL Total n/a 100 µL Briefly vortex and centrifuge the reaction tubes.
Incubate at 50 °C for 2 h.
Add 20 µL of 6× DNA gel loading buffer to each reaction tube and run alongside a well of 1× GeneRuler 1 kb DNA ladder on a 0.7% agarose gel containing 1× SYBRTM Safe DNA gel stain. Electrophorese in 1× TBE buffer at 80 V for 40 min. Expect a result as shown in Figure 4.
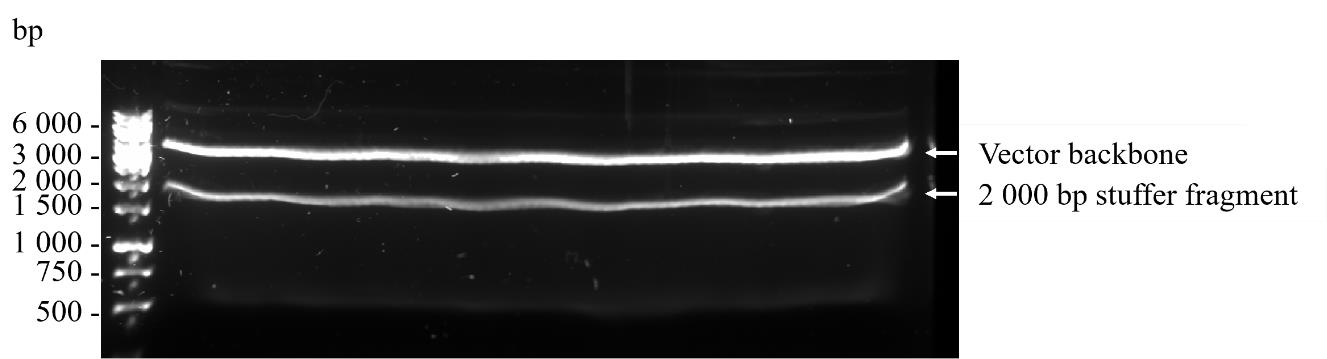
Figure 4. Preparative 0.7% agarose gel containing 1× SYBRTM safe DNA gel stain of the vector backbone (upper band) and stuffer fragment (lower band) after SfiI digestion of pADL23c phagemid vector prior to gel extraction. The sizes in base pairs of a DNA ladder run in parallel are shown.Visualise the digested, higher molecular weight, approximately 3,000 bp vector backbone using a LED Blue transilluminator, excise from the gel using a scalpel knife, and gel extract using 10 spin columns from the Nucleospin Gel and PCR clean-up kit according to the manufacturer’s protocol. Elute each column using 50 µL of NE buffer and pool all the eluates into a single 1.5 mL microcentrifuge tube.
Caution: Use Safe ImagerTM viewing glasses when visualising the DNA during excision of the band from the agarose gel and use caution when using a scalpel knife.
Using the Purelink PCR purification kit, add 4× volume of B3 buffer (2 mL) to the gel extracted product and purify using one Purelink column from the Purelink PCR purification kit eluting in 50 µL E1 buffer. Measure the A280 and expect a concentration between 50 and 80 ng/µL.
Mix 6× DNA gel loading buffer with 120 ng of VHH2 (step C5d) and digest pADL23c backbone (step C6f) to a final concentration of 1× and run alongside a well of 1× HyperLadderTM 1 kb DNA ladder on a 1% agarose gel containing 1× SYBRTM Safe DNA gel stain to determine the purity. Electrophorese in 1× TBE buffer at 80 V for 40 min. Expect a result as shown in Figure 5.
Pause point.
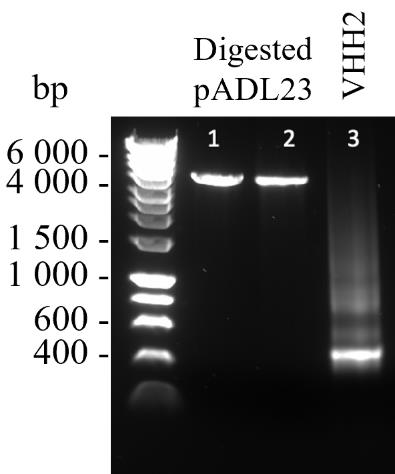
Figure 5. Samples of linearised and purified pADL23c (lanes 1 and 2) and purified VHH2 (lane 3) visualised on a 1% agarose gel containing 1× SYBRTM safe DNA gel stain. The sizes in base pairs of a DNA ladder run in parallel are shown.
Test of library size
Prepare the following in-fusion reaction in a PCR tube (1 × 10 µL) (Table 7).
Table 7. PCR reaction composition
Reagent Final concentration Volume Nuclease-free water n/a 5 µL 5× CEII buffer 1× 2 µL 100 ng/µL VHH2 insert (step C5d) 100 ng 1 µL 20 ng/µL vector DNA (step C6f) 20 ng 1 µL Exnase II 1 µL Total n/a 10 µL Briefly vortex and centrifuge the reaction tube.
Incubate at 42 °C for 30 min using a thermal cycler. Add 40 µL of TE buffer to terminate the reaction.
Using the Purelink PCR purification kit, add 4× volume of B3 buffer (200 µL) to the in-fusion product and purify using one Purelink column eluting in 20 µL nuclease-free water.
Note: Work in a safety cabinet.
Gently mix 5 µL of purified in-fusion reaction with 30 µL of competent TG1 in a 1.5 mL microcentrifuge tube and transfer to a chilled 1 mL electroporation cuvette.
Electroporate at 1.7 kV and immediately add 400 µL of recovery medium to the cuvette. Transfer contents to a 2 mL microcentrifuge tube and incubate for 1 h at 37 °C agitating at 600 rpm. See General note 8.
Using 50 µL of culture, prepare a 1:10, 1:100, and 1:1,000 dilution in 2× YT. Spread 100 µL on 1% agar LB plates containing 100 µg/mL ampicillin until dry. Incubate overnight at 37 °C.
Estimate the library size in terms of colony forming units per millilitre (CFU/mL) using the following formula: number colonies × 10 (to get to mL) × 0.4 (volume) × dilution. See General note 9.
Based on the estimated small-scale library size, set up the required number of reactions considering that 5 µL of in-fusion reaction yielded × size library. Calculate how many microlitres of in-fusion reactions would be needed to make a library with a size of 1 × 106. See General note 10.
To validate the number of full-length VHH clones in the small-scale library, a colony PCR to amplify the ~500 bp VHH gene in 48 randomly selected clones from a plate in step C7g is performed. Prepare the following PCR reaction master mixture in a 2 mL microcentrifuge tube (Table 8).
Table 8. PCR reaction composition
Reagent Final concentration Volume for single reaction Volume for master mixture Nuclease-free water n/a 20.75 µL 1037.5 µL 10 µM PhD seq Fwd primer 0.2 µM 0.5 µL 25 µL 10 µM PhD seq Rev primer 0.2 µM 0.5 µL 25 µL 10× Taq buffer 1× 2.5 µL 125 µL 25 mM dNTP 0.25 mM 0.25 µL 12.5 µL Taq polymerase 1 unit 0.5 µL 25 µL Total n/a 25 µL 1250 µL Briefly vortex and centrifuge the reaction tube.
Pipette 25 µL of the master mixture into 48 wells of a 96-well rigid semi skirted PCR plate. Pick 48 colonies with 48 × 10 µL tips and place into each filled well. Remove the tips using a multichannel pipette. Cover the PCR plate with PCR foil seal.
Set up the thermal cycler as below and perform the PCR (Table 9).
Table 9. PCR cycling conditions
Step Temperature (°C) Duration Number of cycles Denaturation 95 7 min s 1 Annealing 95 15 s 35 Extension 55 30 s Final extension 68 1 min 40 s Hold 68 5 min 1 Denaturation 12 Infinite hold - Add 20 µL of 2× DNA gel loading buffer to each well of the PCR plate and load 10 µL to each well alongside a well of 1× GeneRuler 1 kb DNA ladder on a 1% agarose gel containing 1× SYBRTM safe DNA gel stain. Electrophorese in 1× TBE buffer at 80 V for 40 min. In Figure 6, 46 out of the 48 clones that were subjected to colony PCR possessed the ~500 bp band corresponding to the amplified VHH gene. The two clones that were not positive are indicated by an asterisk.
Pause point.
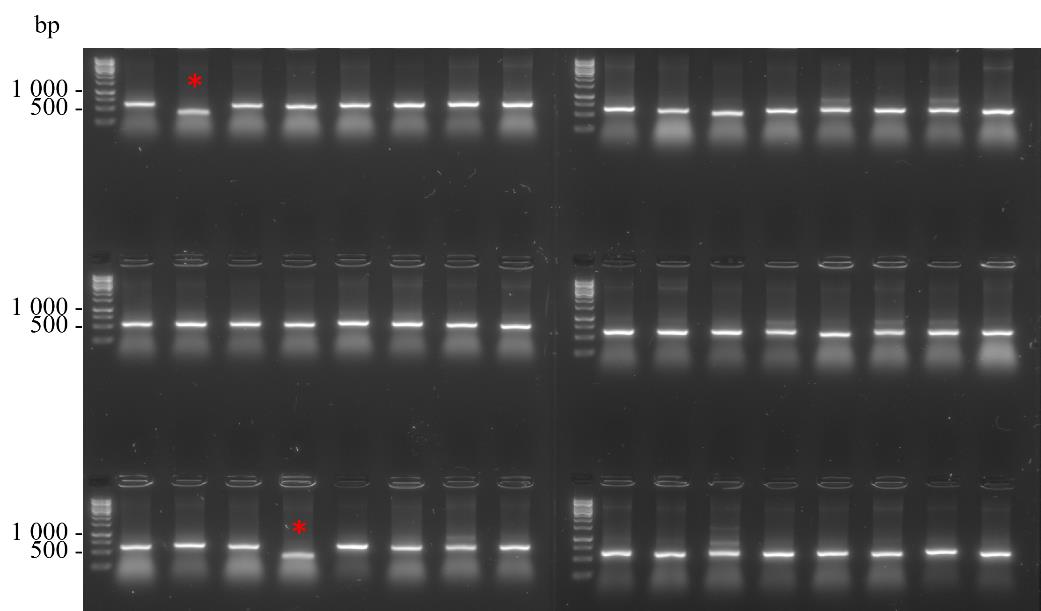
Figure 6. Colony PCR of 48 randomly selected clones from a small-scale library preparation
Scaled library preparation
Prepare the following reaction in PCR tubes (20 × 20 µL) (Table 10).
Table 10. In-Fusion reaction composition
Reagent Final concentration Volume Nuclease-free water n/a 9 µL 5× CEII buffer 1× 4 µL 50 ng/µL VHH2 insert (step C5c) 100 ng 2 µL 5 ng/µL vector DNA (step C6f) 20 ng 4 µL Exnase II 1 µL Total n/a 20 µL Briefly vortex and centrifuge the reaction tube.
Incubate at 42 °C for 30 min in a thermal cycler.
Combine reactions into a single 2 mL microcentrifuge tube. Using the Purelink PCR purification kit, add 4× volume of B3 buffer (1.6 mL) to the in-fusion product and purify using one Purelink column eluting in 50 µL of nuclease-free water. Measure the A280 and expect a concentration of between 35 and 50 ng/µL.
Note: Work in a safety cabinet.
TG1 cells are transformed by electroporation in separate reactions to create the scaled library. Gently mix 6 µL of purified in-fusion reaction with 30 µL of competent TG1 in a 1.5 mL microcentrifuge tube and transfer to a chilled 1 mL electroporation cuvette. Electroporate at 1.7 kV, immediately add 960 µL of recovery medium to the cuvette, and transfer to a 50 mL conical centrifuge tube. See General note 8.
Repeat this a total of eight times with eight separate in-fusion-TG1 mixtures.
Combine two cuvettes worth of electroporated cells into a single 50 mL conical centrifuge tube and incubate at 37 °C for 1 h shaking at 200 rpm. You should have four conical centrifuge tubes in total.
Use 50 µL from one of the conical centrifuge tubes and prepare a 1:10, 1:100, and 1:1,000 dilution in 2× YT. Spread 100 µL of each dilution on 1% agar LB plates containing 100 µg/mL ampicillin until dry. Incubate overnight at 37 °C. Use these plates to estimate library size (CFU/mL) = number colonies × 10 (to get to mL) × 1.92 (volume) × dilution. See General note 9.
Plate the entire culture of each of the four conical centrifuge tubes onto separate bioassay dishes filled with 2% agar LB plates containing 100 µg/mL ampicillin. There should be four plates in total. Spread the entire volume until dry and incubate overnight at 37 °C. Expect a density similar to that shown in Figure 7, which is equivalent to a 1 × 106 CFU/mL sized library. See General note 11.

Figure 7. Expected density on a bioassay plate for a 1 × 106 CFU/mL libraryResuspend the lawn of colonies of each bioassay plate in 6 mL of 2× YT (containing a final concentration of 25% glycerol) and transfer to a single 50 mL conical centrifuge tube. Mix well by serological pipette and prepare 1 mL aliquots in screw cap microtubes, which are stored at -80 °C. This is the VHH library stock, which is used for the first round of panning.
Pause point.
Panning
Below is a suggested timetable for panning, hit identification by ELISA, and small-scale expression to maximise time in the lab and to allow space for overlap if multiple panning campaigns are being performed for multiple proteins by multiple people using shared resources (Table 11).
Table 11. Experimental timetable for the identification of nanobody binders through panning, ELISA, and small-scale expression
Monday Tuesday Wednesday Thursday Friday Week 1 Amplify VHH library (step D3) Recover library phage (step D3) First panning (step D4)
Recover lawn of colonies (step D4)
Amplify sub-library (step D5)
Recovery of sub-library phage (step D5)
Second panning (step D6)
Take plates out of the incubator and place in fridge.
Week 2 Pick 93 colonies and grow overnight (step D7) Prepare master plates (step D7)
Start culture for anti-M13 ELISA (step D8)
Anti-M13 ELISA (step D8) PCR, clean up, sequencing (step D9) Week 3 Overnight culture for small scale (step E1) Small-scale culture to OD and induce with IPTG (step E1) Periplasm isolation (step E1)
Ni-NTA purification (step E2)
Desalting (step E3)
Titration ELISA (step E4)
Off-rate determination (step E5)
Prepare log-phase TG1
Note: Work in a safety cabinet.
Streak TG1 on a 1% agar LB plate without antibiotics and incubate overnight at 37 °C.
Inoculate 20 mL of 2× YT with a single TG1 colony in a 100 mL disposable baffled Erlenmeyer flask and incubate overnight at 37 °C shaking at 200 rpm.
Mix 20 mL of 50% glycerol with the overnight culture and store in 1 mL aliquots at -80 °C. This is the TG1 stock, which we use to prepare log-phase TG1 culture when needed.
To 25 mL of 2× YT in a 125 mL disposable baffled Erlenmeyer flask, add enough TG1 stock (step D1c) so that so that the starting OD600 is approximately 0.05. See General note 12.
Incubate the TG1 culture at 37 °C shaking at 200 rpm until the OD600 reaches 0.4–0.6. This takes approximately 1.5–2 h. This is the log-phase TG1, which should be kept on ice and used within 2 h.
Prepare the CM13K helper phage
Caution: Phage can infect all bacteria. If working in a protein expression lab, care must be taken not to release phage into communal areas. Wipe down surfaces with 5% Chemgene followed by 70% EtOH if the surface is susceptible to corrosion. The use of filtered tips is encouraged to reduce aerosols. All contaminated consumables must be decontaminated with 2% Virkon and autoclaved. If possible, use disposable Erlenmeyer flasks for all cultures outlined in steps D1–D5.
Note: Work in a safety cabinet.
Inoculate 20 mL of 2× YT with 200 µL of log-phase TG1 (step D1e) in a 125 mL disposable baffled Erlenmeyer flask. Add 1 µL of previously prepared 1 × 1013 PFU/mL CM13K helper phage glycerol stock (step D2h) and incubate at 37 °C for 4 h shaking at 250 rpm.
In two 1 L disposable baffled Erlenmeyer flasks, add 250 mL of 2× YT containing 25 µg/mL kanamycin and 10 mL of the prepared culture. Incubate overnight at 25 °C shaking at 250 rpm.
Pellet the cells at 4,500× g for 10 min at 4 °C and transfer approximately 170 mL of the phage containing supernatant into three 250 mL PPCO centrifuge tubes. Add approximately 35 mL of ice-cold PEG/NaCl to each tube, invert, and swirl gently to mix the PEG/NaCl solution with the phage supernatant. Incubate on ice for 1 h.
Centrifuge the tubes at 12,000× g for 15 min at 4 °C and resuspend each of the three pellets in 4 mL of ice-cold sterile PBS. Combine the resuspended pellets into a single 25 mL high-speed PPCO centrifuge tube and add 2.5 mL of ice-cold PEG/NaCl. Swirl and invert gently to mix the PEG/NaCl solution with the phage supernatant. Incubate on ice for 30 min.
Centrifuge at 12,000× g for 10 min at 4 °C and resuspend the pellet in 12 mL of ice-cold sterile PBS. Keep at 4 °C until the phage concentration, expressed as plaque forming units per millilitre (PFU/mL), has been determined either spectrophotometrically or by plaque assay, both of which should yield similar values.
For the spectrophotometric determination of phage concentration, in a 1.5 mL microcentrifuge tube, add 998 µL of PBS and 2 µL of phage (step D2e), transfer to a quartz cuvette, and measure the absorbance at 268 nm. Calculate the PFU/mL considering that an A268 of 1.0 is equivalent to 5 × 1012 PFU/mL [11].
For the determination of phage concentration using the plaque assay, prepare tenfold serial dilutions (10-1 to 10-25) of CM13K helper phage (step D2e) in log-phase TG1 (step D1e) and incubate at RT for 5 min. Plate 100 µL of 10-15 to 10-25 dilutions onto 1% agar LB plates without antibiotics. Add 3 mL of 0.7% agar LB (at 55 °C) per plate and swirl over the entire plate surface. Allow the top agar to solidify and incubate overnight at 37 °C. Calculate the PFU/mL using the following formula: number of plaques (clear areas) × 10 (to get to mL) × dilution.
Dilute the CM13K helper phage (step D2e) to 2 × 1013 PFU/mL using ice-cold sterile PBS, add an equal volume of ice-cold 50% glycerol, and mix gently to achieve a final concentration of 1 × 1013 PFU/mL. Store in 100 µL aliquots in 500 µL microcentrifuge tubes. This is the CM13K helper phage stock. Each time a new 100 µL aliquot is thawed, sub-aliquot 12 µL into 9 × PCR tubes to avoid freeze thawing of the original 100 µL aliquot.
Amplify and recover library
Note: Work in a safety cabinet.
To 50 mL of 2× YT containing 100 µg/mL ampicillin in a 250 mL disposable baffled Erlenmeyer flask, add enough VHH library stock (step C8j) so that the starting OD600 is approximately 0.05. See General note 13.
Incubate the culture at 37 °C shaking at 200 rpm until the OD600 reaches 0.4–0.6. This takes approximately 2 h. See General note 14.
Transfer 10 mL of this culture into a 50 mL conical centrifuge tube, add 10 µL of the CM13K helper phage stock (step D2h), and mix by gentle swirling. Incubate at 37 °C for 1 h without agitation.
Pellet the cells at 2,800× g for 10 min at 18 °C. Resuspend the pellet in 50 mL of 2× YT containing 100 µg/mL ampicillin and 25 µg/mL kanamycin and transfer to a 250 mL disposable baffled Erlenmeyer flask. Incubate overnight at 25 °C shaking at 250 rpm.
Transfer the overnight culture into a 50 mL conical centrifuge tube and pellet the cells at 3,200× g for 10 min at 4 °C.
Pour 40 mL of the phage containing supernatant into a new 50 mL conical centrifuge tube and add 10 mL of ice-cold PEG/NaCl. Invert several times and incubate on ice for 1 h.
Pellet the precipitated phage by centrifugation at 3,200× g for 10 min at 4 °C. Resuspend the phage in 1 mL of ice-cold sterile PBS and transfer to a 2 mL microcentrifuge tube.
After centrifugation at 20,000× g for 1 min at 4 °C, transfer the supernatant containing phage into a new 2 mL microcentrifuge tube. Add 250 µL of ice-cold PEG/NaCl and invert the tube until a homogeneous white suspension appears. Incubate on ice for 30 min.
Pellet the precipitated phage by centrifugation at 20,000× g for 15 min at 4 °C. Resuspend the pelleted phage in 1 mL of ice-cold sterile PBS and transfer to a 2 mL microcentrifuge tube.
After a final centrifugation at 20,000× g for 1 min at 4 °C, transfer the supernatant containing phage into a new 2 mL microcentrifuge tube. This is the isolated library phage; it should be kept either on ice or at 4 °C and is stable for a month.
Titrate 15 µL of the library phage using tenfold serial dilutions from 10-1 to 10-12 with log-phase TG1 cells (step D1e). After incubation at 37 °C for 15 min, spread 50 µL of the 10-4 to 10-12 dilutions on 1% agar LB plates containing 100 µg/mL ampicillin until dry and incubate overnight at 37 °C. Include a TG1-only control. See General note 15.
Use these plates to estimate how many phage can be produced from the library. This is calculated as PFU/mL = number colonies × 10 (to get to mL) × 0.05 (volume) × dilution. Expect 1 × 1010–1 × 1013 PFU/mL. See General note 16.
Panning first round
Note: Work at a lab bench.
In a 2 mL microcentrifuge tube, add 500 µL of StartingBlock buffer and 500 µL of isolated library phage (step D3j). Wrap parafilm around the microcentrifuge tube lid to avoid any accidental spillage and incubate for 30 min at RT using a sample mixer.
Add an appropriate volume of biotinylated antigen to achieve 50 nM final concentration in 1.5 mL (which is achieved by step D4d). See General note 1. Wrap parafilm around the microcentrifuge tube lid to avoid any accidental spillage and incubate for 1 h at RT using a sample mixer.
At this point, one can start growing the log-phase TG1 cells (steps D1d–e), which would ensure that the cells are at the required OD600 once the phage has been eluted (step D4h).
Thirty minutes into the incubation of phage with antigen (step D4b), in a new 2 mL microcentrifuge tube, add 100 µL of streptavidin DynabeadsTM M-280. Wash the beads twice with 500 µL of PBS and then add 500 µL of StartingBlock buffer. Incubate for 30 min at RT using a sample mixer.
Transfer the 1 mL of blocked phage with antigen (step D4b) to the blocked beads (step D4d). The total volume is 1.5 mL, and the final antigen concentration is now 50 nM. Wrap parafilm around the microcentrifuge tube lid to avoid any accidental spillage and incubate for 15 min at RT using a sample mixer.
Place the microcentrifuge tube on a DynaMagTM-2 magnet. Once the beads have been pulled to the side, discard the unbound phage in the supernatant. Wash away loosely bound phage by resuspending the beads in 500 µL of PBST and then using the magnet to pull the beads to the side. Repeat this washing process a total of six times with PBST and then once with PBS.
Add 500 µL of 250 µg/mL trypsin to the beads. Wrap parafilm around the microcentrifuge tube lid to avoid any accidental spillage and incubate for 30 min at RT using a sample mixer.
Collect the eluted phage from the first pan in the supernatant after using the DynaMagTM-2 magnet to pellet the beads.
Note: Work in a safety cabinet.
In a 50 mL conical centrifuge tube, add 500 µL of eluted phage from the first pan (step D4h) to 10 mL of log-phase TG1 cells (step D4c) and incubate at 37 °C for 30 min without agitation.
Pellet the cells at 2,800× g for 10 min at 18 °C and resuspend in 1 mL of 2× YT. Spread the entire volume until dry on one bioassay dish containing 1% agar LB with 100 µg/mL ampicillin and incubate overnight at 37 °C.
Resuspend the lawn of colonies in 6 mL of 2× YT (containing a final concentration of 25% glycerol) and store in 1 mL aliquots in cryotubes at -80 °C. This is the pan 1 sub-library stock.
Amplify and recover sub-library
Note: Work in a safety cabinet.
To 50 mL of 2× YT containing 100 µg/mL ampicillin in a 250 mL disposable baffled Erlenmeyer flask, add enough pan 1 sub-library stock (step D4k) so that the starting OD600 is approximately 0.05. See General note 13.
Incubate the culture at 37 °C shaking at 200 rpm until the OD600 reaches 0.4–0.6. This takes approximately 2 h. See General note 14.
Transfer 10 mL of this culture into a 50 mL conical centrifuge tube, add 10 µL of the CM13K helper phage stock (step D2h), and mix by gentle swirling. Incubate at 37 °C for 1 h without agitation.
Pellet the cells at 2,800× g for 10 min at 18 °C. Resuspend the pellet in 50 mL of 2× YT containing 100 µg/mL ampicillin and 25 µg/mL kanamycin and transfer to a 250 mL disposable baffled Erlenmeyer flask. Incubate overnight at 25 °C shaking at 250 rpm.
Transfer the overnight culture into a 50 mL conical centrifuge tube and pellet the cells at 3,200× g for 10 min at 4 °C.
Pour 40 mL of the phage containing supernatant into a new 50 mL conical centrifuge tube and add 10 mL of ice-cold PEG/NaCl. Invert several times and incubate on ice for 1 h.
Pellet the precipitated phage by centrifugation at 3,200× g for 10 min at 4 °C. Resuspend the phage in 1 mL of ice-cold sterile PBS and transfer to a 2 mL microcentrifuge tube.
After centrifugation at 20,000× g for 1 min at 4 °C, transfer the supernatant containing phage into a new 2 mL microcentrifuge tube. Add 250 µL of ice-cold PEG/NaCl and invert the microcentrifuge tube until a homogeneous white suspension appears. Incubate on ice for 30 min.
Pellet the precipitated phage by centrifugation at 20,000× g for 15 min at 4 °C. Resuspend the pelleted phage in 1 mL of ice-cold sterile PBS and transfer to a 2 mL microcentrifuge tube.
After a final centrifugation at 20,000× g for 1 min at 4 °C, transfer the supernatant containing phage into a new 2 mL microcentrifuge tube. This is the isolated pan 1 phage; it should be kept either on ice or at 4 °C and is stable for a month.
Panning second round
Note: Work in a lab bench.
In a 2 mL microcentrifuge tube, add 500 µL of StartingBlock buffer and 500 µL of isolated pan 1 phage (step D5j). Wrap parafilm around the microcentrifuge tube lid to avoid any accidental spillage and incubate for 30 min at RT using a sample mixer.
Add an appropriate volume of biotinylated antigen to achieve 5 nM final concentration in 1.5 mL (which is achieved by step D6d). See General note 1. Wrap parafilm around the microcentrifuge tube lid to avoid any accidental spillage and incubate for 1 h at RT using a sample mixer.
At this point, one can start growing the log-phase TG1 cells (steps D1d–e), which would ensure that the cells are at the required OD600 once the phage has been eluted (step D6h).
Thirty minutes into the incubation of phage with antigen (step D6b), in a new 2 mL microcentrifuge tube, add 10 µL of streptavidin DynabeadsTM M-280. Wash the beads twice with 500 µL of PBS and then add 500 µL of StartingBlock buffer. Incubate for 30 min at RT using a sample mixer.
Transfer the 1 mL of blocked phage with antigen (step D6b) to the blocked beads (step D6d). The total volume is now 1.5 mL, and the final antigen concentration is now 5 nM. Wrap parafilm around the microcentrifuge tube lid to avoid any accidental spillage and incubate for 15 min at RT using a sample mixer.
Place the microcentrifuge tube on a DynaMagTM-2 magnet. Once the beads have been pulled to the side, discard the unbound phage in the supernatant. Wash away loosely bound phage by resuspending the beads in 500 µL of PBST and then using the magnet to pull the beads to the side. Repeat this washing process a total of six times with PBST and then once with PBS.
Add 500 µL of 250 µg/mL trypsin to the beads. Wrap parafilm around the microcentrifuge tube lid to avoid any accidental spillage and incubate for 30 min at RT using a sample mixer.
Collect the eluted phage from the second pan in the supernatant after using the DynaMagTM-2 magnet to pellet the beads.
Note: Work in a safety cabinet.
To obtain individual colonies for the master plate (step D7), titrate the eluted second pan phage (step D6h) using tenfold serial dilutions from 10-1 to 10-12 with log-phase TG1 cells (step D6c). After incubation at 37 °C for 15 min, spread 50 µL of the 10-4 to 10-12 dilutions on 1% agar LB plates containing 100 µg/mL ampicillin until dry and incubate overnight at 37 °C. Include a TG1 only control. See General note 15 and Troubleshooting 1.
Select the dilution that has approximately 100–300 individual colonies.
Preparation of master plates
Note: Work in a safety cabinet.
In a 1 mL deep-well block, add 180 µL/well of 2× YT containing 100 µg/mL ampicillin. Pick 93 individual clones from selected plate of titrated pan 2 phage in TG1 (step D6j) with 93 × 10 µL tips and place into each filled well. Remove the tips using a multichannel pipette. Leave wells F12, G12, and H12 without a colony, which serve as our controls.
Cover the plate with cell culture adhesive seal and incubate overnight at 37 °C shaking at 600 rpm. This overnight culture will be used to prepare two stock plates, the PCR template (step D7c) and the master plate (step D7d). Prepare the PCR plate first followed by the master plate.
Transfer 20 µL/well of overnight culture to a skirted PCR plate. Cover the plate with PCR foil seal and store at -80 °C. This is the PCR template plate.
To the remaining overnight culture, add 150 µL/well of 50% glycerol and mix by pipetting. Cover the plate with PCR foil seal and store at -80 °C. This is the master plate.
Anti-M13 phage ELISA
Note: Work in a safety cabinet.
In a 1 mL deep-well plate already filled with 150 µL/well of 2× YT, add 10 µL/well of the master plate (step D7d). Cover with cell culture adhesive seal and incubate for 3 h at 37 °C shaking at 600 rpm.
Prepare the CM13K helper phage mixture by adding 2 µL of CM13K helper phage stock (step D2h) to 10 mL of 2× YT. Add 30 µL/well of the prepared CM13K helper phage mixture and incubate for 1 h at 37 °C shaking at 600 rpm.
Pellet the cells at 2,800× g for 10 min at 18 °C and resuspend each well in 400 µL of 2× YT containing 100 μg/mL ampicillin and 25 μg/mL kanamycin.
Cover the plate with cell culture adhesive seal and incubate overnight at 25 °C shaking at 600 rpm.
After centrifugation at 4,000× g for 15 min at 4 °C, transfer the phage containing supernatant to a new 0.5 mL deep-well plate and cover with adhesive film. Store at 4 °C; this is stable for a month. This is the culture that is used for the anti-M13 ELISA (step D8l).
Note: Work in a lab bench.
Coat a 96-well ELISA microplate with 100 µL/well of 10 µg/mL neutrAvidinTM diluted in PBS and incubate overnight at 4 °C.
Using a plate washer, wash the plate five times with 300 µL/well of PBST.
Add 100 µL/well of 50 nM biotinylated target protein diluted in PBS and incubate for 1 h at RT on a microplate shaker. See General note 1.
Using a plate washer, wash the plate five times with 300 µL/well of PBST.
Add 250 µL/well of blocking solution and incubate for 1 h at RT on a microplate shaker.
Using a plate washer, wash the plate five times with 300 µL/well of PBST.
Add 50 µL/well of blocking buffer followed by 50 µL/well of the anti-M13 ELISA culture (step D8e). In the control wells F12, G12, and H12, add 50 µL/well of blocking buffer and 50 µL/well of PBS. Incubate for 1 h at RT on a microplate shaker.
Using a plate washer, wash the plate five times with 300 µL/well of PBST.
Add 100 µL/well of anti-M13-HRP diluted in 0.1% BSA-PBS (at a final concentration of 0.25 µg/mL) and incubate for 1 h at RT on a microplate shaker.
Using a plate washer, wash the plate five times with 300 µL/well of PBST.
Add 100 µL/well of ABTS substrate, which is prepared by mixing solution A and solution B in a 1:1 ratio. Protect from light and measure the absorbance at 405nm. See General note 2.
Wells with absorbances above 1 are considered hits. See Troubleshooting 2 and 3.
PCR and sequencing of ELISA hits
Prepare the following PCR reaction master mixture in a 5 mL tube and set up 96 PCR reactions in a 96-well rigid PCR plate (Table 12). See General note 17. Cover the plate with PCR foil seal.
Table 12. PCR reaction composition
Reagent Final concentration Volume for single reaction Volume for master mixture Nuclease-free water n/a 18.25 µL 1,825 µL 10 µM PhD seq Fwd primer 0.2 µM 0.5 µL 50 µL 10 µM PhD seq Rev primer 0.2 µM 0.5 µL 50 µL 10× Taq buffer 1× 2.5 µL 250 µL 25 mM dNTP 0.25 mM 0.25 µL 50 µL Taq polymerase 1 unit 0.5 µL 50 µL PCR template (step D7c) 2.5 µL - Total n/a 25 µL 2275 µL Set up the thermal cycler as below and perform the PCR (Table 13).
Table 13. PCR cycling conditions
Step Temperature (°C) Duration Number of cycles Denaturation 95 7 min s 1 Annealing 95 15 s 35 Extension 55 30 s Final extension 68 1 min 40 s Hold 68 5 min 1 Denaturation 12 Infinite hold - Add 45 µL of Highprep PCR magnetic beads/well and mix by pipette. Place on DynaMagTM 96-side magnet for 1 min or until the solution is clear and the beads have been pulled against the wall of the PCR tube and pipette off the supernatant.
Add 100 µL of 70% EtOH and mix by pipette. Place on DynaMagTM 96-side magnet for 1 min or until the solution is clear and the beads have been pulled against the wall of the PCR tube and pipette off the supernatant. Repeat this washing step a total of three times.
Leave beads to dry at RT for 10 min and then add 25 µL of EB buffer (from the QIAprep Spin Miniprep Kit) to the beads and mix by pipette. Place on DynaMagTM 96-side magnet for 1 min or until the solution is clear and the beads have been pulled against the wall of the PCR tube and transfer 20 µL of the purified PCR product containing supernatant to a new 96-well skirted PCR plate.
If a nanophotometer that can measure multiple samples at a time (such as the ImplenTM NanoPhotometer® N120) is available, measure the A280 of each well. If a single sample nanophotometer is available, measure the A280 of a random selection of clones. Expect around 15–25 ng/µL. Alternatively, 5 µL of purified PCR product of a random selection of clones can be mixed with 5 µL of 2× DNA gel loading buffer and run alongside a lane of 1× GeneRuler 1 kb DNA ladder on a 1% agarose gel containing 1× SYBRTM Safe DNA gel stain. Electrophorese in 1× TBE buffer at 80 V for 40 min. Image the gel and confirm the presence of amplified ~500 bp VHH genes.
Cover the plate with PCR foil seal and send for sequencing along with the PhD seq Fwd primer. See General note 18.
Small-scale expression and purification
The identification of nanobody clones that are expressed and purified well is achieved by performing a small-scale expression. The addition of a desalting step after purification allows for the confirmation of nanobody binding to the antigen using a titration ELISA or biolayer interferometry (BLI). The off-rate (kdis) of the nanobodies can be determined as well as preliminary epitope binning using BLI. These factors aid in the selection of which nanobodies will be taken forward for large scale expression.
Small-scale expression
Note: Work in a safety cabinet.
In a 1 mL deep-well plate already filled with 180 µL of 2× YT containing 100 µg/mL ampicillin per well, add 10 µL of selected clones based on sequencing results from the master plate (step D7d). Cover with cell culture adhesive seal and incubate overnight at 37 °C shaking at 600 rpm.
Add 10 µL of the overnight culture of each clone to a 24-well plate already filled with 4 mL/well of terrific broth containing 100 µg/mL ampicillin, 0.1% glucose, and 2 mM MgCl2. Cover with cell culture adhesive seal and incubate for 4 h at 37 °C, shaking at 600 rpm.
To each well, add 4 µL of 1 M IPTG (1 mM final concentration), cover with cell culture adhesive seal, and incubate overnight at 25 °C shaking at 600 rpm.
Note: Work in a lab bench.
Pellet the cells at 4,000× g for 15 min at 4 °C and gently resuspend the pellet in 300 µL of 1 mg/mL polymyxin B sulfate. Cover plate with adhesive film and agitate for 1 h at RT using a microplate shaker.
Pellet the cells at 4,000× g for 10 min at 4 °C and transfer the periplasm containing supernatant to a new 1.5 mL microcentrifuge tube.
Mix 10 µL of the isolated periplasm with 10 µL of 2× Laemmli sample buffer, incubate at 95 °C for 5 min, and run on a NuPage 4%–12% bis tris precast gel in 1× MES buffer alongside Mark12TM unstained standard at 200 V for 40 min.
Stain with InstantBlue® Coomassie protein stain. Expect prominent bands at approximately 14 kDa. Refer to the results from Expasy Protparam (Data analysis, step 1d) for the exact size of each clone.
Affinity purification
Note: Work in a lab bench.
Purification of the his-tagged nanobodies using Ni-NTA spin columns is performed according to the manufacturer’s instructions. Briefly, add 600 µL of equilibration buffer to the Ni-NTA spin column and centrifuge at 2,900 rpm (800× g) for 2 min at 4 °C.
Add the periplasm sample to the Ni-NTA spin column and centrifuge at 1,600 rpm (200× g) for 5 min at 4 °C.
Wash the Ni-NTA spin column three times by the addition of 600 µL of equilibration buffer and centrifuging at 2,900 rpm (800× g) for 2 min at 4 °C.
Transfer the Ni-NTA spin column into a new 2 mL microcentrifuge tube and add 100 µL of elution buffer. Incubate at RT for 2 min before centrifugation at 2,900 rpm (800× g) for 2 min at 4 °C. The flowthrough contains the purified nanobody.
Mix 5 µL of the purified nanobody with 5 µL of 2× Laemmli sample buffer, incubate at 95 °C for 5 min, and run on a NuPage 4%–12% bis tris precast gel in 1× MES buffer alongside Mark12TM unstained standard at 200 V for 40 min.
Stain with InstantBlue® Coomassie protein stain. Expect bands at approximately 14 kDa. Refer to the results from Expasy Protparam (Data analysis, step 1d) for the exact size of each clone.
Desalting
Note: Work in a lab bench.
Desalting of the Ni-NTA purified nanobodies using the ZebaTM Spin Desalting Columns is performed according to the manufacturer’s instructions. Briefly, break off the bottom cap of the desalting columns, loosen top cap, and place spin column in a new 2 mL microcentrifuge tube. Centrifuge at 1,500× g for 1 min at 4 °C.
Make a mark on the side of the desalting column where the resin is slanted upwards. Ensure that this mark is facing outwards in all centrifugation steps.
Wash the resin three times by gently adding 300 µL of PBS (or buffer of choice) to the resin bed and centrifuging at 1,500× g for 1 min at 4 °C.
Transfer the desalting column to a new 1.5 mL microcentrifuge tube and gently add 100 µL of purified nanobody (step E2d) to the centre of the resin bed. After the sample has been fully absorbed, gently add 15 µL of PBS (or buffer of choice) to the centre of the resin bed. Centrifuge at 1,500× g for 2 min at 4 °C and collect the desalted nanobody in the flowthrough.
Measure the A280 of the desalted nanobody and determine the molar concentration (µM = A280/ ϵ × 106) as well as the concentration (µg/mL = µM × kDa) using the kDa and extinction coefficient (ϵ) obtained from Expasy Protparam for the individual nanobodies (Data analysis, step 1d).
Titration ELISA
If an Octet R8 (or similar instrument) is not available, a titration ELISA will suffice to confirm binding of the purified nanobodies to the target protein.
Note: Work in a lab bench.
Coat a suitable number of wells of a 96-well ELISA microplate with 100 µL/well of 10 µg/mL neutrAvidinTM diluted in PBS and incubate overnight at 4 °C.
Using a plate washer, wash the plate five times with 300 µL/well of PBST.
Add 100 µL/well of 50 nM biotinylated target protein diluted in PBS and incubate for 1 h at RT on a microplate shaker. See General note 1.
Using a plate washer, wash the plate five times with 300 µL/well of PBST.
Add 250 µL/well of blocking solution and incubate for 1 h at RT on a microplate shaker.
Using a plate washer, wash the plate five times with 300 µL/well of PBST.
Add 200 µL of 25 µg/mL desalted nanobody (step E3d) diluted in blocking solution to the first well. Prepare a serial dilution in the plate, i.e., add 100 µL of blocking solution to the second well and add 100 µL of the solution from the first well. Prepare a total of seven dilutions (25–0.39 µg/mL), leaving the last well with blocking solution only. Incubate for 1 h at RT on a microplate shaker.
Using a plate washer, wash the plate five times with 300 µL/well of PBST.
Add 100 µL/well of anti-camelid VHH-HRP diluted 1:5,000 in 0.1% BSA-PBS and incubate for 1 h at RT on a on a microplate shaker.
Using a plate washer, wash the plate five times with 300 µL/well of PBST.
Add 100 µL/well of ABTS substrate, which is prepared by mixing solution A and solution B in a 1:1 ratio. Protect from light and measure the absorbance at 405 nm. See General note 2.
Off-rate (kdis) determination using Biolayer interferometry (BLI)
Note: Work in a lab bench.
Preheat the Octet R8 at 25 °C overnight or at least 3 h before starting the experiment.
Typical starting concentrations of antigen and nanobody are 50 nM and 100 nM, respectively, prepared in octet dilution buffer. See General note 19.
Prepare all the dilutions in a 96-well black polypropylene plate. See General note 20.
A single replicate per nanobody is sufficient at this stage. Include a no nanobody control to rule out any non-specific interactions with the loaded antigen and to serve as the background.
The experimental run conditions using SA biosensors are as follows:
1,000 rpm shaking for all incubation steps.
Plate definition: Octet dilution buffer to be used in all baseline steps/wells.
Assay definition: Baseline 600 s into octet dilution buffer.
: Loading 300 s into antigen diluted in octet dilution buffer.
: Baseline 30 s in octet dilution buffer.
: Baseline 60 s in octet dilution buffer.
: Association 300 s in desalted nanobody diluted in octet dilution buffer (watch this step, time might need to be extended as we want the curves to reach a plateau).
: Dissociation 300 s into column where baseline 60 s was measured (this step should be the same duration as the association step).
Subcloning of the nanobody into the bacterial and mammalian vectors
We have developed two vector suites for the functionalisation of nanobodies in both bacterial and mammalian expression systems. They have been designed in such a way that any nanobody that has been identified from a library that has been constructed as detailed in section C can be cloned into them using a common forward primer and tag-specific reverse primers as well as using the same restriction sites (Figure 8). The characteristics, tags for functionalisation, and primers for each vector in the bacterial and mammalian toolboxes are detailed in Table 14.
Figure 8. Details of the primers, restriction enzymes, and tags present in each of the vectors in the bacterial and mammalian toolboxes. Abbreviations used: VHH, nanobody; BAP, biotin acceptor peptide/Avi-TagTM; wt, wild-type; huIg1Fc, human Ig1 Fc.Table 14. Features of the vector in the E. coli and mammalian toolboxes. The bp within the primer that is complementary to the vector is indicated in italics.
Expression system Vector Tag Application Restriction enzymes Forward primer (5′–3′) Reverse primer (5′–3′, reverse complement) E. coli pOPINVHH_his His Crystallography NcoI and Bsu36I GCGGCCCAGCCGGCCATGGCCCAGGTGCAGCTGGTGGAG GTGATGGTGGCCTGAGGAGACGGTGACCTGGGTC pOPINVHH_flag_his His and FLAG Epitope detection pOPINVHH_myc_his His and myc Epitope detection pOPINVHH_BAP_his Avi-tagTM and His Biotinylation ATCATTCAAGCCTGAGGAGACGGTGACCTGGGTC pOPINVHH_cys_his Cys and His Chemical conjugation ATGGTGACAGCCTGAGGAGACGGTGACCTGGGTC pOPINVHH_sort_his Sortase Enzymatic labelling CGGCAGGCCGCCTGAGGAGACGGTGACCTGGGTC pOPINVHH_snap_his SNAP-tag® Enzymatic labelling GTCCTTGTCGCCTGAGGAGACGGTGACCTGGGTC Mammalian pOPINTTGneo-huIg1Fc huIg1Fc and His Fc fusions KpnI and PmeI GCGTAGCTGAAACCGGCCAGGTGCAGCTGGTGGAG CAGAACTTCCAGTTTAGGGGAGACGGTGACCTGGGTC pOPINTTGneo
huIg1FcA
(Y407T)huIg1Fc and His Bispecific nanobody generation pOPINTTGneo huIg1FcB
(T334Y)huIg1Fc and His Bispecific nanobody generation pOPINE-eGFP GFP and His Fluorescent fusions NcoI and PmeI AGGAGATATACCATGCAGGTGCAGCTGGTGGAG pOPINE-eYFP YFP and His Fluorescent fusions pOPINE-mCherry mCherry and His Fluorescent fusions Depending on the desired application, the selected nanobody is subcloned into a suitable expression vector. Below we detail the subcloning of a hit nanobody into the pOPINVHH_His vector as an example. The same process is followed to subclone the same nanobody into a mammalian vector for mammalian cell expression.
Restriction digestion of the pOPINVHH_His vector
Note: Work at a lab bench.
Prepare the following digestion mixture in a PCR tube (1 × 100 µL) (Table 15).
Table 15. Restriction digestion reaction composition
Reagent Final concentration Volume Nuclease-free water n/a 80.5 µL 10× rCutSmartTM buffer 1× 10 µL NcoI 2.5 units 2.5 µL Bsu36I 2.5 units 2.5 µL 400 ng/µL vector DNA 18 µg 4.5 µL Total n/a 100 µL Briefly vortex and centrifuge the reaction tube.
Incubate at 37 °C for 1 h and then at 80 °C for 20 min.
Add 4× volume of B3 buffer (400 µL) and purify using 1× Purelink column from the Purelink PCR purification kit according to the manufacturer’s instructions. Elute using 50 µL of E1 buffer. Measure the A280 and expect a concentration between 60 and 100 ng/µL.
Preparation of nanobody insert
Note: Work in a safety cabinet.
From the master plate (step D7d), add 10 µL of the selected nanobody clones to a 24-well plate already filled with 4 mL/well of 2× YT containing 100 µg/mL ampicillin, cover with cell culture adhesive seal, and incubate overnight at 37 °C, shaking at 600 rpm.
Note: Work at a lab bench.
Use 2 mL of the overnight culture to isolate the plasmid using the QIAprep spin miniprep kit according to the manufacturer’s protocol. Elute in a final volume of 50 µL of EB. Measure the A280 and expect a concentration between 100 and 500 ng/µL.
Send an aliquot for sequencing along with the PhD seq Fwd primer. If it matches the sequence obtained by PCR (step D9g), continue to the next step. See General note 18.
Prepare the following reaction in a PCR tube (1 × 25 µL) (Table 16).
Table 16. PCR reaction composition
Reagent Final concentration Volume Nuclease-free water n/a 9 µL 10 µM Common Fwd primer 0.5 µM 1.25 µL 10 µM His FLAG Rev primer 0.5 µM 1.25 µL 2× Phusion flash PCR master mix 1× 12.5 µL 10 ng/µL vector template (step F2b) 10 ng 1 µL Total n/a 25 µL Briefly vortex and centrifuge the reaction tube.
Set up the thermal cycler as below and perform the PCR (Table 17).
Table 17. PCR cycling conditions
Step Temperature (°C) Duration Number of cycles Denaturation 98 10 s 1 Annealing 98 1 s 30 Extension 60 1 s Final extension 72 5 s Hold 72 1 min 1 Denaturation 12 Infinite hold - Add 5 µL of 6× DNA gel loading buffer to the reaction tube and run alongside a well of 1× GeneRuler 1 kb DNA ladder on a 1% agarose gel containing 1× SYBRTM safe DNA gel stain. Electrophorese in 1× TBE buffer at 80 V for 40 min.
Visualise the ~500 bp amplified nanobody band using a LED Blue transilluminator, excise from the gel using a scalpel knife, and gel extract using one spin column from the Nucleospin Gel and PCR clean-up kit according to the manufacturer’s protocol. Elute using 50 µL of NE, measure the A280, and expect a concentration between 30 and 100 ng/µL.
Caution: Use Safe ImagerTM viewing glasses when visualising the DNA during excision of the band from the agarose gel and use caution when using scalpel knives.
Ligation and transformation
Note: Work at a lab bench.
Prepare the following reaction in a PCR tube (1 × 10 µL) (Table 18).
Table 18. In-Fusion reaction composition
Reagent Final concentration Volume Nuclease-free water n/a 5 µL 5× CEII buffer 1× 2 µL 4 ng/µL insert (step F2h) 4 ng 1 µL 19.83 ng/µL vector DNA (step F1d) 19.83 ng 1 µL Exnase II 1 µL Total n/a 10 µL Briefly vortex and centrifuge the reaction tube.
Incubate at 42 °C for 30 min using a thermal cycler. Add 40 µL of TE buffer to terminate the reaction.
Note: Work in a safety cabinet.
Gently mix 5 µL of in-fusion reaction with 25 µL of StellarTM competent cells in a sterile 2 mL microcentrifuge tube. Incubate on ice for 10 min.
Heat shock at 42 °C for 45 s and incubate on ice for 2 min. Add 200 µL of LB medium and incubate for 1 h at 37 °C agitating at 600 rpm.
Spread 50 µL of culture on a 1% agar LB plate containing 100 µg/mL ampicillin, 2 mM IPTG, and 40 µg/mL X-gal until dry. Incubate overnight at 37 °C. Expect many white colonies and few-to-no blue colonies.
Transfer two white colonies from the transformation plate to two wells of a 24-well plate already filled with 4 mL/well of 2× YT containing 100 µg/mL ampicillin. Cover with cell culture adhesive seal and incubate overnight at 37 °C, shaking at 600 rpm.
Note: Work at a lab bench.
Use 2 mL of the overnight culture to isolate the plasmid using the QIAprep spin miniprep kit according to the manufacturer’s protocol. Elute in a final volume of 50 µL of EB. Measure the A280 and expect a concentration between 100 and 500 ng/µL.
Send an aliquot for sequencing along with the PhD seq Fwd primer. If it matches the sequence obtained by PCR (step D9g), continue to the next step. See General note 18.
Proceed to large-scale expression using WK6 E. coli or Expi293TM as detailed in Le Bas et al. [12] or use in imaging experiments using other mammalian cell lines (see section G) depending on the expression vector that was selected.
Nanobody functionalisation and application
By using a selection of vectors from the E. coli and mammalian toolboxes, we demonstrate how nanobodies can be used as a reagent for pull downs using streptavidin and the pOPINVHH_BAP vector, for imaging by chemical conjugation using the pOPINVHH_Cys vector and by transfection with the pOPINE-GFP and pOPINE-YFP for in vivo and live cell imaging.
Confirmation of biotinylation of anti-GFP nanobody
The anti-GFP nanobody was cloned into the pOPINVHH_BAP vector, expressed at large scale in BL21(DE3)-R3-pRARE2-BirA to allow for in vivo biotinylation, and purified using Ni-NTA and size exclusion chromatography as detailed in section F and in Le Bas et al. [12] with some modifications. Briefly, LB agar plates contained 34 µg/mL chloramphenicol, 50 µg/mL spectinomycin, and 50 µg/mL carbenicillin, cultures of terrific broth were supplemented with 50 µg/mL spectinomycin and 50 µg/mL carbenicillin, and a final concentration of 0.2 mM biotin was added alongside 0.1 mM IPTG. Below, we detail how we confirm that the anti-GFP nanobody has been biotinylated.
Note: Work at a lab bench.
Add 2 µg of anti-GFP expressed in pOPINVHH_BAP or pOPINVHH_His and 1.5 µL of 50 µg/mL Streptavidin Alexa FluorTM 488 conjugate to a 1.5 mL microcentrifuge tube and make up the volume to 10 µL with PBS. Incubate for 30 min at 37 °C.
Add 10 µL of 2× Laemmli sample buffer, incubate at 95 °C for 5 min, and run on a NuPage 4%–12% bis tris precast gel in 1× MES buffer alongside Mark12TM unstained standard and BenchMarkTM fluorescent protein standard at 200 V for 40 min.
Image the in-gel fluorescence using the ProQ emerald 488 image acquisition preset on the ChemiDocTM imaging system (blue Epi excitation, 532/28 emission filter). Stain with InstantBlue® Coomassie protein stain (Figure 9).
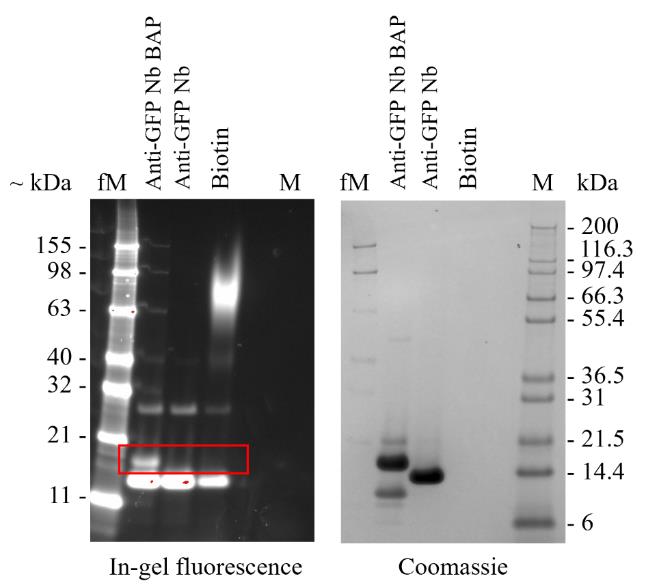
Figure 9. Confirmation of nanobody biotinylation after expression in pOPINVHH_BAP and co-expression with BirA by analysis on SDS-PAGE. Samples were visualised by in-gel fluorescence and stained with InstantBlue® (Coomassie). Protein molecular weight markers in kDa were run in parallel.
Biotin-mediated pull downs
Below, we detail the utilisation of purified biotinylated anti-GFP nanobody in the purification of GFP from spiked E. coli lysate.
Note: Work at a lab bench.
Wash 100 µL of streptavidin DynabeadsTM M-280 twice with 1 mL of PBS.
Add 100 µL of 10 µg biotinylated anti-GFP nanobody diluted in PBS to the prepared beads and incubate for 30 min at RT using a sample mixer.
Collect the unbound sample and wash the beads three times with 1 mL of PBS.
Add 10 µg of pure GFP to E. coli lysate and make up to a final volume of 100 µL using PBS. Add this to the washed beads and incubate for 30 min at RT using a sample mixer.
Collect the unbound sample and wash the beads three times with 1 mL of PBS.
Add 20 µL of 2× Laemmli sample buffer to the beads.
To 10 µL of each loaded and unbound sample, add 10 µL of 2× Laemmli sample buffer.
Incubate all Laemmli-treated samples at 95 °C for 5 min and run on a NuPage 4%–12% bis tris precast gel in 1× MES buffer alongside Mark12TM unstained standard at 200 V for 40 min.
Stain with InstantBlue® Coomassie protein stain (Figure 12A).
Fluorophore conjugation
The anti-vimentin (VB3) nanobody was cloned into the pOPINVHH_Cys vector, expressed at large scale in WK 6 E. coli, and purified using Ni-NTA and size exclusion chromatography as detailed in section F. Below, we detail how to prepare fluorescently labelled VB3 nanobody (VB3-AF647) and its application in in vivo imaging.
Note: Work at a lab bench.
To 300 µg of purified VB3 nanobody, add 1 mM TCEP pH 8.0 to make up to a final volume of 100 µL and incubate for 20 min at 4 °C using a sample mixer.
Add 1 µL of Alexa FluorTM 647 C2-maleimide to the VB3 nanobody solution and incubate for 1 h at 4 °C using a sample mixer.
Remove any unbound Alexa FluorTM 647 from the fluorescently labelled VB3 nanobody using a ZebaTM dye and biotin removal spin column as per the manufacturer’s protocol. This sample is stored at 4 °C for two weeks protected from light but can be flash frozen and stored at -80 °C for an extended period.
Mix 10 µL of the VB3-AF647 with 10 µL of 2× Laemmli sample buffer, incubate at 95 °C for 5 min, and run on a NuPage 4%–12% bis tris precast gel in 1× MES buffer alongside PageRulerTM prestained protein ladder at 200 V for 40 min.
Image the in-gel fluorescence using the Oriele image acquisition preset on the ChemiDocTM imaging system (UV excitation, 590/110 emission filter). Stain with InstantBlue® Coomassie protein stain (Figure 10).
Measure the absorbance of the VB3-AF647 from 200 to 800 nm with baseline correction at 750 nm using the Nanodrop. Expect a peak at 280 and 651 nm, which correspond to VB3 nanobody and to Alexa FluorTM 647, respectively (Figure 10).
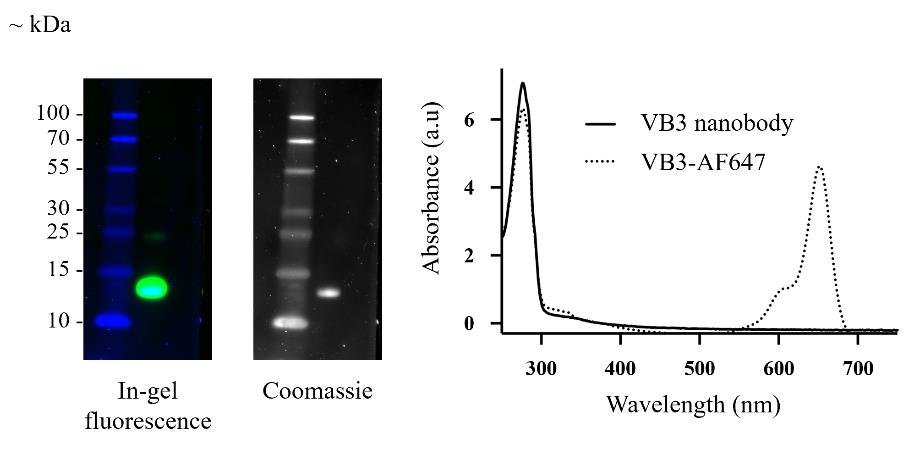
Figure 10. Conjugation of Alexa FluorTM 647 to VB3 nanobody (VB3-AF647) expressed in pOPINVHH_Cys as determined by in-gel fluorescence (left), InstantBlue® staining (middle), and absorbance profile (right)
In vivo visualisation
Note: Work in a safety cabinet.
Place a single coverslip into the wells of a 6-well culture plate. Seed 1 mL of 10,000 HeLa cells/well and culture in phenol red–free DMEM medium supplemented with 10% FBS, 1× GlutaMAXTM, and 1× penicillin-streptomycin at 37 °C and 5% CO2 in a CO2 incubator.
Note: Work at a lab bench.
After 24 h, wash the cells three times with 2 mL of PBS and fix with 2 mL of 4% paraformaldehyde for 20 min at RT.
Wash the cells three times with 2 mL of PBS and block the cells with 2 mL of confocal blocking buffer for 1 h at RT.
During the blocking step, prepare the fluorescently labelled VB3 nanobody solution for labelling vimentin in HeLa cells. In a 1.5 mL microcentrifuge tube, add 5 µL of VB3-AF647 (step G3c) to 1 mL of confocal dilution buffer PBS and mix gently by inversion.
Remove the blocking buffer from the cells, add the prepared VB3-AF647 (step G4d), and incubate for 2 h at 4 °C using a gyratory rocker.
Remove the VB3-AF647 solution and wash the cells three times with 2 mL of PBS.
For each coverslip, add a small drop of Fluoroshield mounting media with DAPI onto a microscope slide. Using Dumont tweezers, gently remove the coverslip from the well and place cell side down onto the mounting media. Remove excess liquid by gently pressing the coverslip slide down towards the microscope slide and secure the coverslip by adding nail varnish around the perimeter of the coverslip.
The stained cells on the prepared slides are imaged using a Leica SP8 confocal microscope at 63× objective with type F immersion liquid. Use Ex405 and Em430–550nm for DAPI detection and Ex633 and Em650 for Alexa FluorTM 647 detection.
Live cell imaging
The VB3 nanobody was cloned into the pOPINE-eGFP and -eYFP vectors as detailed in section F. The produced vectors are referred to as pOPINE-VB3-eGFP and pOPINE-VB3-eYFP. Below, we detail how to transiently transfect HeLa cells with the pOPINE-VB3-eGFP and pOPINE-VB3-eYFP vectors for live-cell imaging of vimentin filaments.
Note: Work in a safety cabinet.
In an 8-well chambered coverslip, seed 200 µL containing 25,000 HeLa cells/well and culture in phenol red–free DMEM medium supplemented with 10% FBS, 1× GlutaMAXTM, and 1× penicillin-streptomycin at 37 °C and 5% CO2 in a CO2 incubator.
After 24 h, prepare the transfection mixture as follows. In a 500 µL microcentrifuge tube, add 10 µL of serum-free DMEM medium and gently mix in 2 µL of 100 ng/µL pOPINE-VB3-eGFP and pOPINE-VB3-eYFP vectors. Add 0.5 µL of Fugene transfection reagent and incubate for 10 min at RT.
Remove the old media from each of the wells in the 8-well microscope chambered coverslip and add 270 µL/well of DMEM medium supplemented with 10% FBS. Add each DNA-Fugene complex dropwise, by gently depressing the plunger of the pipette, into each well and incubate the chambered coverslip at 37 °C and 5% CO2 in a CO2 incubator for 48 h.
Live-cell imaging is captured using a Leica SP8 confocal microscope at 63× objective with immersion liquid. Use Ex405 and Em430–550nm for DAPI detection and Ex488 and Em518-558 for GFP and YFP detection.
Data analysis
Analysis of sequencing data (section D9)
Drag and drop the .seq files into IMGT/V-QUEST. Make the following selections on the site: Species: Vicugna Pacos (alpaca); Receptor type or locus: Ig; choose to get the results in excel format. Submit. See General note 21.
There are multiple tabs in the produced Excel file. The fifth tab (AA-sequences) is the one that we will work in. Sort the sequences by productive, non-productive, and no results.
Then, sort the CDR3-IMGT of the productive sequences to visualise the CDR3 clusters. Highlighting the different CDR3 clusters in different colour aids in the selection of representative clones, which will be used for small-scale expression.
Open the .seq files of the selected clones in SnapGene to obtain the amino acid sequences. Copy the single letter amino acid sequence and paste into Expasy - ProtParam tool. Delete the pelB sequence (MKYLLPTAAAGLLLLAAQPAMA), submit, and note the expected kDa, pI, and extinction coefficient (ϵ), assuming that all Cys residues are reduced.
Analysis of BLI data for off-rate determination (section E5)
In the Preprocess data tab: Subtract the background of the no nanobody well to all the other tested nanobody wells. Correct the Y axis to the baseline step and the interstep to the dissociation step. Apply the Savitzky-Golay filtering.
In the Kinetic analysis tab: Analyse both the association and dissociation steps. Use a 1:1 binding model. Use a local (individual) fitting. Fit the full association and dissociation steps and note the calculated off-rate (kdis).
Validation of protocol
Identification of nanobodies to the G protein of Nipah virus
The improved workflow for nanobody generation was exemplified by the identification of nanobodies to the receptor-binding G protein of the Nipah virus (NivG). The extracellular region of the NivG protein (residues 183–602) with N-terminal hexahistidine tag was expressed in Expi293TM cells and purified by a combination of immobilised metal affinity chromatography (IMAC) and size exclusion (Figure 11A). Comparison of pre- and post-immune sera in a seroconversion ELISA of biotin-tagged NivG confirmed that an immune response had been generated to the antigen (Figure 11B). Following two rounds of panning of the VHH library 13, 93 phage clones were picked and tested in a phage ELISA. The sequences of phage binders were amplified by PCR and sequenced and the translated VHHs were clustered by CDR3 sequence identity. A representative clone from each of the three major clusters was selected, expressed at small scale (4 mL cultures), and purified by IMAC (panel C). Binding to NivG was confirmed by titration ELISA (panel D). Analysis of binding off-rates by BLI confirmed that the three clones (A8, B7, and D5) bound to NivG (panel E) and were prioritised for large-scale expression.
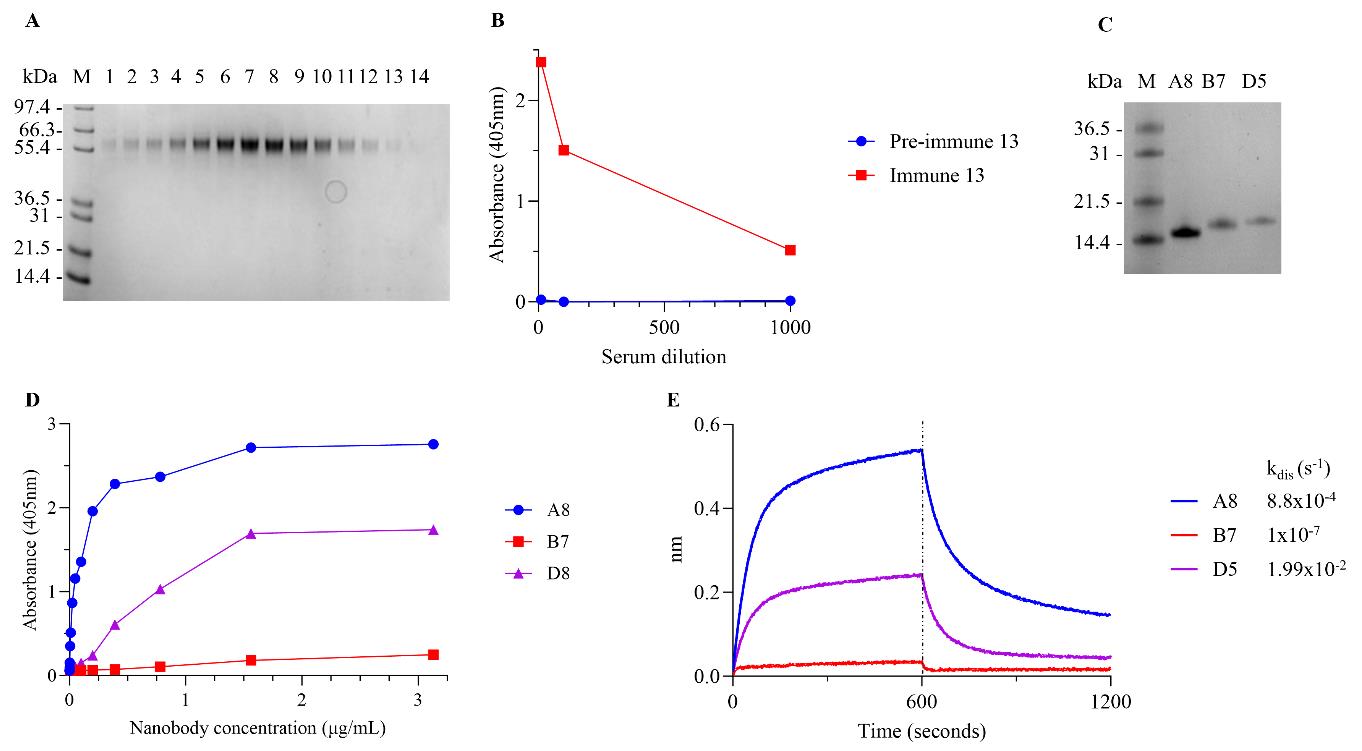
Figure 11. Generation of NivG-binding nanobodies from an immunised llama. (A) SDS-PAGE of fractions from gel filtration of purified NivG, (B) seroconversion ELISA of pre- and post-immune sera following immunisation with NivG, (C) SDS-PAGE of purified anti-NiVG nanobodies from small-scale expression, (D) binding of purified nanobodies to NivG 183-602 by titration ELISA, (E) BLI traces of nanobody association to and dissociation from NivG 183-602; off-rates for the three nanobodies are shown.
Functionalisation of nanobodies for purification and imaging
The functionalisation of nanobodies using three of the vectors presented in Table 1 and their application was exemplified using an anti-GFP [9] and an anti-vimentin (VB3) [10] nanobody. The anti-GFP nanobody was expressed in the pOPINVHH_BAP vector, biotinylated in vivo, and used for pull-down of GFP from spiked E. coli lysate with magnetic-coupled streptavidin beads (Figure 12A). The anti-VB3 nanobody was expressed in E. coli using the pOPINVHH-cys vector, purified, and coupled to AlexaFluorTM 647 via a maleimide linker. HeLa cells were fixed and permeabilised and the vimentin filaments were stained with the anti-VB3 nanobody conjugate (Figure 12B). The same nanobody was genetically fused to either YFP or GFP by cloning into pOPINE-YFP and pOPINE-GFP vectors, respectively. Live HeLa cells were transfected with these vectors and imaged by confocal microscopy to visualise vimentin filaments (Figure 12C).
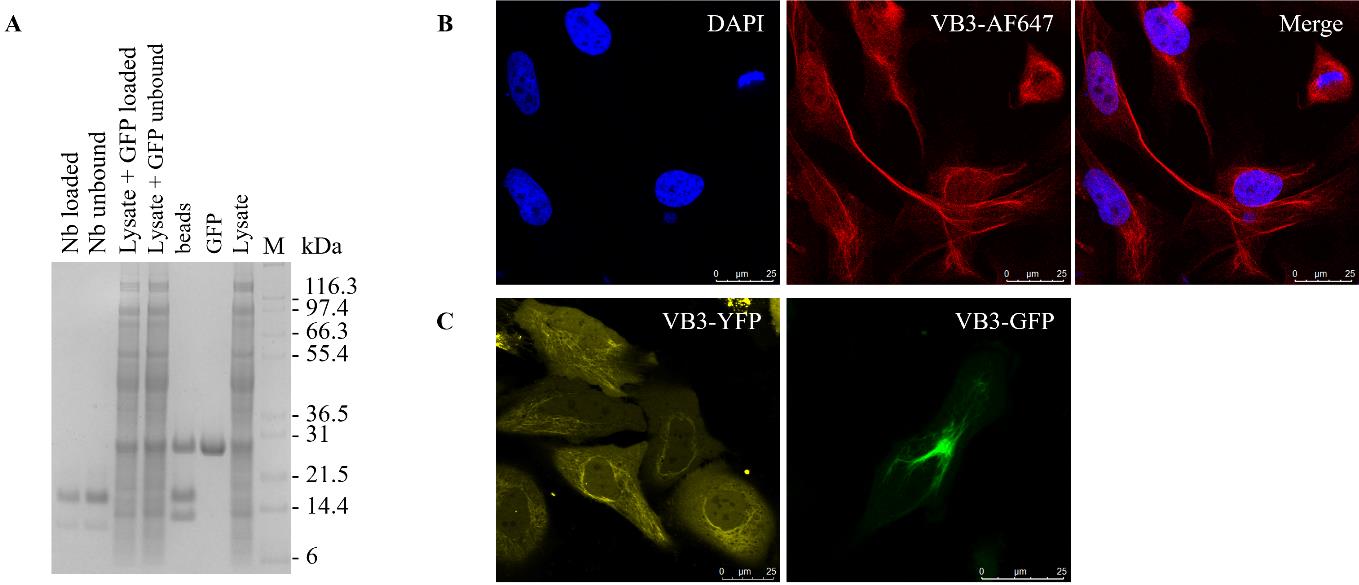
Figure 12. Functionalisation of nanobodies. (A) Pull-down of GFP spiked into an E. coli lysate by biotinylated anti-GFP nanobody coupled to streptavidin magnetic beads. Detection of vimentin in HeLa cells using (B) fluorescently labelled anti-vimentin nanobody (VB3-AF647) and (C) transiently transfected with VB3-eGFP and VB3-eYFP.
General notes and troubleshooting
General notes
Preparation of biotinylated proteins can be achieved in two ways: chemical conjugation or by in vitro biotinylation of a protein with the Avi-tag® sequence and co-expression with BirA.
Expect colour development within 5–15 min of substrate addition. If signals are low, the longest incubation before absorbance measurement is 1 h.
If we receive 170 mL of llama blood in 17 × 10 mL blood tubes without anticoagulant, the addition of an equal volume of PBS will result in a total of 340 mL of diluted blood. Fifteen millilitres of diluted blood are required for each 50 mL tube with 15 mL Histopaque®-1007 already added; therefore, a total of 22.6 tubes are required. As such, the 23rd tube will have less than 15 mL of diluted blood.
Holding the 50 mL conical centrifuge tube at a 45° angle allows for the gentle addition of the diluted blood over the Histopaque®-1007 to prevent the disturbance of the Histopaque®-1007 layer. During the process of gently layering the blood on top of the Histopaque®-1007 over the many 50 mL conical centrifuge tubes, the lower layer will turn red as the heavier red blood cells fall to the bottom of the tube. This is to be expected.
Mixing the trypan blue with the PMNCs too vigorously may damage the PMNCs, resulting in low viability values that may not be a true representation of the actual viability.
If no PCR workstation or dedicated RNase free space is available, keeping boxes of tips and bags microcentrifuge tubes that are used only for RNA work will be sufficient if gloves are changed regularly and RNaseZapTM is used.
RNA should be stored at -20 °C under ethanol where it is stable for extended periods of time. Once it is resuspended in water, the stability is reduced and, as such, for the best possible results, we recommend that cDNA synthesis is carried as soon as RNA has been resuspended in water (step C3i).
For the electroporations, expect ms values greater than 4.5.
Note that the estimated library sizes for the small-scale library will vary slightly from that of the scaled library.
An example on how to calculate how many microlitres of in-fusion reactions would be needed to make a library with a size of 1 × 106: We purify the 10 µL in-fusion reaction using a Purelink column, eluting in 20 µL nuclease-free water. We use 5 µL of this for the electroporation, which translates to 2.5 µL of the original in-fusion reaction being added to 30 µL of competent TG1. Hence, if 2.5 µL of in-fusion library yielded 373 CFU at 1:100 dilution, the small-scale library size would be 1.492 × 105 CFU/mL. To create a 1 × 106 CFU/mL library, 6.7 × 2.5 µL of in-fusion reactions would be required, which is just over 1 × 10 µL of in-fusion reaction. If enough VHH2 and digested pADL23c backbone has been prepared, we routinely use more in-fusion reactions than necessary, i.e., 10 × 10 µL, to ensure that we have the best chance of producing a 1 × 106 CFU/mL library.
If there is a large discrepancy between the library size as calculated by titre and by plate density, use the plate density to guide your decision if the library is big enough. See Figure 7.
Begin by using 200 µL of the TG1 stock when preparing the starting culture.
Begin by using 20 µL of the isolated library phage and pan 1 sub-library stock when preparing the starting culture.
In some cases, it might take longer than 2 h for the culture to reach an OD600 of 0.5. Continue culturing until the required OD600 is reached. As long as the TG1 control plate in either step D3k or D6i are negative, you can be assured that this might just be variance in the library and/or sub-library.
A TG1-only control functions to show that the TG1s used have not been infected with phage prior to this titration step. See Troubleshooting 1 and 2.
Steps C8h–i show how diverse the library is. The titration of the amplified and recovered library (step D3l) gives an indication of how many phage particles we can make using our diverse library.
Adjust the volume of the prepared PCR master mixture according to the number of actual ELISA positive clones.
Volumes and concentrations of the DNA and primers might be supplier specific. Check the requirements of your local sequencing company.
Use enough antigen to load the biosensors to result in a 1 nm signal increase.
Polypropylene plates must be used as they do not bind protein.
The IMGT site can only process 48 sequences at a time. If more than 48 clones have been sequenced, combine the two generated IMGT Excel files together before sorting by V-DOMAIN functionality and CDR3-IMGT.
Troubleshooting
Problem 1: Growth on TG1 control plates.
Possible cause: Indication of phage contamination of the E. coli before our isolated/eluted phage was added.
Solution: Prepare fresh log-phase TG1 from fresh aliquot of TG1 in the -80 °C. Repeat the titration of the phage in question.
Problem 2: Growth on TG1 control plates.
Possible cause: Ampicillin stock is old.
Solution: Using a new ampicillin aliquot, prepare fresh 1% agar LB plates with ampicillin. Take a colony from the “contaminated” TG1 control plate as well as the glycerol stock used to grow the log-phase TG1 and streak it on the new plates. If no growth is observed, this indicates that the ampicillin stock is old and that the TG1 was not infected with phage prior to this.
Problem 3: Only a single hit is found on the anti-M13 ELISA plate.
Possible cause: Only a few binders are present after the rounds of panning.
Solution: Another 93 colonies can be picked to prepare a second master plate to test in an anti-M13 ELISA.
Problem 4: None or very few anti-M13 ELISA hits present after the second round of panning using 5 nM of target protein.
Possible cause: Might not have enrichment.
Solution: Go back to 50 nM phage and pan using 10 nM antigen. If there are still not hits, try Troubleshooting 3. If there are still no hits, consider looking at another library. Use the seroconversion ELISA data to guide this decision.
Acknowledgments
This work was supported by the Rosalind Franklin Institute, with funding delivery partner the Engineering and Physical Sciences Research Council UK (EPSRC) and grants from the Biotechnology and Biological Sciences Research Council UK (BBSRC) for nanobody discovery (ref. BB/V018523/1) and the Wellcome Trust for technology development (ref. 223733/Z/21/Z). Graphical overview created with BioRender.
Competing interests
The authors declare that they have no competing interests with respect to the work described.
Ethical considerations
Immunisations and handling of the llamas were performed under the authority of the UK Home Office project license PA1FB163A.
References
- Hamers-Casterman, C., Atarhouch, T., Muyldermans, S., Robinson, G., Hammers, C., Songa, E. B., Bendahman, N. and Hammers, R. (1993). Naturally occurring antibodies devoid of light chains. Nature 363(6428): 446–448.
- Bao, G., Tang, M., Zhao, J. and Zhu, X. (2021). Nanobody: a promising toolkit for molecular imaging and disease therapy. EJNMMI Res. 11(1): 6.
- Muyldermans, S. (2020). A guide to: generation and design of nanobodies. FEBS J. 288(7): 2084–2102.
- Parker, J. L., Deme, J. C., Wu, Z., Kuteyi, G., Huo, J., Owens, R. J., Biggin, P. C., Lea, S. M. and Newstead, S. (2021). Cryo-EM structure of PepT2 reveals structural basis for proton-coupled peptide and prodrug transport in mammals. Sci. Adv. 7(35): eabh3355.
- Girt, G. C., Lakshminarayanan, A., Huo, J., Dormon, J., Norman, C., Afrough, B., Harding, A., James, W., Owens, R. J., Naismith, J. H., et al. (2021). The use of nanobodies in a sensitive ELISA test for SARS-CoV-2 Spike 1 protein. R. Soc. Open Sci. 8(9): e211016.
- Huo, J., Mikolajek, H., Le Bas, A., Clark, J. J., Sharma, P., Kipar, A., Dormon, J., Norman, C., Weckener, M., Clare, D. K., et al. (2021). A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19. Nat. Commun. 12(1): 5469.
- Akkermans, O., Delloye-Bourgeois, C., Peregrina, C., Carrasquero-Ordaz, M., Kokolaki, M., Berbeira-Santana, M., Chavent, M., Reynaud, F., Raj, R., Agirre, J., et al. (2022). GPC3-Unc5 receptor complex structure and role in cell migration. Cell 185(21): 3931–3949.e26.
- Pardon, E., Laeremans, T., Triest, S., Rasmussen, S. G. F., Wohlkönig, A., Ruf, A., Muyldermans, S., Hol, W. G. J., Kobilka, B. K., Steyaert, J., et al. (2014). A general protocol for the generation of Nanobodies for structural biology. Nat. Protoc. 9(3): 674–693.
- Kirchhofer, A., Helma, J., Schmidthals, K., Frauer, C., Cui, S., Karcher, A., Pellis, M., Muyldermans, S., Casas-Delucchi, C. S., Cardoso, M. C., et al. (2010). Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol. 17(1): 133–138.
- Maier, J., Traenkle, B. and Rothbauer, U. (2015). Real-time analysis of epithelial-mesenchymal transition using fluorescent single-domain antibodies. Sci. Rep. 5(1): 13402.
- Tonikian, R., Zhang, Y., Boone, C. and Sidhu, S. S. (2007). Identifying specificity profiles for peptide recognition modules from phage-displayed peptide libraries. Nat. Protoc. 2(6): 1368–1386.
- Le Bas, A., Mikolajek, H., Huo, J., Norman, C., Dormon, J., Naismith, J. and Owens, R. (2022). Production and Crystallization of Nanobodies in Complex with the Receptor Binding Domain of the SARS-CoV-2 Spike Protein. Bio Protoc. 12(9): e4406.
Article Information
Copyright
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Eyssen, L. E. A., Ramadurai, S., Abdelkarim, S., Buckle, I., Cornish, K., Lin, H., Jones, A. K., Stephens, G. J. and Owens, R. J. (2024). From Llama to Nanobody: A Streamlined Workflow for the Generation of Functionalised VHHs. Bio-protocol 14(6): e4962. DOI: 10.21769/BioProtoc.4962.
Category
Biological Engineering
Biochemistry > Protein > Expression
Microbiology > Microbial biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link







