- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
14C-paraquat Efflux Assay in Arabidopsis Mesophyll Protoplasts
(*contributed equally to this work) Published: Vol 12, Iss 18, Sep 20, 2022 DOI: 10.21769/BioProtoc.4512 Views: 1641
Reviewed by: Yingnan HouAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Maize Seedlings Colonization with Serendipita indica and Its Colonization Efficiency Analysis
Om Prakash Narayan [...] Atul Kumar Johri
Oct 20, 2023 2267 Views

Botrytis cinerea in vivo Inoculation Assays for Early-, Middle- and Late-stage Strawberries
Piao Yang [...] Ye Xia
Oct 20, 2023 2762 Views
Abstract
Weeds compete with crops for growth resources, causing tremendous yield losses. Paraquat is one of the three most common non-selective herbicides. To study the mechanisms of paraquat resistance, we need to trace the movement of paraquat in plants and within the cell. 14C is a radioactive carbon isotope widely used to trace substances of interest in various biological studies, especially in transport analyses. Here, we describe a detailed protocol using 14C-paraquat to demonstrate paraquat efflux in Arabidopsis protoplasts.
Keywords: 14C-paraquatBackground
Paraquat (PQ), a non-selective broad-spectrum herbicide, has been widely used in weed control. It mainly acts on plant green tissue and is relatively safe for soil microorganisms and plant roots (Huang et al., 2019). It competes for electrons from the photosynthesis system I (PS I) and reacts with oxygen, producing a large quantity of reactive oxygen species (ROS) that destroy the photosynthesis system and ultimately kill plants (Apel and Hirt, 2004). However, with the heavy use of PQ, resistant weeds have emerged (Hawkes, 2014). Through extensive studies of PQ-resistant weeds and Arabidopsis, several molecular mechanisms of PQ resistance have been unraveled. These include disrupted PQ transport that blocks it from reaching its target, PQ sequestration into the vacuole and apoplast, enhanced ability to scavenge ROS, and PQ detoxification (Chen et al., 2009; Fujita et al., 2012; Xi et al., 2012; Li et al., 2013; Luo et al., 2016; Lv et al., 2021; Xia et al., 2021; Huang et al., 2022; Nazish et al., 2022).
To study the function of the MATE transporter DTX6, we designed an experiment using 14C-paraquat and Arabidopsis protoplasts to confirm its PQ-exporting activity from cytosol to apoplast, which assisted us in revealing the PQ resistance mechanism via sequestration.
Materials and Reagents
50 mL centrifuge tube with round bottom
2 mL centrifuge tube with round bottom
70 μm Nylon mesh (EMD Millipore, CLS431751)
3–4 weeks old Arabidopsis plants
Scintillation fluid (Perkin-Elmer, Waltham, catalog number: 6013321)
14C-paraquat (American Radiolabeled Chemicals Inc. ARC 1311). Store at -20 °C
NaCl (Sinopharm Chemical Reagent Co., Ltd., CHINA, CAS: 7647-14-5)
CaCl2 (Sinopharm Chemical Reagent Co., Ltd., CHINA, CAS: 10043-52-4)
KCl (Sinopharm Chemical Reagent Co., Ltd., CHINA, CAS: 7447-40-7)
MES (SIGMA, USA CAS: 1266615-59-1)
W5 solution (see Recipes)
Equipment
Centrifuge with temperature control (Allegra X-22R Benchtop Centrifuge, Beckman Coulter, Indianapolis, USA)
Automated cell counter (IC1000, Shanghai RuiYu Biotech Co. Ltd., Shanghai, CHINA)
Ventilation cabinet (YKD-DAT004F, Wuxi Safoo Safety Equipment Co. Ltd., Wuxi, CHINA)
Scintillation vial (Perkin-Elmer, Waltham, USA)
Liquid scintillation counter (Tri-Carb 2910 TR, Perkin-Elmer, Waltham, USA)
-20 °C freezer
Procedure
Isolate protoplasts according to Yoo et al. (2007) and suspend in cold W5 solution (see Recipes).
Count the protoplasts with a cell counter to estimate their concentration. Centrifuge at 40 × g and 4 °C for 2 min. Decant the supernatant and resuspend the protoplasts in cold W5 solution to approximately 3 × 106 protoplasts/mL. This protoplast preparation is ready for 14C-paraquat loading (Figure 1).
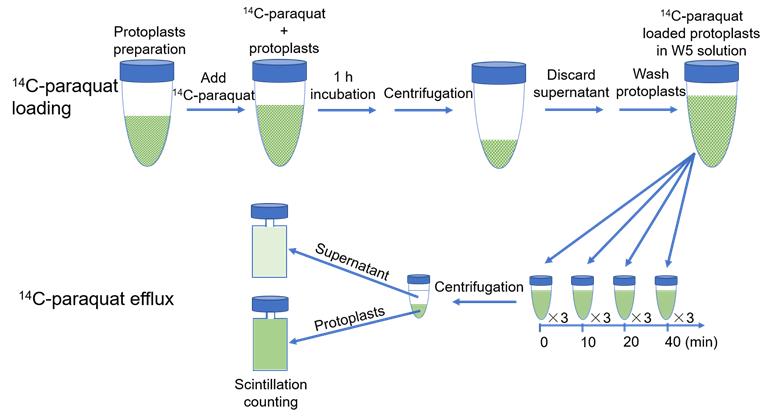
Figure 1. Experimental scheme of 14C-paraquat loading into and efflux out of Arabidopsis mesophyll protoplasts.Thaw out 14C-paraquat (3.1 mM, 0.1 μCi/μL) and dilute 10 times with W5 solution at room temperature.
Add the same volume of diluted 14C-paraquat solution to the protoplast preparation, to obtain a final concentration of 3 μM 14C-paraquat. Incubate for 1 h at room temperature to load paraquat into the protoplasts.
Centrifuge the protoplasts at 40 × g and 4 °C for 2 min. Discard the supernatant with the unloaded 14C-paraquat and wash the protoplasts with 15 mL of cold W5 solution 2–3 times.
Resuspend the protoplasts in 13 mL of W5 solution at room temperature and immediately initiate the paraquat efflux assay.
At different time points (0, 10, 20, and 40 min) after W5 solution resuspension, transfer 1 mL of protoplasts suspension into a 2 mL round-bottom centrifuge tube. Take at least three replicates at each time point.
Immediately centrifuge the aliquoted samples (40 × g, 2 min, 4 °C). Transfer the supernatant to a scintillation vial. Resuspend the protoplast pellet in 1 mL of W5 solution and transfer it to a scintillation vial. Then add 5 mL of scintillation fluid to each sample. Detect radioactivity on a liquid scintillation counter.
Analyze and present data as shown in Figure 2.
Note: 14C-paraquat waste was disposed according to the University’s safety management of isotopes.
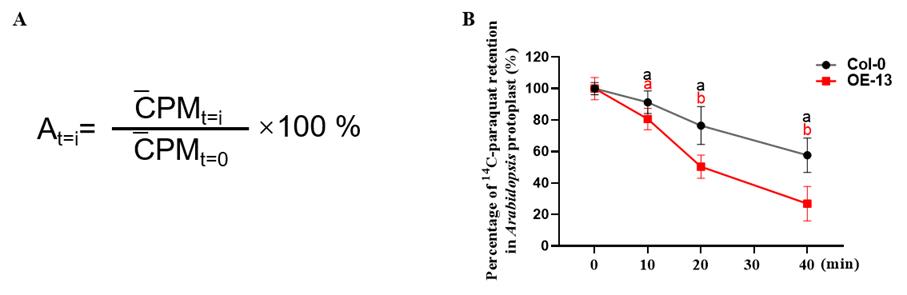
Figure 2. Data analyses and presentation. (A) Calculation of 14C-paraquat retention in protoplasts at different time points. t=i: time of 14C-paraquat protoplasts efflux (0, 10, 20, and 40 min). CPMt=i: mean value of 14C-paraquat radioactivity retention in protoplasts at different time points. CPMt=0: mean value of 14C-paraquat radioactivity retention in protoplasts at the initial time. At=i: percentage of 14C-paraquat radioactivity retention in protoplasts at different time points relative to the initial time. (B) Paraquat retention in the protoplasts, measured in the Col-0 and DTX6 overexpression line (OE-13) (Xia et al., 2021). Paraquat retention was defined as a percentage of the 14C-paraquat loaded in the protoplasts. Values are mean ± SD (n = 3). Different letters indicate significant differences (P < 0.05; one-way ANOVA).
Recipes
W5 solution
154 mM NaCl
125 mM CaCl2
5 mM KCl
2 mM MES
pH 5.7
Acknowledgments
This protocol was adapted from previous work (Zhang et al., 2014; Qin et al., 2021); we would like to thank the authors of these studies for their protocols.
Competing interests
No conflict of interest declared.
References
- Apel, K. and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373-399.
- Chen, R., Sun, S., Wang, C., Li, Y., Liang, Y., An, F., Li, C., Dong, H., Yang, X., Zhang, J. and Zuo, J. (2009). The Arabidopsis PARAQUAT RESISTANT2 gene encodes an S-nitrosoglutathione reductase that is a key regulator of cell death. Cell Res 19(12): 1377-1387.
- Fujita, M., Fujita, Y., Iuchi, S., Yamada, K., Kobayashi, Y., Urano, K., Kobayashi, M., Yamaguchi-Shinozaki, K. and Shinozaki, K. (2012). Natural variation in a polyamine transporter determines paraquat tolerance in Arabidopsis. Proc Natl Acad Sci U S A 109(16): 6343-6347.
- Hawkes, T. R. (2014). Mechanisms of resistance to paraquat in plants. Pest Manag Sci 70(9): 1316-1323.
- Huang, Y., Zhan, H., Bhatt, P. and Chen, S. (2019). Paraquat Degradation From Contaminated Environments: Current Achievements and Perspectives. Front Microbiol 10: 1754.
- Huang, Y. J., Huang, Y. P., Xia, J. Q., Fu, Z. P., Chen, Y. F., Huang, Y. P., Ma, A., Hou, W. T., Chen, Y. X., Qi, X., et al. (2022). AtPQT11, a P450 enzyme, detoxifies paraquat via N-demethylation.J Genet Genomics S1673-8527(22)00127-8.
- Li, J., Mu, J., Bai, J., Fu, F., Zou, T., An, F., Zhang, J., Jing, H., Wang, Q., Li, Z., et al. (2013). Paraquat Resistant1, a Golgi-localized putative transporter protein, is involved in intracellular transport of paraquat. Plant Physiol 162(1): 470-483.
- Luo, C., Cai, X. T., Du, J., Zhao, T. L., Wang, P. F., Zhao, P. X., Liu, R., Xie, Q., Cao, X. F., and Xiang, C. B. (2016). PARAQUAT TOLERANCE3 is an E3 ligase that switches off activated oxidative response by targeting histone-modifying PROTEIN METHYLTRANSFERASE4b.PLoS Genet 12: e1006332.
- Lv, Z., Zhao, M., Wang, W., Wang, Q., Huang, M., Li, C., Lian, Q., Xia, J., Qi, J., Xiang, C., et al. (2021). Changing Gly311 to an acidic amino acid in the MATE family protein DTX6 enhances Arabidopsis resistance to the dihydropyridine herbicides. Mol Plant 14(12): 2115-2125.
- Nazish, T., Huang, Y. J., Zhang, J., Xia, J. Q., Alfatih, A., Luo, C., Cai, X. T., Xi, J., Xu, P. and Xiang, C. B. (2022). Understanding paraquat resistance mechanisms in Arabidopsis thaliana to facilitate the development of paraquat-resistant crops. Plant Commun 3(3): 100321.
- Qin, P., Zhang, G., Hu, B., Wu, J., Chen, W., Ren, Z., Liu, Y., Xie, J., Yuan, H., Tu, B., et al. (2021). Leaf-derived ABA regulates rice seed development via a transporter-mediated and temperature-sensitive mechanism. Sci Adv 7(3): eabc8873.
- Xi, J., Xu, P. and Xiang, C. B. (2012). Loss of AtPDR11, a plasma membrane-localized ABC transporter, confers paraquat tolerance in Arabidopsis thaliana. Plant J 69(5): 782-791.
- Xia, J. Q., Nazish, T., Javaid, A., Ali, M., Liu, Q. Q., Wang, L., Zhang, Z. Y., Zhang, Z. S., Huang, Y. J., Wu, J., et al. (2021). A gain-of-function mutation of the MATE family transporter DTX6 confers paraquat resistance in Arabidopsis. Mol Plant 14(12): 2126-2133.
- Yoo, S. D., Cho, Y. H. and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2(7): 1565-1572.
- Zhang, H., Zhu, H., Pan, Y., Yu, Y., Luan, S. and Li, L. (2014). A DTX/MATE-type transporter facilitates abscisic acid efflux and modulates ABA sensitivity and drought tolerance in Arabidopsis. Mol Plant 7(10): 1522-1532.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Xia, J. Q., Liu, Q. Q. and Xiang, C. B. (2022). 14C-paraquat Efflux Assay in Arabidopsis Mesophyll Protoplasts. Bio-protocol 12(18): e4512. DOI: 10.21769/BioProtoc.4512.
Category
Plant Science > Plant physiology > Biotic stress
Cell Biology > Cell signaling > Development
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










