- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Serial Room Temperature Crystal Loading, Data Collection, and Data Processing of Human Glutaminase C in Complex with Allosteric Inhibitors
Published: Vol 12, Iss 18, Sep 20, 2022 DOI: 10.21769/BioProtoc.4509 Views: 1514
Reviewed by: Joana Alexandra Costa ReisLaura RotilioLuca Iacovino
Abstract
Cancer cells often overexpress glutaminase enzymes, in particular glutaminase C (GAC). GAC resides in the mitochondria and catalyzes the hydrolysis of glutamine to glutamate. High levels of GAC have been observed in aggressive cancers and the inhibition of its enzymatic activity has been shown to reduce their growth and survival. Numerous GAC inhibitors have been reported, the most heavily investigated being a class of compounds derived from the small molecule BPTES (bis-2-(5-phenylacetamido-1,3,4-thiadiazol-2-yl)ethyl sulfide). X-ray structure determination under cryo-cooled conditions showed that the binding contacts for the different inhibitors were largely conserved despite their varying potencies. However, using the emerging technique serial room temperature crystallography, we were able to observe clear differences between the binding conformations of inhibitors. Here, we describe a step-by-step protocol for crystal handling, data collection, and data processing of GAC in complex with allosteric inhibitors using serial room temperature crystallography.
Graphical abstract:

Workflow for serial room temperature crystallography. Diagram showing the processing and scaling routine for crystals analyzed using serial room temperature crystallography.
Background
We solved a number of X-ray crystal structures of glutaminase C (GAC) bound to different BPTES (bis-2-(5-phenylacetamido-1,3,4-thiadiazol-2-yl)ethyl sulfide)-class allosteric inhibitors under cryogenic crystallographic conditions (Huang et al., 2018; Milano et al., 2022). Although they vary in potency for GAC, the structures show that the contacts between the small molecule inhibitors and GAC are largely conserved. In order to visualize differences at the binding interface between GAC and the inhibitors, we employed fixed-target serial room temperature synchrotron crystallography (Wierman et al., 2019; Milano et al., 2022). An advantage of using serial room temperature X-ray crystallography is to help overcome any cryo-induced structural biases, such as “trapping” the protein/ligand in a distinct conformation as a result from crystal freezing. Although this technique requires substantially more crystals than cryogenic crystallography due to radiation damage to the sample, it can yield structures that are more dynamic and fluid. Consequently, structures solved using room temperature X-ray crystallography are capable of revealing differences in proteins and protein/ligand complexes that perhaps are more physiologically relevant. We recently used this technique to solve the structure of apo GAC (Illava et al., 2021). We hypothesized that structures of the GAC/inhibitor complexes at room temperature would make it possible to visualize subtle changes in the binding characteristics of the small molecules that were not evident from the low-temperature cryo-structures. Indeed, we were able for the first time to observe clear structural differences in the binding poses between two allosteric inhibitors of widely different potencies in the GAC/inhibitor complexes solved at room temperature. Here, we describe a detailed protocol for crystal handling, data collection, and data processing when employing serial room temperature crystallography (Figure 1).
Materials and Reagents
Pipette tips (USA Scientific, TipOne, catalog number: 1111-3700)
GAC/inhibitor crystals (room temperature)
Equipment
Forceps (Fisher Scientific, catalog number: 09-753-50)
Micro tools set (Hampton Research, catalog number: HR4-811)
Pipettes (Rainin, catalog number: 17008649)
Light microscope (Olympus)
Hex key
Crystallography sample supports (MiTeGen)
Sample loading box: humidity controlled and includes optical microscope
Sample loading station: connected to a vacuum pump (controlled by a foot pedal) for sample wicking
Sample supports: include a frame containing the support film and a goniometer base
Sample Mylar seals
Software
XDS Package (https://xds.mr.mpg.de)
CCP4 Program Suite (https://www.ccp4.ac.uk)
Filtering program cut_XDS and script to run it cut_XS.csh (https://www.chess.cornell.edu/macchess/mx/mx_software)
Procedure
Handling and loading of GAC/inhibitor crystals (Figure 2)
All crystals are grown in 24-well plates at room temperature using the hanging drop vapor diffusion method. Crystals are handled in a humidity-controlled environment to help prevent crystal dehydration.
Assemble sample support by inserting the frame into the goniometer base and tighten with a hex key.
Remove a coverslip containing GAC crystals from one of the wells of the crystal tray.
Under the microscope and using the micro tools, pick up crystals and place them onto the film of the sample support. The landscape of the film enables the crystals to settle in random orientations, maximizing data collection. Pre-wet the film with well solution if necessary. The crystals can lie anywhere on the film.
After 20 or more crystals (100 × 100 × 100 μM3 in size) are on the film, transfer the sample support into the loading box and place on sample loading station.
Looking through the optical scope, use the foot-pedal controlled vacuum to wick away excess liquid on the sample film.
Remove the white covering around the sample frame (on both sides) to expose the adhesive gasket.
Apply the Mylar sealing film on both sides of the frame to cover and protect the crystals from drying.
The completed sample support containing the crystals is now ready to be mounted in the beamline hutch and exposed to X-rays.
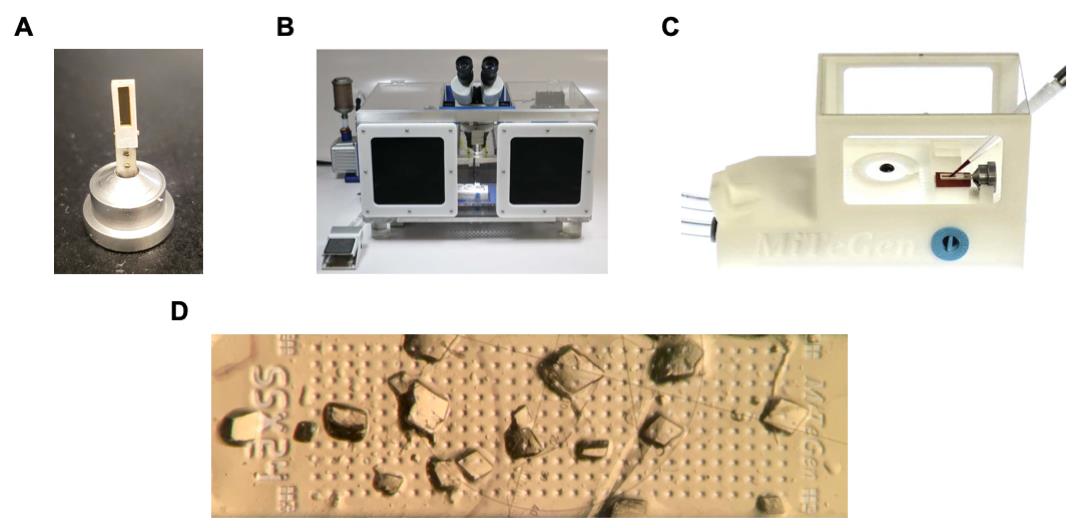
Figure 2. Serial room temperature crystallography sample handling system. (A) Sample support containing a thin film within a rectangular frame mounted into a goniometer base. The frame is surrounded by an adhesive gasket protected by a white covering. (B) Humidity-controlled sample loading box containing an optical microscope. (C) Sample loading station that connects to a vacuum pump for liquid wicking. (D) GAC crystals on the film of the sample support.Data collection and processing
Data were collected at the Cornell High Energy Synchrotron Source (CHESS) at Cornell University on beamline ID7B2 (FlexX).
Sample supports containing GAC crystals are mounted in the beamline hutch on a goniometer and raster scanned in 20 μm steps, recording 5° of oscillation at each position.
Each step of the raster scanning is completed in 0.75–0.5 s, with 0.25 s for data acquisition (25 frames, 0.2°, and 10 ms/frame), corresponding to a 1.3 Hz raster rate.
Individual oscillation frame sets are processed with XDS (processing indicated that GAC crystallized in both orthorhombic and monoclinic crystal forms. Overall, the monoclinic crystal form produced the best data quality for GAC. We selected for this polymorph by filtering the data based on unit cell dimensions and angles in XDS and reprocessed the datasets).
The data are then scaled using XSCALE (part of the XDS software package).
Datasets that poorly agree with each other are removed through filtering methods.
For GAC, data is filtered based on agreement of each 25-frame set with the entire set for a support, using a script (cut_XS.csh) to remove discrepant sets (with the program cut_XDS) and rescaled (with XSCALE). R-factors on intensities between an individual set and the average of all sets were used for filtering discrepant data.
Lists of the accepted 25-frame sets from the different sample supports are then combined using a text editor such as gedit, producing a control file for XSCALE.
XSCALE is then used with this control file to rescale the combined data to generate a merged dataset.
The merged dataset can be further filtered, as in step 6, with various values of the parameter (R-factor) defining discrepant data. The datasets are filtered in order to achieve satisfactory data completeness and statistical values.
Using CCP4 programs pointless, aimless, and ctruncate, the merged dataset is then converted into an mtz file for structure determination.
Acknowledgments
This work was supported by grants to R.A.C. (GM122575, CA201402), L.A.M. [University of Pittsburgh Medical Center Competitive Medical Research Fund (CMRF)], and to CHEXS (DMR-1829070), and MacCHESS (P30GM126166). We also thank MiTeGen for supplying key resources for the serial room temperature X-ray crystallography experiments. Parts of this protocol were adapted from previously described methods (Illava et al., 2021; Milano et al., 2022).
Competing interests
The authors declare that they have no conflicts of interest with the contents of this article.
References
- Huang, Q., Stalnecker, C., Zhang, C., McDermott, L. A., Iyer, P., O'Neill, J., Reimer, S., Cerione, R. A. and Katt, W. P. (2018). Characterization of the interactions of potent allosteric inhibitors with glutaminase C, a key enzyme in cancer cell glutamine metabolism. J Biol Chem 293(10): 3535-3545.
- Milano, S. K., Huang, Q., Nguyen, T. T., Ramachandran, S., Finke, A., Kriksunov, I., Schuller, D. J., Szebenyi, D. M., Arenholz, E., McDermott, L. A., et al. (2022). New insights into the molecular mechanisms of glutaminase C inhibitors in cancer cells using serial room temperature crystallography. J Biol Chem 298(2): 101535.
- Wierman, J. L., Pare-Labrosse, O., Sarracini, A., Besaw, J. E., Cook, M. J., Oghbaey, S., Daoud, H., Mehrabi, P., Kriksunov, I., Kuo, A., et al. (2019). Fixed-target serial oscillation crystallography at room temperature. IUCrJ 6(Pt 2): 305-316.
- Illava, G., Jayne, R., Finke, A. D., Closs, D., Zeng, W., Milano, S. K., Huang, Q., Kriksunov, I., Sidorenko, P., Wise, F. W., et al. (2021). Integrated sample-handling and mounting system for fixed-target serial synchrotron crystallography. Acta Crystallogr D Struct Biol 77(Pt 5): 628-644.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Milano, S. K., Szebenyi, D. M. and Cerione, R. A. (2022). Serial Room Temperature Crystal Loading, Data Collection, and Data Processing of Human Glutaminase C in Complex with Allosteric Inhibitors. Bio-protocol 12(18): e4509. DOI: 10.21769/BioProtoc.4509.
- Milano, S. K., Huang, Q., Nguyen, T. T., Ramachandran, S., Finke, A., Kriksunov, I., Schuller, D. J., Szebenyi, D. M., Arenholz, E., McDermott, L. A., et al. (2022). New insights into the molecular mechanisms of glutaminase C inhibitors in cancer cells using serial room temperature crystallography. J Biol Chem 298(2): 101535.
Category
Biophysics > X-ray crystallography
Drug Discovery > Drug Design
Biological Sciences > Biological techniques
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










