- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
High-Efficiency Retroviral Transduction for Type 1 Regulatory T Cell Differentiation
Published: Vol 12, Iss 17, Sep 5, 2022 DOI: 10.21769/BioProtoc.4499 Views: 2684
Reviewed by: David PaulVishal Nehru

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Functional Phenotyping of Lung Mouse CD4+ T Cells Using Multiparametric Flow Cytometry Analysis
Céline M. Maquet [...] Bénédicte D. Machiels
Sep 20, 2023 4201 Views

T-Cell-Based Platform for Functional Screening of T-Cell Receptors Identified in Single-Cell RNA Sequencing Data Sets of Tumor-Infiltrating T-Cells
Aaron Rodriguez Ehrenfried [...] Rienk Offringa
Apr 20, 2024 6397 Views

Novel Experimental Approach to Investigate Immune Control of Vascular Function: Co-culture of Murine Aortas With T Lymphocytes or Macrophages
Taylor C. Kress [...] Eric J. Belin de Chantemèle
Sep 5, 2025 3548 Views
Abstract
Type 1 regulatory T (Tr1) cells are an immunoregulatory CD4+ Foxp3- IL-10high T cell subset with therapeutic potential for various inflammatory diseases. Retroviral (RV) transduction has been a valuable tool in defining the signaling pathways and transcription factors that regulate Tr1 differentiation and suppressive function. This protocol describes a method for RV transduction of naïve CD4+ T cells differentiating under Tr1 conditions, without the use of reagents such as polybrene or RetroNectin. A major advantage of this protocol over others is that it allows for the role of genes of interest on both differentiation and function of Tr1 cells to be interrogated. This is due to the high efficiency of RV transduction combined with the use of an IL10GFP/Foxp3RFP dual reporter mouse model, which enables successfully transduced Tr1 cells to be identified and sorted for functional assays. In addition, this protocol may be utilized for dual/multiple transduction approaches and transduction of other lymphocyte populations, such as CD8+ T cells.
Keywords: Retrovirus TransductionBackground
Type 1 regulatory T (Tr1) cells are an immunoregulatory Foxp3- CD4+ T cell subset that is able regulate inflammation and induce tolerance, making them a promising candidate for immunotherapies (Mobs et al., 2010; Gol-Ara et al., 2012; Roncarolo et al., 2014; Bohm et al., 2015). Therefore, signaling pathways and transcription factors regulating Tr1 differentiation and suppressive function are of great clinical interest. RV transduction of CD4+ T cells has been an effective tool in this endeavor (Wu et al., 2013; Mascanfroni et al., 2015; Karwacz et al., 2017). Many studies use Tr1 differentiation as an endpoint because detection of IL-10 and Foxp3 require terminal methods, such as fixation of cells. As a result, signaling pathways and transcription factors regulating the suppressive function of Tr1 cells are less understood than those required for differentiation. This protocol was developed to investigate the role of signaling pathways and transcription factors downstream of TCR/ITK, which regulate both Tr1 differentiation and suppressive function (Huang et al., 2017). The use of an IL10GFP/Foxp3RFP dual reporter mouse model, high efficiency of RV transduction, and bicistronic retroviral vectors that enable transduced cells to be identified via expression of a reporter gene allows sufficient numbers of live transduced Tr1 cells to be isolated for functional assays. Another advantage of this protocol is that it does not make use of chemical transduction enhancers, such as RetroNectin or polybrene, which may be cost-prohibitive and cause cytotoxicity (Kurachi et al., 2017; Eremenko et al., 2021). In addition, the protocol described here may also be utilized for dual transduction strategies and other lymphocyte populations, such as CD8+ T cells.
Materials and Reagents
Eppendorf® Safe-Lock microcentrifuge 1.5 mL tubes (Sigma-Aldrich, catalog number: T9661)
70 μm Cell Strainer (Vita Scientific, catalog number: NSTF90051)
CytoOne T75 filter cap TC flask (USA Scientific, catalog number: CC7682-4875)
100 mm Dish, Tissue Culture-Treated (VWR, catalog number: 25382-428)
Corning® 96-well Clear Flat Bottom Polystyrene TC-treated Microplates (Corning, catalog number: 3596)
0.45 µm Sterile Polyethersulfone Syringe Filter (VWR, catalog number: 28145-493)
10 mL BD Luer-LokTM Syringe (BD, catalog number: 309605)
Axygen 96 Well Clear V-Bottom 550 µL Polypropylene Deep Well Plate (Axygen, catalog number: P-96-450V-C)
IL10GFP/Foxp3RFP dual reporter mouse model (generated as we previously reported in Huang et al., 2017)
Rag-/- mouse model (Jackson Laboratory, Strain #: 002216)
Note: All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at the Louisiana State University (protocol #: 18-047 and 21-039).
MS Columns (Miltenyibiotec, catalog number: 130-042-201)
LS Columns (Miltenyibiotec, catalog number: 130-042-401)
InVivoMAb anti-mouse CD3ϵ [clone: 145-2C11] (Bio X Cell, catalog number: BE0001-1)
InVivoMAb anti-mouse CD28 [clone: 37.51] (Bio X Cell, catalog number: BE0015-1)
Mouse IL-27 (NS0-expressed) Protein (R&D Systems, catalog number: 2799-ML-010)
Retro-XTM Concentrator (Takara Bio, catalog number: 631456)
Opti-MEMTM, Reduced Serum Medium (Thermo Fisher Scientific, catalog number: 31985070)
Mirus Bio TransIT®-293 Transfection Reagent (VWR, catalog number: 10766-968)
Plat-E cells, a retrovirus packaging cell line described in Morita et al. (2000)
Bicistronic retroviral vector (e.g., pGC-IRES-Thy1.1, pMI-IRES-hCD2, pGC-IRES-BFP2. See Acknowledgments for details).
RPMI 1640 Medium (Fisher Scientific, catalog number: 11-875-119)
DMEM (Fisher Scientific, catalog number: 11-965-118)
Fetal Bovine Serum (Thermo Fisher Scientific, catalog number: 10082147)
1 M HEPES solution (Cytiva, catalog number: SH30237.01)
100× Nonessential Amino Acids (Corning, catalog number: 25-025-ci)
100 mM Sodium Pyruvate (Hyclone, catalog number: sh30239.01)
200 mM L-Glutamine (Hyclone, catalog number: SH30034.02)
100× Penicillin/Streptomycin Solution (Hyclone, catalog number: SV30010)
Puromycin dihydrochloride (Sigma-Aldrich, catalog number: P7255)
Blasticidin S.HCl Solution in HEPES buffer 10 mg/mL (A.G. Scientific, catalog number: B-1247-SOL)
Red Blood Cell (RBC) Lysis Buffer (10×) (Tonbo Biosciences, catalog number: TNB-4300)
Anti-biotin Microbeads (Milteny Biotec, catalog number: 130-090-485)
Mitomycin C (CAS 50-07-7) (Santa Cruz Biotechnology, catalog number: SC-3514B)
3% Acetic Acid with Methylene Blue (StemcellTM Technologies; catalog number: 07060)
Biotin Anti-human/mouse CD45R (B220) [clone: RA3-6B2] (Tonbo Biosciences, catalog number: 30-0452)
Biotin Anti-CD19 monoclonal antibody [clone: HIB19] (Thermo Fisher Scientific, catalog number: 13-0199-82)
Biotin Anti-CD8α rat monoclonal antibody [clone: 53-6.7] (Biolegend, catalog number: 100704)
Biotin Anti-mouse TER-119 [clone: TER-119] (Tonbo Bioscience, catalog number: 30-5921)
Biotin Anti-CD49b (pan-NK cells) rat monoclonal antibody [clone: DX5] (Biolegend, catalog number: 108904)
Biotin Anti-NK1.1 [clone: PK136] (Tonbo Bioscience, catalog number: 30-5941)
Biotin Anti-TCRγδ [clone: GL 3] (eBioscience, catalog number: 13-5711-85)
Biotin Anti-Ly-6G/Ly-6C (Gr-1) rat monoclonal antibody [clone: RB6-8C5] (Biolegend, catalog number: 108404)
Biotin Anti-mouse CD122 (IL-2Rβ) [clone: 5H4] (Biolegend, catalog number: 105904)
Biotin Anti-CD25 rat monoclonal antibody [clone: PC61] (Biolegend, catalog number: 102004)
Biotin Anti-CD44 rat monoclonal antibody [clone: IM7] (Biolegend, catalog number: 103003)
Biotin Anti-CD11b rat monoclonal antibody [clone: M1/70] (Biolegend, catalog number: 101204)
Biotin anti-CD11c monoclonal antibody [clone: 3.9] (Thermo Fisher Scientific, catalog number: 13-0116-82)
Biotin Anti-mouse F4/80 [clone: BM8] (Biolegend, catalog number: 123106)
Ghost DyeTM Violet 510 (Tonbo Biosciences, catalog number: 13-0870-T100)
FITC Anti-mouse TCRβ [clone: H57-597] (Tonbo, catalog number: 35-5961)
Phycoerythrin (PE) Anti-mouse CD25 monoclonal antibody [clone: PC61.5] (Tonbo Biosciences, catalog number: 50-0251)
Allophycocyanin (APC) Anti-CD4 rat monoclonal antibody [clone: GK1.5] (Biolegend, catalog number: 100412)
PE-Cy7 Anti-CD62L rat monoclonal antibody [clone: MEL-14] (Tonbo Bioscience, catalog number: 60-0621-U100)
Pacific BlueTM Anti-human CD2 (hCD2) antibody [clone:TS1/8] (Biolegend, catalog number: 309216)
PE Anti-rat CD90/mouse CD90.1 (Thy-1.1) antibody [clone: OX-7] (Biolegend, catalog number: 202523)
Full DMEM (see Recipes)
Full DMEM + Puromycin/Blasticidin (see Recipes)
Full RPMI (see Recipes)
1× RBC Lysis Buffer (see Recipes)
Primary biotin antibodies master mix (see Recipes)
Magnetic anti-biotin microbeads + sorting antibody master mix (see Recipes)
Equipment
LSR Fortessa X-20 (BD Biosciences)
FACS Aria II Cell Sorter (BD Biosciences)
Avanti J-15R Centrifuge (Beckman Coulter)
Eppendorf Centrifuge 5420 (Eppendorf AG)
MACS® MultiStand (Miltenyi Biotec, catalog number: 130-042-303)
FormaTM Series 3 Water Jacketed CO2 Incubator (Thermo Fisher, catalog number: 4110)
Software
FlowJo (FlowJo, LLC)
FACS DIVA software version 9.0 (BD Biosciences)
GraphPad Prism (GraphPad Software)
Procedure
Retrovirus Production
Note: All handling of retroviral production and transduction occurred under a BSL-2 hood. RV produced from Plat-E are replication incompetent and ecotropic. However, many commonly used genes of interest have been identified as oncogenes, e.g., HRasG12V (Huang et al., 2017).
Maintain Plat-E cells in a T75 flask with full DMEM + Puromycin/Blasticidin (see Recipes).
Seed 4 × 106 Plat-E cells into a 100 mm dish with full DMEM media without Puromycin/Blasticidin.
Transfect Plat-E cells at ~70%–80% confluent, normally ~18–24 h after seeding.
Warm OPTI-MEM and TransIT®-293 to room temperature.
Pipette 1 mL of OPTI-MEM into a polypropylene 1.5 mL Eppendorf tube.
Add 10–15 μg bicistronic retroviral vector plasmid DNA to OPTI-MEM. Gently pipette to mix.
Note: Adjust total plasmid for optimal transfection efficiency. In our experience, 10–15 μg is appropriate for most plasmids.
Before use, vortex TransIT®-293 for 0.5 seconds. Pipette 30–45 μL TransIT®-293 to the OPTI-MEM + plasmid mixture. Gently pipette to mix.
Note: A ratio of 1 μg plasmid DNA to 3 μL TransIT®-293 works well for most plasmids. Adjust total amount of plasmid DNA and ratio DNA:TransIT®-293 reagent to optimize.
Incubate transfection mixture at room temperature for 20 min.
Add transfection mixture dropwise to cells, evenly distributed across the plate. Rock gently back and forth to spread evenly.
Return cells to incubator.
At 24 h post-transfection, collect culture media, and replace with fresh prewarmed full DMEM media.
Centrifuge the collected media at 600 × g and 4 °C for 5 min, then pass through a 0.45 μm filter into a clean 50 mL tube. Store filtered media at 4 °C.
Repeat steps 11 and 12 at 48- and 60-h post transfection, and combine filtered media into a single 50 mL tube.
Add one volume of Retro-X concentrator per three volumes of filtered media.
Gently invert tube to mix.
Incubate at 4 °C overnight.
Centrifuge at 1,500 × g and 4 °C for 45 min.
Discard supernatant. Resuspend pellet in 1/10th of the original volume, in full RPMI media.
Use concentrated RV media immediately, or store at -80 °C in single use aliquots. Under these conditions, RV may be stored up to 2 years after production.
Isolation of Naïve CD4+ T cells
Collect spleen and lymph nodes (superficial cervical, brachial, and inguinal are easily accessible) from IL10GFP/Foxp3RFP dual reporter mouse model.
Homogenize tissues through a 70 μm cell strainer via grinding, and transfer to a 15 mL centrifuge tube.
Centrifuge at 600 × g for 5 min.
Discard supernatant and resuspend cells in 1 mL of 1× RBC lysis buffer (see Recipe 4). Add RBC Lysis Buffer to 5 mL. Incubate at room temperature for 5 min.
Add 2 mL of full RPMI media to neutralize the lysis reaction. Centrifuge at 600 × g for 5 min.
Discard supernatant, resuspend in 1 mL of full RPMI media, and transfer to a 1.5 mL tube.
Centrifuge at 1,500 × g for 1 min, aspirate supernatant, and resuspend in primary biotin antibody master mix (see Recipe 5), using 200 μL per mouse. Remove irregular clumps if any are present.
Incubate on ice for 15 min, shaking every 5 min.
During incubation, prepare magnetic anti-biotin microbeads + sorting antibody master mix (see Recipe 6).
Add 1 mL of full RPMI media to cells. Centrifuge at 1,500 × g for 1 min, aspirate supernatant, and resuspend in magnetic anti-biotin microbeads + sorting antibody master mix.
Incubate cells on ice for 15 min, shaking every 5 min.
While cells are incubating, prepare MS or LS magnetic separation columns on the MACS Multistand. Pre-wet columns with full RPMI media at room temperature. Load a sterile collection tube under the magnetic separation column.
MS column holds one spleen. Pre-wet with 0.5 mL of full RPMI media.
LS column holds four spleens. Pre-wet with 2 mL of full RPMI media.
Add 1 mL of full RPMI media to cells. Centrifuge at 1,500 × g for 1 min, and aspirate supernatant.
Resuspend cells in 550 μL of full RPMI media. Take 500 μL from the top of the tube, and load into magnetic separation columns. Allow media to flow through.
To increase yield, add 550 μL of RPMI to the 1.5 mL tube. Take 500 μL from the top of the tube, and load into magnetic columns.
Allow cells to flow through the column into the sterile collection tube. The naïve T cells are in the flow-through and are ready for sorting (Figure 1A).
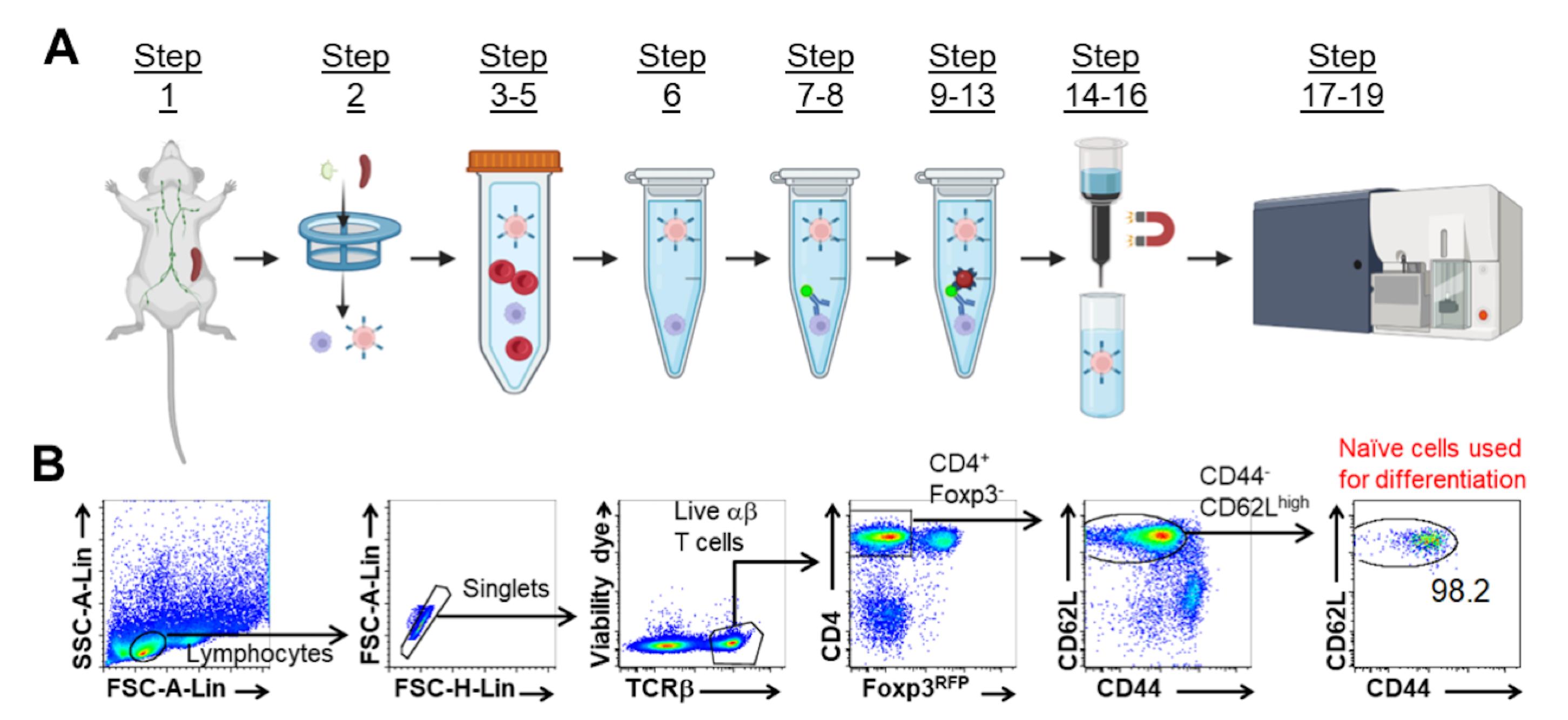
Figure 1. Naïve CD4+ T Cell Isolation Gating Strategy. Schematic representation of naïve CD4+ T cell isolation (A). Representative FACS plots of gating strategy for isolating naïve CD4+ T cells by flow sorting for in vitro differentiation (B). Adapted from Huang et al. (2017).Prepare flow sorting collection tubes by adding 1 mL of cold full RPMI media to 15 mL tubes.
To isolate naïve CD4+ T cells, sort CD44loCD62LhiCD25-Foxp3RFP-TCRβ+CD4+ cells via FACS Aria II Cell Sorter (BD Biosciences). An example of the gating strategy is presented in Figure 1B (Huang et al., 2017).
Centrifuge purified naïve CD4+ T cells at 600 × g for 5 min. Discard supernatant. Resuspend at 1 × 106 cells/mL in full RPMI media.
Tr1 Cell Differentiation
To obtain antigen presenting cells (APCs), homogenize spleen of Rag-/- mouse through a 70 μm strainer into 5 mL of full RPMI media.
Add Mitomycin C to a final concentration of 50 μg/mL. Incubate at 37 °C for 30 min.
Centrifuge at 600 × g for 5 min. Discard supernatant.
Resuspend cells in 1 mL of full RPMI media. Transfer to a 1.5 mL tube.
Wash cells by centrifuging at 1,500 × g for 1 min. Aspirate supernatant. Resuspend in 1 mL of full media. Repeat wash step a total of four times.
Aliquot 10 μL of cells into 90 μL of 3% Acetic Acid with Methylene Blue. Mix by pipetting, and count APCs using a hemocytometer and light microscope. Resuspend at 3 × 106 cells/mL.
Mix equal volumes of Mitomycin C treated Rag-/- APCs and purified naïve CD4+ T cells. The final ratio of naïve CD4+:APCs is 1:3 at 0.5 × 106/mL naïve CD4+ T cells, with 1.5 × 106/mL APCs.
Add anti-CD3ϵ, anti-CD28, and IL-27 to the CD4+:APCs mixture at a final concentration of:
1 μg/mL anti-CD3ϵ (145-2C11)
1 μg/mL anti-CD28 (37.51)
25 ng/mL IL-27
Plate 200 mL of cells per well in a 96-well flat bottom plate.
Incubate at 37 °C and 5% CO2.
Transduction
Note: All handling of retroviral production and transduction occurred under a BSL-2 biosafety cabinet.
Transduce CD4+ T cells after ~18–24 h in culture.
Centrifuge the 96-well plate at 1,500 × g and 37 °C for 5 min.
Carefully remove 170 μL of media, starting from the top of well to avoid disturbing the cells.
Add 20 μL of the concentrated RV (Section A) to the wells.
Empty parent bicistronic retroviral vectors are used as controls (i.e., pGC-IRES-Thy1.1 for gene of interest-IRES-Thy1.1).
Centrifuge the plate at 1,500 × g and 37 °C for 2 h.
Place the 96 well plate back in the incubator at 37 °C for 2 h.
Add 150 μL of prewarmed media to each well with original culture conditions (described in step C8).
Analyze Tr1 cell differentiation, or sort cells for functional assays, 48–72 h post transduction.
Analysis of Tr1 Cell Differentiation
Prepare flow cytometry staining master mix:
30 μL of PBS per sample
1:200 dilution of APC Anti-CD4 Rat [clone: GK1.5], Ghost DyeTM Violet 510, and anti-reporter antibody (e.g., Pacific Blue Anti-hCD2 [clone:TS1/8], PE anti-Thy1.1 [clone: OX-7])
Transfer cells into V-bottom staining plate.
Centrifuge cells at 1,000 × g for 1 min.
Invert plate to remove supernatant.
Add 30 μL of staining master mix to each well, and pipette up and down to resuspend. Incubate at room temperature, protected from light for 20 min.
Add 230 μL of PBS to each well. Centrifuge cells at 1,000 × g for 1 min. Invert plate to remove supernatant.
Resuspend pellet in 280 μL of PBS. Transfer to flow cytometry tubes for analysis.
Use gating strategy in Figure 2, to successfully identify transduced (RV Reporter+) and nontransduced (RV Reporter-) cells. Collect an adequate number of each cell for proper analysis.
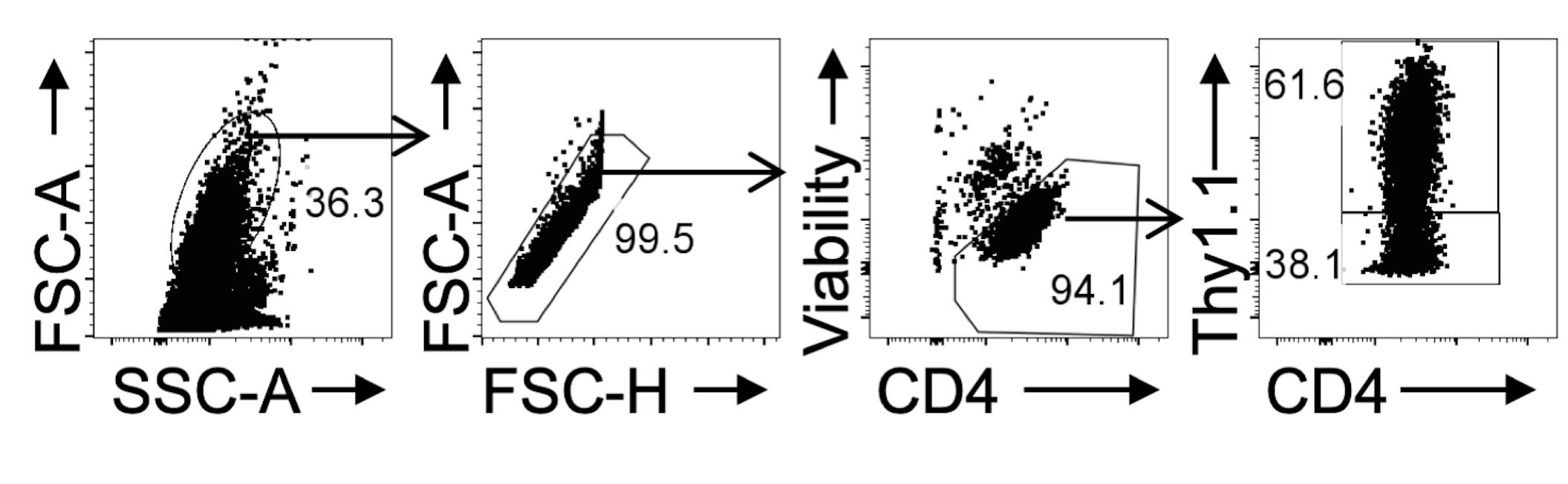
Figure 2. Data Collection Gating Strategy. Naïve IL10GFP/Foxp3RFP reporter CD4+ cells were cultured under Tr1 differentiating conditions and RV transduced with control-Thy1.1. Representative FACS plots of gating strategy for collection of RV transduced Thy1.1+ (Reporter+) and nontransduced Thy1.1- (Reporter-) CD4+ cells.
Sorting Transduced Tr1 Cells for Functional Assays
Combine the cells 48–72 h post transduction in a 15 mL tube.
Centrifuge at 600 × g for 5 min. Aspirate supernatant
Resuspend in full RPMI media with 1:200 dilution of:
Ghost DyeTM Violet 510
APC Anti-CD4 Rat [clone: GK1.5]
Anti-Reporter antibody (e.g., Pacific Blue Anti-hCD2 [clone:TS1/8], PE anti-Thy1.1 [OX-7])
Incubate on ice for 15 min, shaking every 5 min.
Transfer to 1.5 mL tube. Incubate on ice for 15 min, shaking every 5 min.
Add 1 mL of full RPMI media to cells. Centrifuge at 1,500 × g for 1 min, aspirate supernatant.
Resuspend cells in 1 mL of full RPMI media, and transfer to tube for sorting.
Using the gating strategy from Huang et al. (2017) to sort for Lymphocyte/Singlet/Live/CD4+/ RV Reporter+/IL10GFP+Foxp3RFP-.
Note: Including Reporter-IL10GFP+Foxp3RFP- cells is useful as another control.
Proceed to use purified cells in functional assays.
Data analysis
Use gating strategy lymphocyte/singlets/live cells/CD4+/Reporter+CD4+ cells for analysis of Tr1 differentiation (Figure 3).
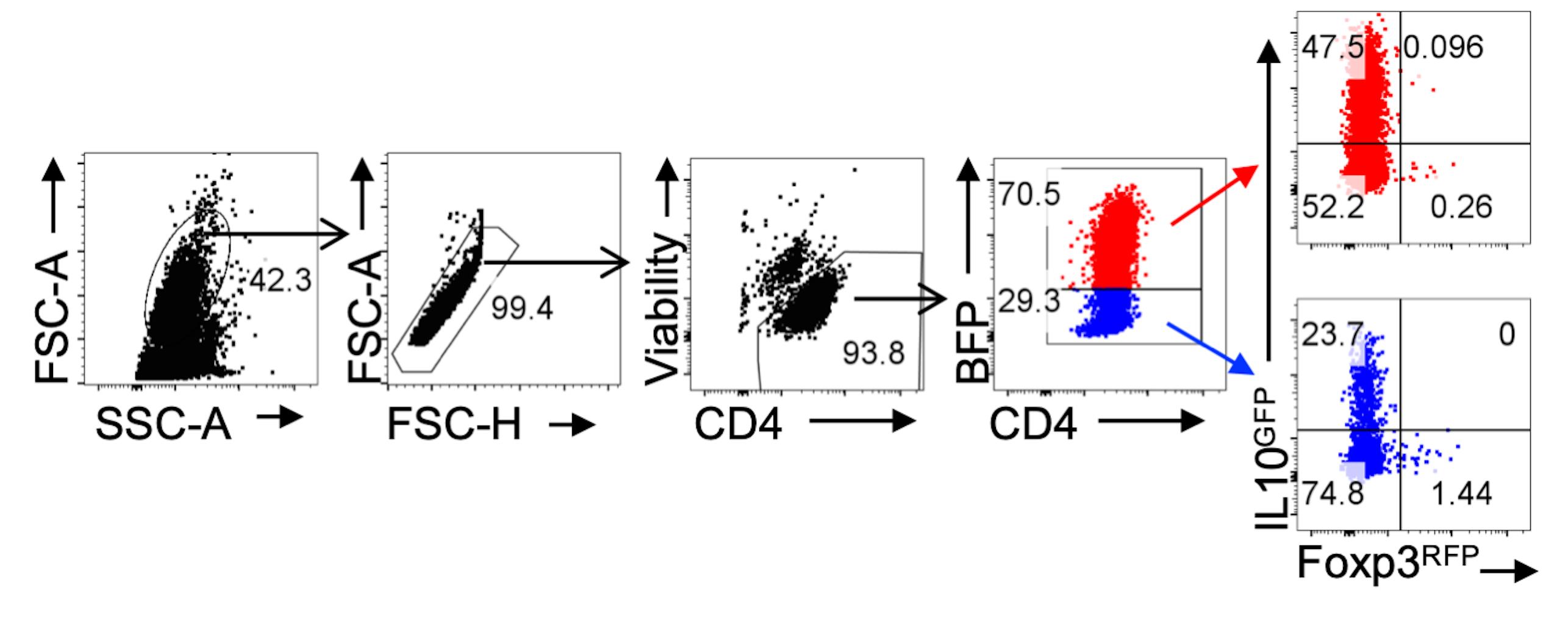
Figure 3. Gating Strategy for Analysis of RV Transduced Tr1 cells. CD4+ T cells were transduced with HRasG12V-IRES-BFP2 RV particles. Representative FACS plots of gating strategy for analysis of RV transduced (BFP+) and nontransduced (BFP-) IL10GFP+ Foxp3RFP- Tr1 cells.The frequency of IL10GFP+Foxp3RFP-CD4+ T cells can be compared via t-test or one-way ANOVA with an appropriate post hoc test in GraphPad Prism, depending on the number of experimental groups. Reporter- cells can be analyzed as an internal control.
Notes
Including the classic Tr1 markers LAG3 and CD49b (Gagliani et al., 2013) provides further evidence of Tr1 differentiation and is highly recommended.
Overexpression of transcription factors in the Reporter+ cells should be verified via nuclear staining. The eBioscienceTM Foxp3/Transcription Factor Staining Buffer Set (Thermo Fisher, 00-5523-00) is recommended.
In our hands, the protocol described here produces high efficiency of transduction with several different bicistronic retroviral vector backbone plasmids, such as pGC-IRES-Thy1.1, pMI-IRES-hCD2, and pGC-IRES-BFP2.
The efficiency of Plat-E cell transfection (Section A) is a critical step in achieving optimal RV titers and downstream RV transduction efficiency. This can be easily assessed via flow cytometry of Plat-E cells after RV media has been harvested, via gating on viable/Reporter+ cells.
The size and biological function of an insert affect both Plat-E transfection efficiency and RV transduction efficiency. In our hands, transduction efficiency drops substantially with an insert ~2,500 bp, unless it promotes T cell activation (e.g., constitutively active STAT5).
The high efficiency of transduction allows this protocol to be used for dual transduction experiments and transduction of CD8+ T cells under IL-10 inducing conditions (Figure 4).
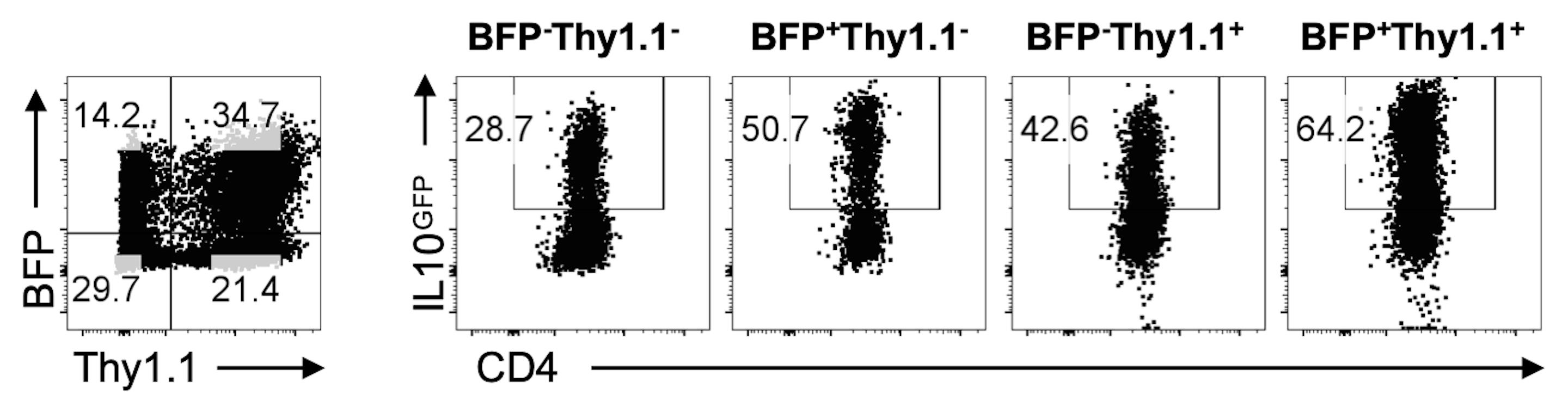
Figure 4. Gating Strategy for Analysis of dual RV Transduced Tr1 cells. Representative FACS plots of IL10GFP+ Foxp3RFP- Tr1 cells transduced with HRasG12V-IRES-BFP2 and caSTAT5-IRES-Thy1.1.
Recipes
Full DMEM
DMEM
10% FBS
10 mM HEPES solution (pH 7.5)
1× Nonessential amino acids
1 mM Sodium pyruvate
2 mM L-Glutamine
1× Penicillin/Streptomycin Solution
Full DMEM + Puromycin/Blasticidin
DMEM
10% FBS
10 mM HEPES solution (pH 7.5)
1× Nonessential amino acids
1 mM Sodium pyruvate
2 mM L-Glutamine
1× Penicillin/Streptomycin Solution
1 μg/mL Puromycin
10 μg/mL Blasticidin
Full RPMI
RPMI 1640 Medium
10% FBS
10 mM HEPES solution (pH 7.5)
1× Nonessential amino acids
1 mM Sodium Pyruvate
2 mM L-Glutamine
1× Penicillin/Streptomycin Solution
1× RBC Lysis Buffer
50 mL of 10× RBC Lysis Buffer
450 mL of ddH2O
Primary biotin antibodies master mix
200 μL of full RPMI media per mouse
Biotin antibodies to a final concentration of:
5 μg/mL: Anti- B220, CD19, and CD8
2.5 μg/mL: Anti- Ter119, DX5/CD49b, NK1.1 1, TCRγδ, Gr-1 1, CD122, CD25 CD11c, CD11b, and F4/80
0.25 μg/mL: Anti-CD44
Magnetic anti-biotin microbeads + sorting antibody master mix
80 μL of full RPMI Media per mouse
20 μL of anti-biotin microbeads per mouse (Milteny Biotec anti-biotin microbeads)
1:200 Ghost DyeTM Violet 510
1:200 FITC Anti-TCRβ [clone: H57-597]
1:200 PE Anti-CD25 [clone: PC61.5]
1:200 APC Anti-CD4 [clone: GK1.5]
1:200 PE-Cy7 Anti-CD62L [clone: MEL-14]
Acknowledgments
This work is generously supported by National Institutes of Health (R21-AI129422, R56-AI146226, R01-AI138570, and R01-AI151139). M.C.M. is supported by a Careers in Immunology Fellowship from the American Association of Immunologists. W.H. is fellow recipient of the Research Publication Grant in Engineering, Medicine, and Science from the American Association of University Women. We would like to express our gratitude to Drs J. Sun, C. Li, and Y. M. Son for the Plat-E cell line and helpful discussions. We also thank Drs A. Pernis and S. Gupta (pGC) and T. Malek and A. Yu (pMI) for retroviral plasmid backbones. pLKO.3 Thy1.1 (gift from Drs C. Benoist and D. Mathis; Addgene plasmid # 14749) and mTagBFP2-Lifeact-7 (gift from Dr. M. Davidson; Addgene plasmid # 54602) were used to clone Thy1.1 and BFP2 into the respective plasmid backbones, respectively. The protocol presented here was adapted from previously published work (Huang et al., 2017).
Competing interests
M.C.M. declares no conflict of interest. A patent disclosure was filed by W.H. and A.A. on the application of RAS signaling in Tr1 cell differentiation and function at Cornell University, when the original research on RAS signaling was published in 2017. W.H. received funding from MegaRobo Technologies, Ltd and A.A. received funding from 3M, which were not relevant to the research presented in this work.
Ethics
All experimental procedures are approved by the Institutional Biosafety and Animal Care and Use Committees at the Louisiana State University under protocol 18015 (12/10/2018 – present) for biosafety level 2 research, and protocols IACUCAM 18-047 (5/23/2018 – 5/22/2021) and IACUCAM 21-039 (4/26/2021 – 4/25/2024) for animal use.
References
- Bohm, L., Maxeiner, J., Meyer-Martin, H., Reuter, S., Finotto, S., Klein, M., Schild, H., Schmitt, E., Bopp, T. and Taube, C. (2015). IL-10 and regulatory T cells cooperate in allergen-specific immunotherapy to ameliorate allergic asthma. J Immunol 194(3): 887-897.
- Eremenko, E., Taylor, Z. V., Khand, B., Zaccai, S., Porgador, A. and Monsonego, A. (2021). An optimized protocol for the retroviral transduction of mouse CD4 T cells. STAR Protoc 2(3): 100719.
- Gagliani, N., Magnani, C. F., Huber, S., Gianolini, M. E., Pala, M., Licona-Limon, P., Guo, B., Herbert, D. R., Bulfone, A., Trentini, F., et al. (2013). Coexpression of CD49b and LAG-3 identifies human and mouse T regulatory type 1 cells.Nat Med 19(6): 739-746.
- Gol-Ara, M., Jadidi-Niaragh, F., Sadria, R., Azizi, G. and Mirshafiey, A. (2012). The role of different subsets of regulatory T cells in immunopathogenesis of rheumatoid arthritis. Arthritis 2012: 805875.
- Huang, W., Solouki, S., Koylass, N., Zheng, S.-G. and August, A. (2017). ITK signalling via the Ras/IRF4 pathway regulates the development and function of Tr1 cells. Nature Communications 8: 15871.
- Karwacz, K., Miraldi, E. R., Pokrovskii, M., Madi, A., Yosef, N., Wortman, I., Chen, X., Watters, A., Carriero, N., Awasthi, A., et al. (2017). Critical role of IRF1 and BATF in forming chromatin landscape during type 1 regulatory cell differentiation. Nat Immunol 18(4): 412-421.
- Kurachi, M., Kurachi, J., Chen, Z., Johnson, J., Khan, O., Bengsch, B., Stelekati, E., Attanasio, J., McLane, L. M., Tomura, M., et al. (2017). Optimized retroviral transduction of mouse T cells for in vivo assessment of gene function.Nat Protoc 12(9): 1980-1998.
- Mascanfroni, I. D., Takenaka, M. C., Yeste, A., Patel, B., Wu, Y., Kenison, J. E., Siddiqui, S., Basso, A. S., Otterbein, L. E., Pardoll, D. M., et al. (2015). Metabolic control of type 1 regulatory (Tr1) cell differentiation by AHR and HIF1-α. Nat Med 21(6): 638-646.
- Mobs, C., Slotosch, C., Loffler, H., Jakob, T., Hertl, M. and Pfutzner, W. (2010). Birch pollen immunotherapy leads to differential induction of regulatory T cells and delayed helper T cell immune deviation. J Immunol 184(4): 2194-2203.
- Morita, S., Kojima, T. and Kitamura, T. (2000). Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther 7(12): 1063-1066.
- Roncarolo, M. G., Gregori, S., Bacchetta, R. and Battaglia, M. (2014). Tr1 cells and the counter-regulation of immunity: natural mechanisms and therapeutic applications. Curr Top Microbiol Immunol 380: 39-68.
- Wu, C., Pot, C., Apetoh, L., Thalhamer, T., Zhu, B., Murugaiyan, G., Xiao, S., Lee, Y., Rangachari, M., Yosef, N., et al. (2013). Metallothioneins negatively regulate IL-27-induced type 1 regulatory T-cell differentiation. Proc Natl Acad Sci U S A 110(19): 7802-7807.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
McGee, M. C., August, A. and Huang, W. (2022). High-Efficiency Retroviral Transduction for Type 1 Regulatory T Cell Differentiation. Bio-protocol 12(17): e4499. DOI: 10.21769/BioProtoc.4499.
Category
Immunology > Immune cell function > Lymphocyte
Biological Engineering > Biomedical engineering
Cell Biology > Cell engineering > Lentiviral delivery
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








