- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Human Skin Explant Preparation and Culture
Published: Vol 12, Iss 18, Sep 20, 2022 DOI: 10.21769/BioProtoc.4514 Views: 3926
Reviewed by: Giusy TornilloDongsheng JiangGiovanna Piovani

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Ex vivo Culture and Lentiviral Transduction of Benign Prostatic Hyperplasia (BPH) Samples
Urszula Lucja McClurg [...] Olivier Binda
Nov 5, 2018 6158 Views

Method for Prolonged Incubation of Brain Slices
Orsolya Kekesi and Yossi Buskila
Jul 20, 2020 5205 Views

In-vitro GLP-1 Release Assay Using STC-1 Cells
Liu Qi [...] Sifuentes-Dominguez Luis
Aug 20, 2020 6288 Views
Abstract
The ex vivo experimentation with surgically discarded human skin represents a unique methodology amenable for mechanism and pharmacologic agent studies without the involvement of human subjects. Here, we describe a protocol that includes preparation, culture, and stimulation of human skin explants, and subsequent analyses by quantitative reverse transcription PCR and immunostaining. This protocol may also be applied for ex vivo studies of murine skin, reducing animal numbers and potentially harmful treatments. In our hands, this protocol has been used for wound healing, viral infection, and hair growth–related studies.
Graphical abstract:
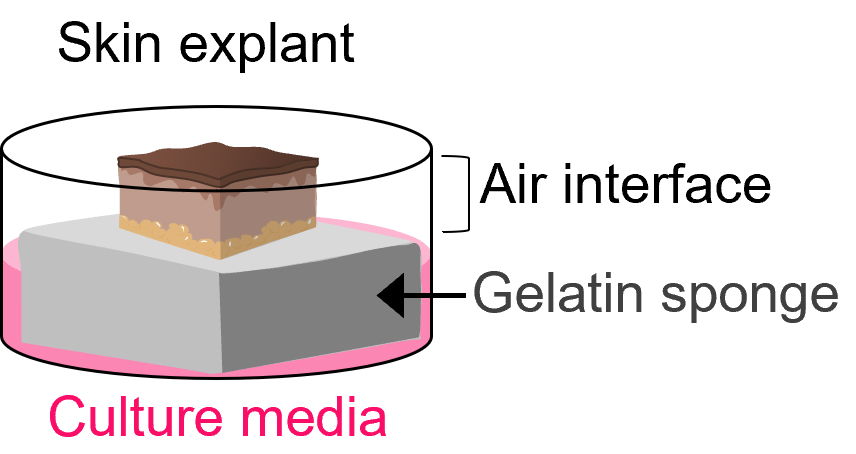
Cartoon of explant skin culture. Skin explant sits on top of a gelatin surgical sponge saturated with culture medium at an air–liquid interface.
Background
Genetic animal models are powerful in recapitulating complex tissue environment with intact circulation. Mice are by far the most frequently used animal for gene function, disease modeling, and pharmacological studies. However, murine skin and human skin are architecturally different and show differences in their responses to injury (Zomer and Trentin, 2018). It is not surprising that findings using murine skin often need corroboration in human skin models that harbor tissue-resident immune cells (Zomer and Trentin, 2018).
Significant technical advances have been achieved in epidermal keratinocyte culture in vitro, allowing procurement and expansion of primary keratinocytes. However, monolayer cell culture does not always recapitulate in vivo processes and often lacks tissue-resident immune cells. The organotypic cell culture model generated with primary or immortalized keratinocytes allows three-dimensional epidermal stratification and incorporation of certain dermal cells such as fibroblasts (Smits et al., 2017). Each of these methods has limitations in recapitulating skin biology.
Our lab has adapted a protocol for using surgically discarded human skin samples for ex vivo studies (Shannon et al., 2022), such as wound assays, cytokine stimulation, and viral infections. The protocol described below includes necessary materials, reagents, equipment, and procedures.
Materials and Reagents
1.7 mL microtube (Genesee Scientific, catalog number: 22-281LR)
SURGIFOAM® absorbable gelatin sponge (Ethicon, catalog number: 1969)
12-well cell culture plates, flat bottom wells, TC-treated (GenClone 25-106)
Sterile 1× phosphate buffered saline (PBS) (Gibco, catalog number: 14190144)
Penicillin–streptomycin–glutamine (100×) (Gibco, catalog number: 10378016)
EpiLifeTM CF kit (Gibco, catalog number: MEPICF500)
Note: This kit contains 500 mL of EpiLifeTM calcium-free media and 500 μL of 0.06 M CaCl2 (see Recipes).
Human keratinocyte growth supplement (HKGS, 100×) (Gibco, catalog number: S0015)
Adwin scientific Tissue-Tek Cryomold, intermediate (Fisher Scientific, catalog number: NC9511236)
Tissue-Tek optimum cutting temperature (OCT) compound (VWR, catalog number: 25608-930)
Rat anti-human Integrin α6 CD49f antibody (BioLegend, catalog number: 313602, diluted 1:500)
Alexa Fluor 647-conjugated goat anti-rat secondary antibody (BioLegend, catalog number: 313609). Diluted 1:400
Prolong Gold antifade reagent (ThermoFisher Scientific, catalog number: P36930)
TRIzolTM reagent (ThermoFisher Scientific, catalog number: 15596026)
Hoechst 33342, trihydrochloride, trihydrate (ThermoFisher Scientific, catalog number: H3570)
iScripTM cDNA synthesis kit (Bio-Rad, catalog number: 1708891)
Culture medium (see Recipes)
Wash buffer (see Recipes)
70% ethanol (see Recipes)
Equipment
CO2 incubator (Panasonic, model: MCO-170AICUVL-PA)
Technocut® disposable scalpels, sterile, MYCO Medical, No. 20 (VWR, catalog number: 10148-894)
Forceps (VWR, catalog number: 89259-946)
Surgical scissors (VWR, catalog number: 76192-134)
Pipettes (ThermoFisher Scientific, model: FinnpipetteTM F2 variable volume pipettes, catalog number: 4701070)
Olympus IX73 fluorescent microscope (Olympus Corporation Microscopy Technologies)
Nanodrop 1000
-80 °C freezer
Leica Cryocut 1800
Procedure
Solution Preparation
Note: The following steps should be completed in a biosafety cabinet with laminar flow to maintain sterility. Wipe all containers and reagents with 70% ethanol prior to placement in the laminar flow tissue culture hood and use sterile technique during all steps.
Wash buffer (see Recipe 1)
Prepare wash buffer by diluting 100× penicillin–streptomycin–glutamine (100×) 1:100 in 1× PBS.
Culture medium (see Recipe 2)
Skin Preparation
Note: Surgically excised skin should be maintained at 4 °C and used as soon as possible after removal.
Remove subcutaneous fat from skin using forceps and wash with PBS containing 1× concentration of penicillin–streptomycin–glutamine (Shannon et al., 2022).
Use a scalpel to carefully trim off laser-cut edges from surgical excised skin sample.
Prepare 12-well culture plates with 0.5–1.0 mL of EpiLifeTM CF media (supplement free) with 2 cm × 2 cm surgical gelatin sponge. Allow foam to absorb media (MacLeod et al., 2013; Shannon et al., 2022). The culture media should completely saturate the surgical sponge and fill the well by approximately 25%.
Cut tissue into 1 cm × 1 cm squares and place skin, dermis side-down, on gelatin sponge squares pre-saturated with culture media (prepared in step B3). Ensure the epidermis maintains an air interface and that the dermis has contact with the media-soaked sponge. The tissue should not be submerged in media.
Incubate tissue at 37 °C and 5% CO2.
Change media every 2–3 days by aspirating media from the well using a 1 mL sterile serological pipette. Avoid any contact between the aspiration tip and the gelatin sponge by gently tilting the plate to pool media from the bottom. Add fresh media to the well without disturbing the gelatin sponge and skin explant and ensure that the skin maintains in air–liquid interface.
Skin explants can be maintained in this fashion for up to four weeks (Companjen et al., 2001; Steinstraesser et al., 2009).
Note: Successful use of skin explant for wound healing studies has been reported up to two weeks, though epidermal thickening is evident in ex vivo skin cultures within the first week (Xu et al., 2012; Neil et al., 2020), and epidermal barrier function is reported to remain intact for up to four weeks (Steinstraesser et al., 2009). However, tissue viability decreases over time. It is important to optimize culture conditions and experimental parameters.
Ex vivo Wounding
To generate ex vivo wounds, hold scissors vertically to inflict 20–30 cuts to skin tissue. Do not mince the tissue: these wounds should be shallow, and each cut should be approximately half the length of the explant.
Note: Incisional wounds may not be immediately obvious by the naked eye.
Wounded samples should be collected within 24 h using Trizol reagent for RNA extraction or snap frozen in OCT compound for immunofluorescence staining.
Cytokine Stimulation
If cytokine stimulation is necessary, prepare media supplemented with cytokines of choice [e.g., IL-1β (Companjen et al., 2001), IL-15, or IL-27 (Suwanpradid et al., 2021)] and add 0.5–1.0 mL per well as described in step B3.
Carefully transfer explant to the freshly prepared 12-well plate without allowing media to touch the epidermal surface.
Skin can be collected and carefully removed from the gelatin sponge within two days after cytokine treatment.
Tissue Collection
Immunostaining
Cut tissue into 2 mm × 2 mm squares and place the skin with dermis side down in the bottom of an intermediate Tissue-Tek cryomold.
Slowly add OCT compound to cover the tissue sample, avoiding bubbles.
Pop any bubbles with a 10 µL pipette tip if necessary.
Allow the OCT to settle on a flat surface at room temperature for 10 min.
Snap freeze the tissue by resting cryomold on dry ice on a flat surface until frozen.
Store the frozen OCT blocks in a -80 °C freezer until use.
For immunostaining, 7–10 µm thick cryo-sections containing both epidermis and dermis are obtained from the OCT blocks using a Cryostat.
RNA analysis
Mince tissue thoroughly using scissors and transfer minced tissue to a clean microcentrifuge tube containing at least 800 μL of Trizol reagent for each explant.
Vortex sample and incubate at room temperature for 5–15 min to allow tissue lysis by Trizol.
Centrifuge tubes containing the lysed tissues at 12,000 × g for at least 10 min at 4 °C. Carefully transfer supernatant to a clean tube for standard RNA isolation techniques, such as ethanol precipitation or commercially available kits designed for RNA extraction.
Example results
Immunostaining
Following the methods described above, generate ex vivo wounds on human tissue for 24 h and freeze in OCT compound. Warm 8 µm sections to room temperature prior to fixation with 4% paraformaldehyde for 15 min and permeabilization with 0.1% Triton for 15 min.
Incubate sections with the rat anti-human Integrin α6 CD49f antibody overnight at 4 °C in a moist chamber.
Wash slides with PBS with 0.1% Triton, and incubate for 1 h with Alexa Fluor 647-conjugated goat anti-rat secondary antibody (diluted 1:400).
After subsequent washing, stain nuclei using Hoechst diluted 1:10,000 for 10 min.
Mount slides using Prolong Gold antifade reagent and cover with a coverslip prior to imaging on a fluorescent microscope (Figure 1).
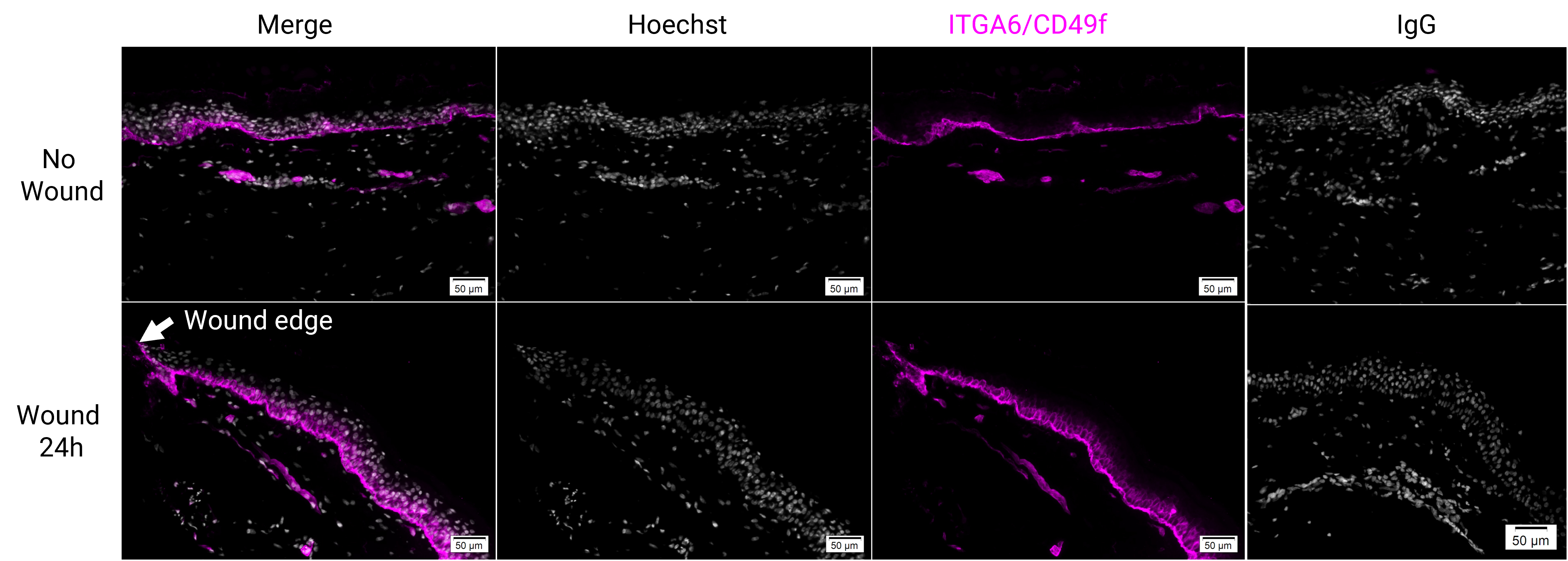
Figure 1. Immunostaining for integrin α6 (ITGA6/CD49f, magenta) in human skin explants that were wounded ex vivo and cultured for 24 h. Tissues were sectioned to 8 μm thickness. Scale bars = 50 μm.RT-qPCR
Isolate total RNA from Trizol reagent per the manufacturer’s instructions, followed by treatment with DNase I.
Quantify RNA using Nanodrop 1000 and generate cDNA using cDNA synthesis kit. Amplification is detected using Fast SYBR Green Master Mix (Applied Biosystems) or SYBR green blue (PCR biosystems); 10 ng cDNA is used per qPCR reaction.
Perform qPCR with the following primers:
Gene Forward (5’-3’) Reverse (3’-5’) S100a7 CCTGCTGACGATGATGAA TGGCTCTGCTTGTGGTAG GAPDH ATGGGAAGGTGAAGGTCGGA CAGCGTCAAAGGTGGAGGAGT Use GAPDH expression as internal control (see primer sequences).
Normalize fold changes calculated for gene expression in human samples to untreated or unwounded controls, as indicated. Data are represented as fold change or using ΔΔCt method as indicated (Figure 2).
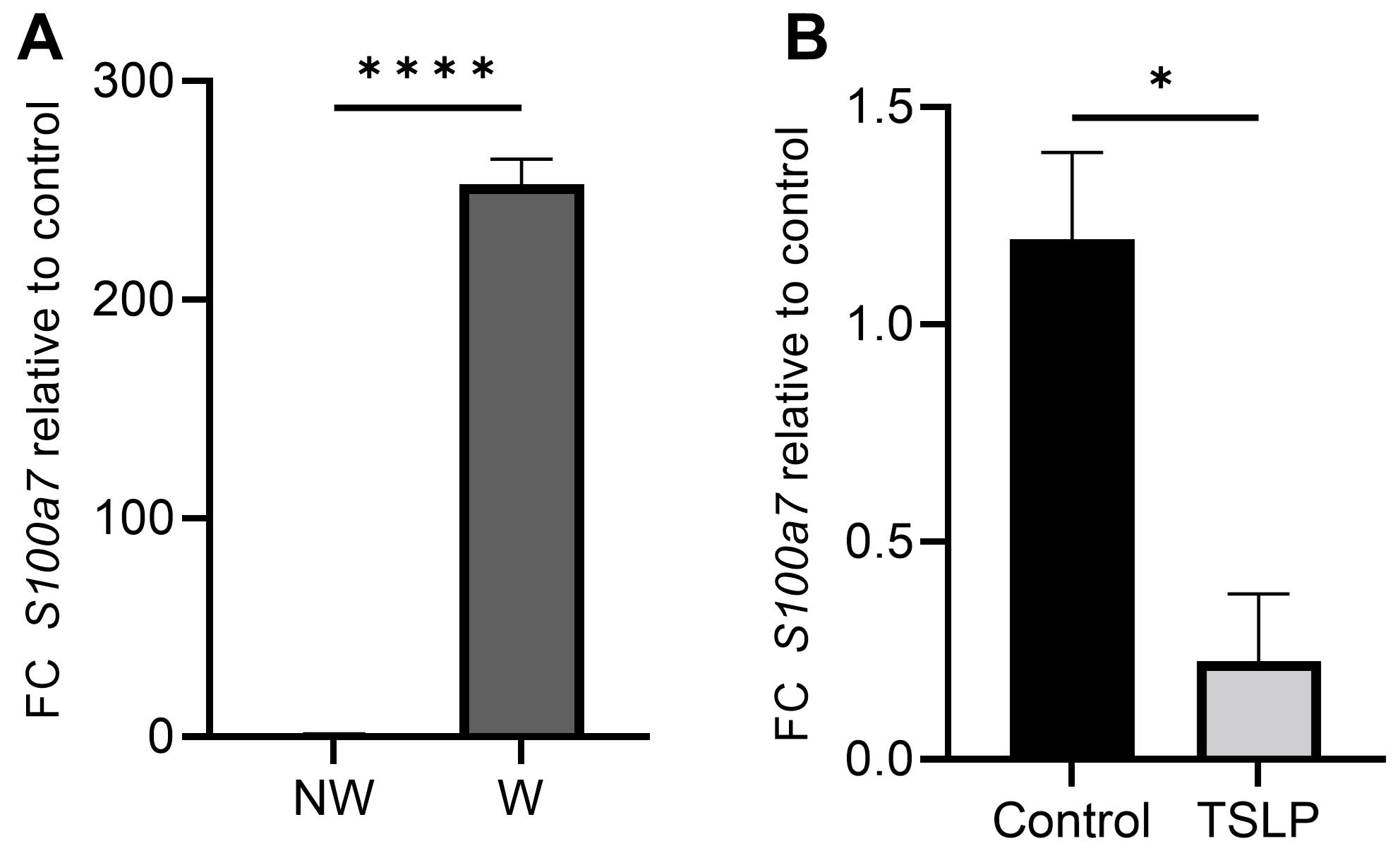
Figure 2. qPCR showing relative mRNA levels of S100a7 in response to ex vivo wounding (W) 24 h after injury (A) or topical treatment with 100 ng of thymic stromal lymphopoietin (TSLP) for 16 hours (B). Data presented are from four independent experiments using two different human donors in technical duplicate. Error bars represent ± SEM. Statistical analysis was performed using the two-tailed unpaired Student’s t-test, under the untested assumption of normality. A p-value of <0.05 was considered statistically significant; **: p < 0.01 and ****: p < 0.0001.
Recipes
Wash Buffer
Reagent Final concentration Amount 1× phosphate buffered saline 1× 495 mL 100× penicillin–streptomycin–glutamine 1× 5 mL Total n/a 500 mL Culture Medium
Thaw one vial containing 5 mL of 100× HKGS to room temperature and gently swirl to mix.
Swirl 0.5 mL of 0.06 M calcium solution to mix (included in EpiLifeTM CF kit).
Wipe the outside of the containers with 70% ethanol to sanitize prior to moving to tissue culture hood.
Draw 5 mL of 100× HKGS and add to 495 mL of EpiLifeTM calcium free medium.
Slowly add calcium stock to 500 mL of medium to reach a final concentration of 0.06 mM Ca2+.
Store unused solution at 4 °C for up to one month.
70% ethanol
Reagent Final concentration Amount Ethanol (absolute) 70% 700 mL H2O n/a 300 mL Total n/a 1,000 mL
Acknowledgments
We would like to thank Jutamas Suwanpradid and Yingai Jane Jin of Duke University for protocol development. This work is in part supported by NIH/NIAID grant R01-AI139207 and NIH/NIAMS grant R01-AR068991 and the Department of Dermatology at Duke University. This protocol was adapted from methods described in previous publications (Macleod et al., 2013; Shannon et al., 2022).
Competing interests
All authors declare no competing interests.
Ethics
Normal skin samples were obtained from otherwise discarded and deidentified tissues of adult male and female patients undergoing abdominoplasty at Duke University Medical Center. All human samples for this study were obtained according to the protocols approved by the Institutional Review Board at Duke University.
References
- Companjen, A. R., van der Wel, L. I., Wei, L., Laman, J. D. and Prens, E. P. (2001). A modified ex vivo skin organ culture system for functional studies. Arch Dermatol Res 293(4): 184-190.
- MacLeod, A. S., Hemmers, S., Garijo, O., Chabod, M., Mowen, K., Witherden, D. A. and Havran, W. L. (2013). Dendritic epidermal T cells regulate skin antimicrobial barrier function. J Clin Invest 123(10): 4364-4374.
- Neil, J. E., Brown, M. B. and Williams, A. C. (2020). Human skin explant model for the investigation of topical therapeutics. Sci Rep 10(1): 21192.
- Shannon, J. L., Corcoran, D. L., Murray, J. C., Ziegler, S. F., MacLeod, A. S. and Zhang, J. Y. (2022). Thymic stromal lymphopoietin controls hair growth. Stem Cell Reports 17(3): 649-663.
- Smits, J. P. H., Niehues, H., Rikken, G., van Vlijmen-Willems, I., van de Zande, G., Zeeuwen, P., Schalkwijk, J. and van den Bogaard, E. H. (2017). Immortalized N/TERT keratinocytes as an alternative cell source in 3D human epidermal models. Sci Rep 7(1): 11838.
- Steinstraesser, L., Rittig, A., Gevers, K., Sorkin, M., Hirsch, T., Kesting, M., Sand, M., Al-Benna, S., Langer, S., Steinau, H. U., et al. (2009). A human full-skin culture system for interventional studies. Eplasty 9: e5.
- Suwanpradid, J., Lee, M. J., Hoang, P., Kwock, J., Floyd, L. P., Smith, J. S., Yin, Z., Atwater, A. R., Rajagopal, S., Kedl, R. M., et al. (2021). IL-27 Derived From Macrophages Facilitates IL-15 Production and T Cell Maintenance Following Allergic Hypersensitivity Responses. Front Immunol 12: 713304.
- Xu, W., Jong Hong, S., Jia, S., Zhao, Y., Galiano, R. D. and Mustoe, T. A. (2012). Application of a partial-thickness human ex vivo skin culture model in cutaneous wound healing study. Lab Invest 92(4): 584-599.
- Zomer, H. D. and Trentin, A. G. (2018). Skin wound healing in humans and mice: Challenges in translational research. J Dermatol Sci 90(1): 3-12.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Shannon, J. L., Kirchner, S. J. and Zhang, J. Y. (2022). Human Skin Explant Preparation and Culture. Bio-protocol 12(18): e4514. DOI: 10.21769/BioProtoc.4514.
Category
Cell Biology > Tissue analysis > Tissue culture
Biological Sciences > Biological techniques
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link