- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Isolation and ex vivo Expansion of Limbal Mesenchymal Stromal Cells
Published: Vol 12, Iss 14, Jul 20, 2022 DOI: 10.21769/BioProtoc.4471 Views: 2985
Reviewed by: Vivien Jane Coulson-ThomasMingxia SunIsabel Moreno

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Thrombopoietin-independent Megakaryocyte Differentiation of Hematopoietic Progenitor Cells from Patients with Myeloproliferative Neoplasms
Chloe A. L. Thompson-Peach [...] Daniel Thomas
Jan 20, 2023 2372 Views

Isolation, Purification, and Culture of Embryonic Melanoblasts from Green Fluorescent Protein–expressing Reporter Mice
Melissa Crawford [...] Lina Dagnino
Sep 5, 2023 2002 Views

Identification and Sorting of Adipose Inflammatory and Metabolically Activated Macrophages in Diet-Induced Obesity
Dan Wu [...] Weidong Wang
Oct 20, 2025 2271 Views
Abstract
Limbal mesenchymal stromal cells (LMSC), a cellular component of the limbal stem cell niche, have the capability of determining the fate of limbal epithelial progenitor cells (LEPC), which are responsible for the homeostasis of corneal epithelium. However, the isolation of these LMSC has proven to be difficult due to the small fraction of LMSC in the total limbal population, and primary cultures are always hampered by contamination with other cell types. We recently published the efficient isolation and functional characterization of LMSC from the human corneal limbus using CD90 as a selective marker. We observed that flow sorting yielded a pure population of LMSC with superior self-renewal capacity and transdifferentiation potential, and supported the maintenance of the LEPC phenotype. Here, we describe an optimized protocol for the isolation of LMSC from cadaveric corneal limbal tissue by combined collagenase digestion and flow sorting with expansion of LMSC on plastic.
Graphical abstract:
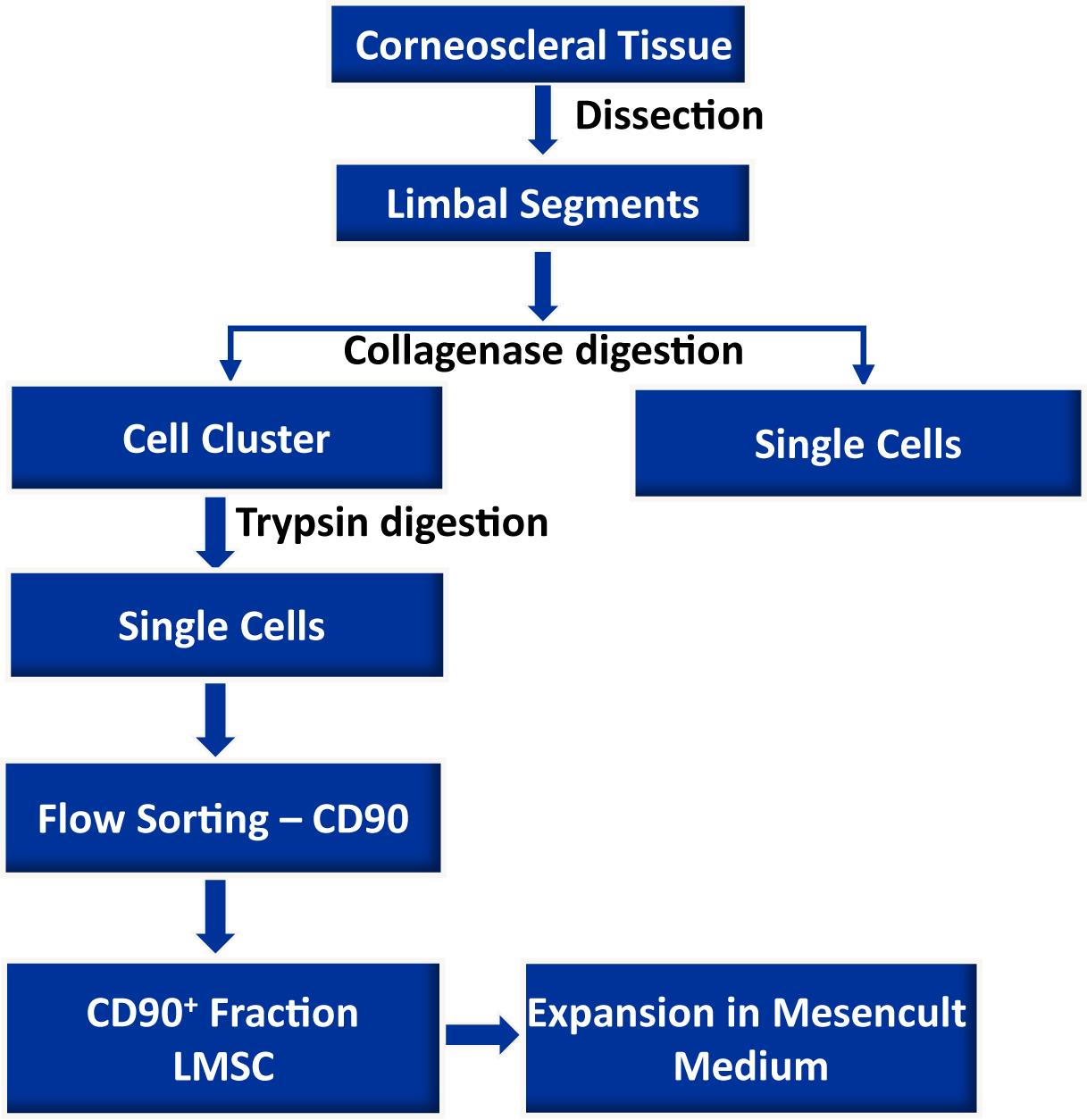
Background
Homeostasis of the corneal epithelium is regulated by limbal epithelial stem/progenitor cells (LEPC) located at a specific anatomic location referred to as the limbal stem cell niche (Gonzalez et al., 2018). It is characterized by limbal vasculature, a specific extracellular matrix composition (ECM), and surrounding non-epithelial limbal niche cells (LNCs) (Shortt et al., 2007; Ordonez et al., 2012; Polisetti et al., 2016) (Figure 1). Limbal mesenchymal stromal cells (LMSC), a cellular component of the limbal stem cell niche, have been shown to support corneal epithelial regeneration during wound healing, and the maintenance of the LEPC phenotype, both in vitro and in vivo (Dziasko et al., 2014; Nakatsu et al., 2014; Li et al., 2018; Polisetti et al., 2016). In addition, LMSC were shown to have potent immunomodulatory, anti-inflammatory, and anti-angiogenic properties, making them potentially attractive tools for clinical use (Funderburgh et al., 2016; Veréb et al., 2016; Al-Jaibaji et al., 2019; Polisetti et al., 2021). Thus, the co-cultivation of LEPC with LMSC might represent an improved strategy to generate cell transplants for patients suffering from limbal stem cell deficiency (Rama et al., 2017; Ghareeb et al., 2020). Previously, LMSC have been isolated either by enzymatic digestion of limbal tissue (dispase, collagenase, or in combination) or by explant culture of limbal tissue followed by enrichment using cell type-specific media (Chen et al., 2011; Xie et al., 2012; Li et al.,2012; Li et al., 2014; Chen et al., 2015; González et al., 2013; Polisetty et al., 2008; Xiao et al., 2020). The main disadvantage of these methods is the contamination by other cell types (Polisetty et al., 2008; Li et al., 2012). Collagenase digestion of limbal tissue results in cell clusters consisting of 80% epithelial cells and 20% LMSC, whereas a combination of dispase and collagenase yields clusters composed of approximately 95% LMSC (Li et al., 2012). Thus, current protocols for LMSC purification require culturing of at least one passage to eliminate contaminating cells. Thus, research on freshly isolated LMSC has been hampered by the lack of a good protocol for isolating this cell type.
Here, we present an optimized protocol for the isolation of LMSC from organ-cultured corneal samples by means of fluorescence-activated cell sorting (FACS), using CD90 as a selective marker (Polisetti et al., 2022).
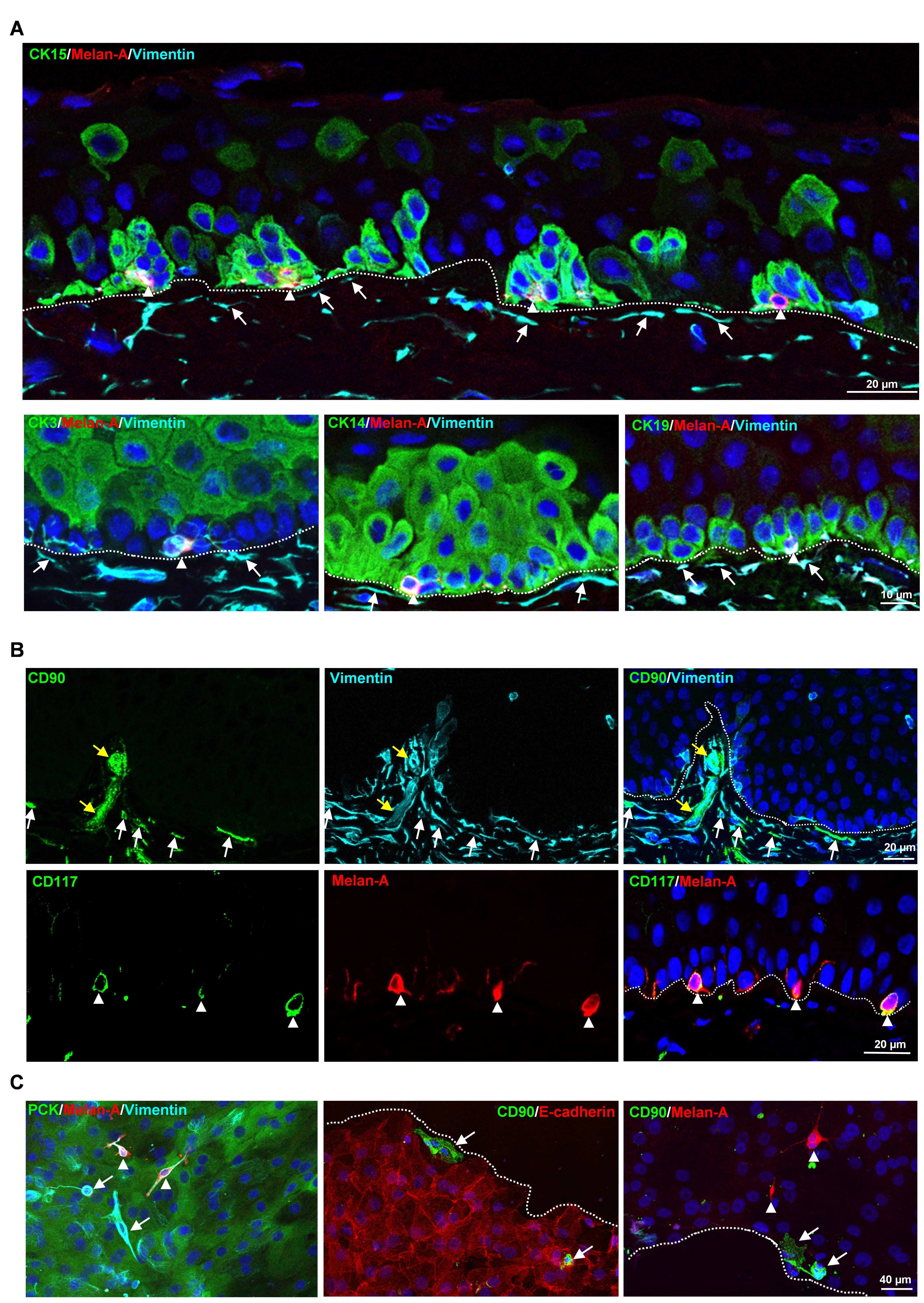
Figure 1. Localization of limbal niche cells in situ. A. Triple immunostaining analysis of limbal tissue sections showing melanocytes [melan-A+ (red) vimentin+ (cyan) cells, arrow heads] in close contact with clusters of cytokeratin (CK)15+, CK14+, CK19+ (green) limbal epithelial progenitor cells (LEPC), whereas sub-epithelial stromal cells [vimentin+ cells (cyan), arrows] were in close association with basal limbal epithelial cells, and not with more superficial CK3+ cells (green). Dashed line represents the basement membrane (BM), and nuclear counterstaining was done with 4′,6-diamidino-2-phenylindole (DAPI, blue). B. Double immunostaining of limbal sections showing the co-localization of CD90 (green) and vimentin (cyan) in the sub-epithelial stromal cells (white arrows), which were in close association with basal layers of limbal epithelium (dotted line represents the BM), as well as blood vessels of the limbal stroma (yellow arrows). The limbal sections also show the co-localization of CD117 (green) and melan A (red) in the melanocytes (arrow heads) at the basal layer of the limbal epithelium. Nuclear counterstaining with DAPI (blue). C. Immunofluorescence analysis of cultured limbal clusters showing the expression of keratins (PCK, green) and vimentin (cyan) in epithelial cells, melan-A (red) and vimentin (cyan) expression in melanocytes (arrow heads), and only vimentin expression in stromal cells (arrows). Double immunostaining of cultured limbal clusters showing the CD90+ stromal cells (green, arrow) at the edge of clusters and also in between E-cadherin+ epithelial cells (red, dashed line represents the edge of the cluster), whereas melan-A+ melanocytes were located between the cells (red, arrow heads). Nuclear counterstaining with DAPI (blue). Reprinted from Polisetti et al. (2022), licensed under a CC BY 4.0.
Materials and Reagents
12-well plate (Corning, Costar®, catalog number:3513)
Micropipette tips (0.5–20 µL, 100–200 µL, 1,000 µL) (Greiner Bio-One)
60 mm cell culture dish (Corning, Falcon®, catalog number: 353004)
100 mm cell culture dish (Corning, Falcon®, catalog number: 353003)
Syringe filter 0.2 μm (VWR, catalog number: 28145-501)
Disposable Scalpel blades No. 10 (pfm Medical ag, Feather®, catalog number: 201000010)
Serological pipettes (5 mL, 10 mL) (Corning, StripetteTM)
15 mL conical tubes (Greiner Bio-One, catalog number:188271)
50 mL conical tubes (Greiner Bio-One, catalog number:227261)
T75 flasks (Corning, catalog number: CLS430641)
Reversible cell strainers (Stem Cell Technologies, catalog number:27215)
Cell filter 20 µm (Cell TricsTM, Sysmex Partec GmbH, catalog number:04-004-2325)
FACS tubes (5 mL polystyrene round-bottom tube, Falcon, catalog number: 352058)
Collagenase A (Sigma-Aldrich, Roche Diagnostics, catalog number: 10103578001)
Dulbecco’s Phosphate Buffered Saline (DPBS) (no calcium, no magnesium) (Thermo Fisher Scientific, GibcoTM, catalog number: 14190094)
0.25% Trypsin-EDTA (Thermo Fisher Scientific, Gibco®, catalog number: 25200056)
Dulbecco’s Modified Eagle Medium (DMEM) high glucose (Thermo Fisher Scientific, GibcoTM, catalog number: 11960044)
Fetal Bovine Serum (FBS) (Thermo Scientific, GibcoTM, catalog number: 10082147)
Penicillin-Streptomycin (Sigma-Aldrich, catalog number: P4333)
MesenCultTM MSC Basal Medium (Human) (Stemcell Technologies, catalog number: 05401)
MesenCultTM MSC Stimulatory Supplement (Human) (Stemcell Technologies, catalog number: 05402)
70% Ethanol
0.5 M EDTA (Invitrogen, catalog number: AM9260G)
CD11c-PE (Biolegend, catalog number: 337205)
CD14-PE (Biolegend, catalog number: 301805)
CD19-PE (Biolegend, catalog number: 302207)
CD44-PE (Biolegend, catalog number: 397503)
CD45-PE (Biolegend, catalog number: 304007)
CD73-PE (Biolegend, catalog number: 344003)
CD90-PE (Abcam, catalog number: 328109)
CD90-APC (BD biosciences, catalog number: 559869)
CD105-PE (Biolegend, catalog number: 323205)
Mouse IgG2a, k Isotype-APC (eBioscience, catalog number: 17-4724-81)
Mouse IgG2a, k Isotype-PE (Biolegend, catalog number: 400212)
Cytokeratin AE1/AE3 (DAKO, catalog number:M3515)
Melan-A (Abcam, catalog number: EPR20380)
Vimentin (R&D Systems, catalog number: MAB2105)
4′,6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich, catalog number: MBD0015)
Collagenase solution (see Recipes)
Mesencult complete medium (see Recipes)
FACS Buffer (see Recipes)
Equipment
Pipette aid (BRAND, accu-jet® pro)
Micropipette (Eppendorf Research plus, P20, P200, P1000)
Forceps (Dumont, 5-Dumoxel®-H)
Hemocytometer (MARIENFELD, Neubauer, catalog number: 0640130)
Biosafety cabinet (Thermo ScientificTM, Type S2020 1.2)
CO2 incubator (Thermo ScientificTM, HeracellTM 240i)
Centrifuge (Thermo ScientificTM, Heraeus Multifuge 1S-R)
Phase contrast inverted microscope with a camera (ZEISS, Objectives 4×, 10×, 20×)
Freezer -20°C (Liebherr)
Refrigerator 2–8°C (Siemens)
FACS Aria II sorter (BD Biosciences)
Water bath (GFL®, catalog number: 1013)
Software
CapturePro 2.10.0.1 (JENOPTIC Optical systems GmbH)
FACSDiva software (BD Pharmingen, BD Biosciences)
FlowJo software (Tree Star)
Procedure
The dissection of limbus and preparation of limbal clusters is similar to the previously published procedure (Polisetti et al., 2019).
Dissection of limbus
Organ-cultured corneoscleral tissues, which are not suitable for transplantation due to low corneal endothelial cell density (<2,200 cells/mm2), or the presence of neurological disorders or malignancies in the donor, were obtained from the cornea bank with appropriate research consent and ethical approval. Donor cornea remnants after corneal endothelial transplant preparation are also a valuable source, if appropriate research consent has been obtained.
Place the organ-cultured corneoscleral tissue in a 60 mm culture dish, and wash twice with DPBS. Cut the tissue into four equal quarters, using a scalpel blade and forceps (see Video 1).
Note: For single preparation, use 4–6 corneoscleral tissues to get enough LMSC for downstream applications. Organ-cultured corneoscleral tissue used in this study was light pigmented donor limbal tissue obtained from donors with a mean age of 69.8 ± 10.7 years, and the culture duration was 24.0 ± 4.9 days, after the postmortem time of 33.54 ± 17.4 h.
Make incisions at 1 mm before and beyond the anatomical limbus to get limbal segments (see Video 1). The limbal segments are further dissected into 2–3 segments, as shown in the video.
Video 1. Limbal cell isolationIsolation of limbal mesenchymal stromal cells
Place the limbal segments in a 60-mm dish containing 5 mL of collagenase A (2 mg/mL), and cut the limbal segments into smaller pieces (2–3 pieces) with a scalpel blade. Incubate at 37°C with 5% CO2 overnight, to digest the stromal collagen and obtain limbal cell clusters.
After incubation, triturate the suspension inside the dish with an up and down motion using a 1-mL pipette (P1000), and observe for the presence of cell clusters and single cells in the microscope (Figure 2B). The cell clusters are supposed to consist of limbal epithelial cells, stromal, and melanocyte niche cells (as shown in Figure 1C).
Note: In case of incomplete digestion of limbal segments after overnight incubation and trituration, re-incubate in the same solution at 37°C with 5% CO2 for an additional 2 h to achieve complete digestion. On the contrary, over-digestion of tissue (more than 20 h) might adversely affect cell viability and the quality of cells.
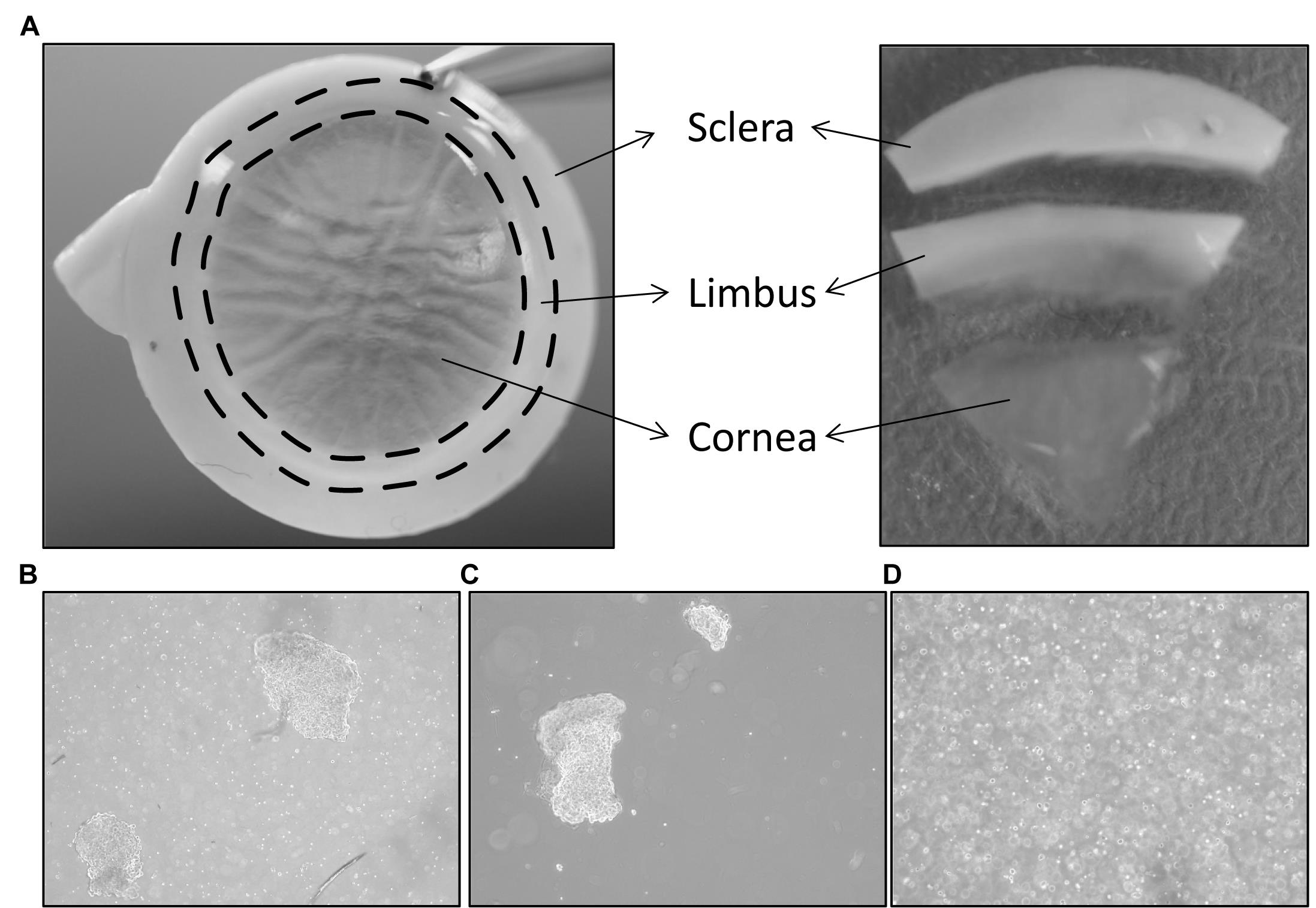
Figure 2. Isolation of limbal cluster cells. A. The corneal scleral rim (left) was cut into sectors, and each sector was trimmed off 1 mm before and after the limbal region (right). Reprinted from Polisetti et al. (2019), licensed under a CC BY 4.0. B. Different sizes of limbal clusters and single cells (left) formed after overnight incubation of limbal segments in collagenase solution (x40 magnification). C. Limbal clusters separated from single cells after filtration. D. Single cell suspension of limbal cells after digestion of limbal clusters with trypsin-EDTA (x40 magnification).Separate limbal cell clusters from single cells using cell filters with a pore size of 20 µm that enable single cells to pass through the cell filter and the clusters to be retained. Wash the filters twice with DPBS, to remove any remaining single cells. Reverse the strainer and place it on a 60 mm dish. Add 0.25% trypsin-EDTA (5 mL) to flush clusters (Figure 2C) into a petri dish, and incubate at 37°C for 10–15 min, to dissociate the clusters into single cells.
Note: In place of 20 µm cell filters, 37 µm reversible cell strainers can be used. The single cell suspension obtained after filtration can either be discarded or used for other purposes, such as to obtain limbal fibroblasts.
After incubation, triturate the cell suspension with an up and down motion with a 1-mL pipette. Observe the cell suspension under the microscope (Figure 2D). Inhibit trypsin digestion by adding 5 mL of pre-warmed DMEM (37°C in water bath) containing 10% FBS. Transfer the cell suspension into a 15-mL Falcon tube, and centrifuge at 200 × g for 5 min.
Note: In case of incomplete dissociation of clusters after 15 min incubation and trituration, re-incubate in the same solution at 37°C with 5% CO2 for an additional 5 min to achieve complete dissociation. On the contrary, prolonged dissociation of clusters might adversely affect cell viability and the quality of cells.
After centrifugation, resuspend the cell pellet by pipetting up and down using a P200 pipette in 200 µL of FACS buffer (see Recipe 3).
Fluorescence Activated Cell Sorting (FACS)
Transfer the cell suspension to FACS tubes (100 µL/tube) and add a mouse APC-conjugated anti-human CD90 antibody (5 µL/106 cells) to one tube, and an IgG2a-Isotype APC to another tube at 4°C. Gently vortex the samples, and incubate on ice for 45 min, tapping at 15 min intervals.
Note: If the cell number is high, i.e., more than 106 cells, adjust the volumes and concentration of antibody accordingly.
After incubation, add 1 mL of FACS buffer to each FACS tube, and centrifuge the cells at 400 × g for 5 min. Repeat the washing twice.
After washing, add 500 µL of FACS buffer containing DAPI (1:5000), to exclude dead cells, and proceed to flow sorting using a FACS Aria II sorter (Polisetti et al., 2022).
The gating strategy is shown below (Figure 3).
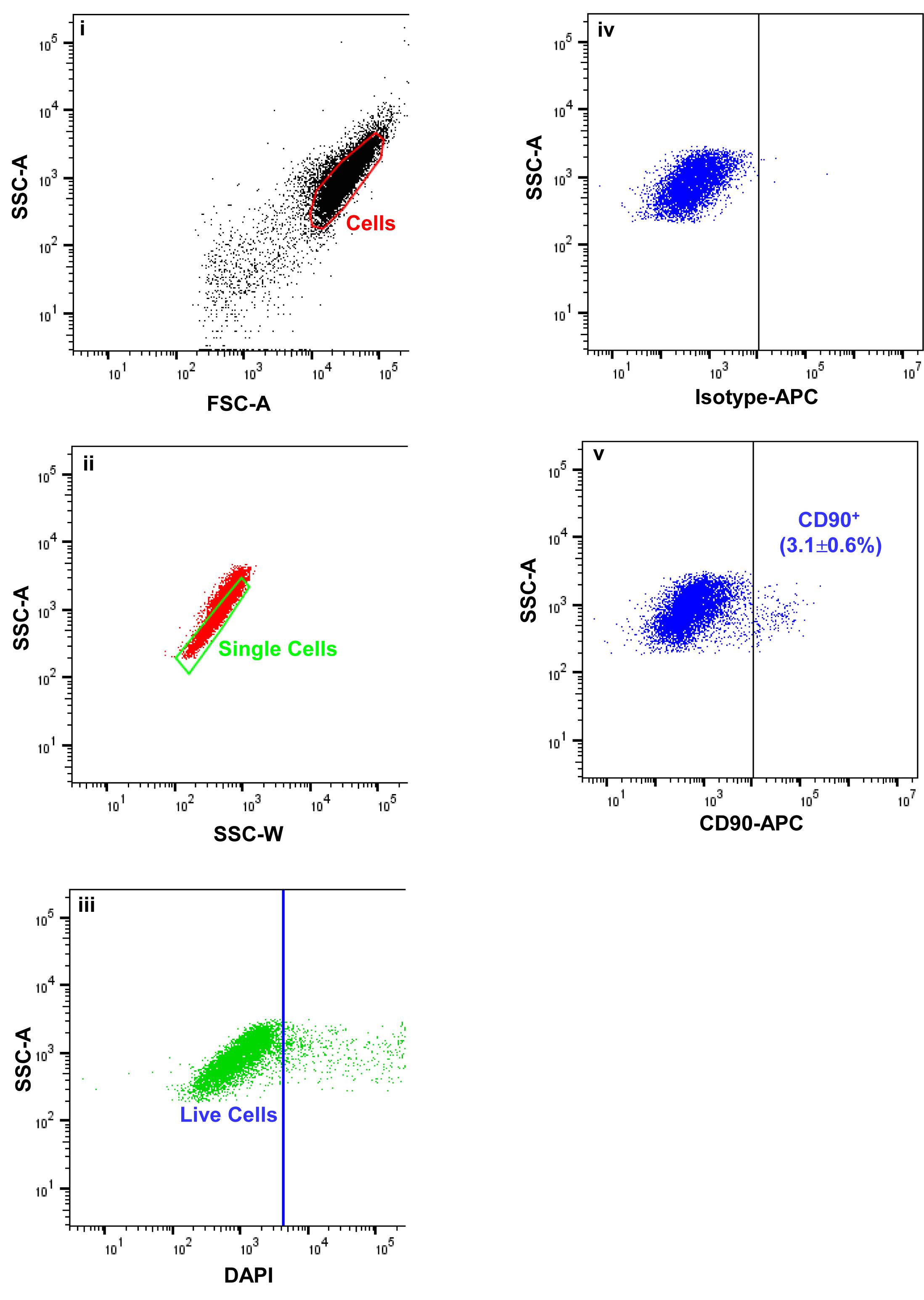
Figure 3. Fluorescence activated cells sorting (FACS) images demonstrating the gating strategy used to isolate limbal mesenchymal stromal cells. Forward scatter (FSC-A) vs. side scatter (SSC-A) graph showing the cells of interest selected on the basis of size and granularity (i). Side scatter area vs. width graph showing the selection of single cells by excluding doublets or clumps, (ii) followed by dead cell exclusion using 4′,6-diamidino-2-phenylindole DAPI (iii). The isotype control graph shows the set of gates (iv) used to select the CD90+ cells (iv). Percentages (%) of positive cells are expressed as the means ± SEM.Expansion of limbal mesenchymal stromal cells
Seed the sorted CD90+ LMSC onto a well of a 12-well plate.
Note: The number of CD90+ cells per limbus (150–900) varies from sample to sample. The CD90- populations mainly contain LEPC, and can be used to enrich LEPC, using cell type-specific media (Polisetti et al., 2020).
Cultivate the LMSC at 37°C with 5% CO2 in Mesencult complete medium to expand LMSC. Change media every 2 days.
Visualize the morphology of LMSC by phase-contrast microscopy. LMSC appear as spindle-shaped, elongated cells with prominent nucleoli (Figure 4).
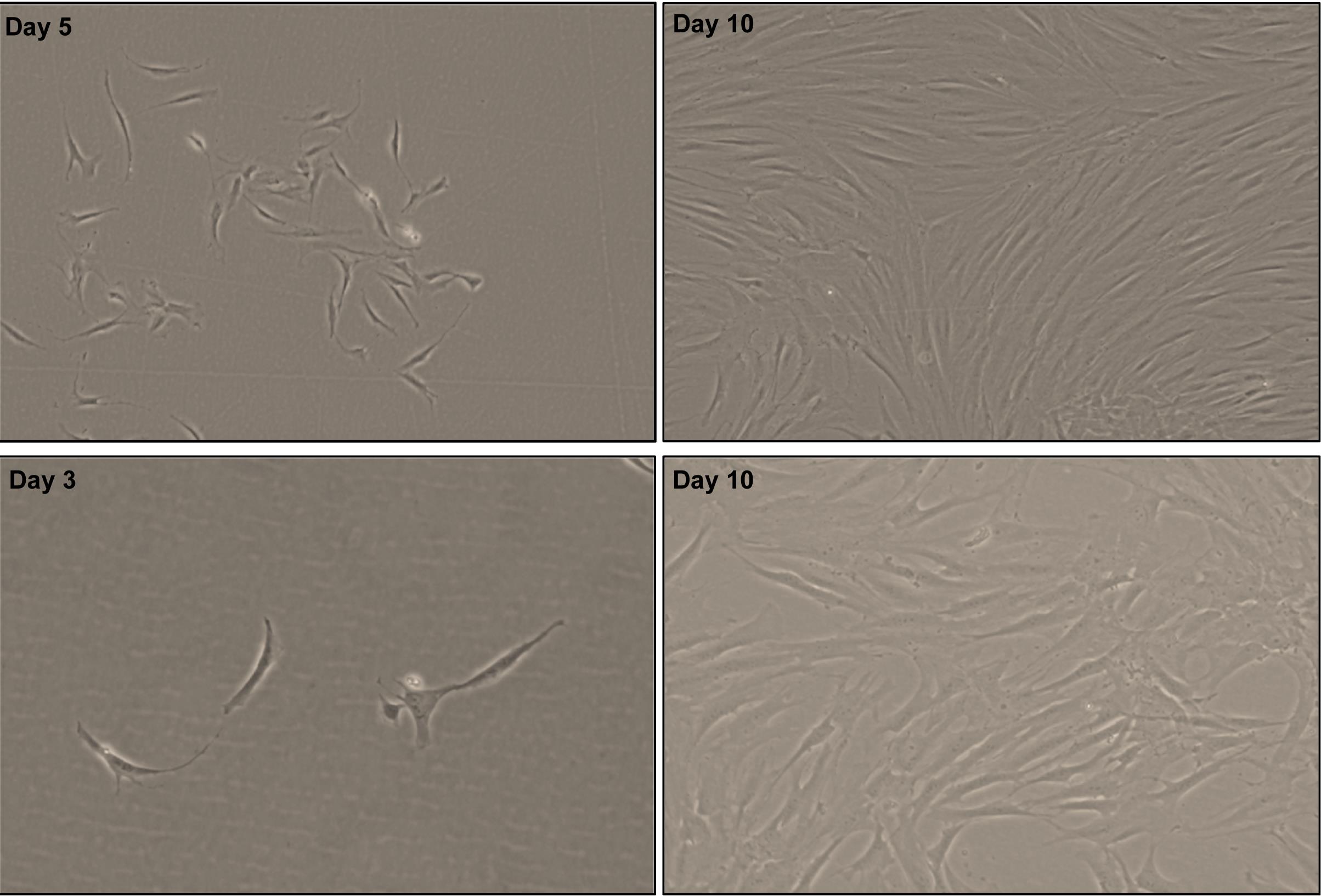
Figure 4. Phase contrast images showing the spindle shaped, elongated with prominent nucleolus of CD90+ cells after day 3, 5, and 10 of seeding [×40 magnification (upper row); ×100 magnification (bottom row)].Sub-cultivation of limbal mesenchymal stromal cells
Remove the media from the culture vessel at 70 to 80% confluency.
Wash the cells using DPBS, and add 1 mL of trypsin-EDTA (0.25%; pre-warmed at 37°C in a water bath). Incubate at 37°C with 5% CO2 for 5 min.
After incubation, add 2 mL of DMEM containing 10% FBS to inhibit trypsin action, and mix well.
Transfer the cell suspension to a 15-mL tube, and centrifuge at 200 × g for 5 min. Resuspend the cell pellet in Mesencult medium, and count the total cell number using a hemocytometer.
Use cells for the application of choice or for subculturing.
Note: Over-confluence (more than 80%) and prolonged trypsin digestion (more than 5 min) adversely affect cell viability and the quality of cells during sub-culturing. Always passage cells at 70 to 80% confluence. Avoid prolonged incubations in trypsin.
To evaluate the LMSC characteristics, phenotypic profile, colony forming efficiency, growth characteristics and differentiation potential have been tested (Figure 5). Please refer to the published article for the detail protocols (Polisetti et al., 2022).
Data analysis
The conditions provided in this protocol have been optimized to isolate and expand LMSCs. A detailed analysis of the isolation and expansion of the LMSCs can be found in Polisetti et al. (2022).
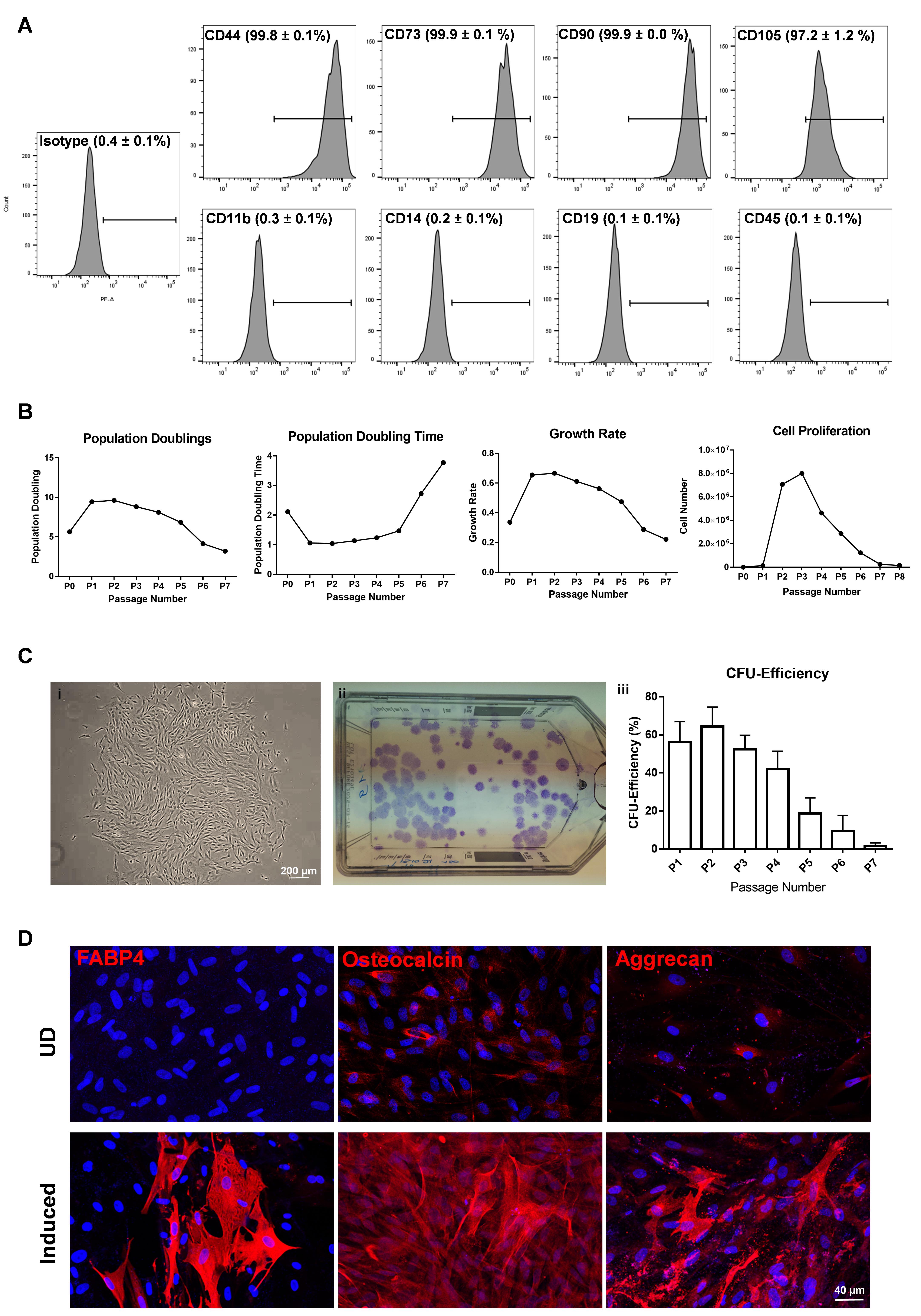
Figure 5. Phenotypic profile and functional characterization of CD90+ (LMSC) cells. A. Flow cytometry analysis showing the expression of CD markers. Percentage of cells expressed as mean ± SEM of four individual experiments. B. Graphs showing the population doublings [log10(y/x)/log102, where y is the final density of the cells, and x is the initial seeding density of the cells], population doubling time [(t − t0)log2/(logy – logx), where t, t0 represents the time at cell counting, y equals the number of cells at time t, and x equals the number of cells at time t0], growth rate [ln(Nt/No)/t, where Nt represents final cell number, No represents the initial cell number, and t equals the number of days in culture], and proliferation potential of LMSC over the passages. Data are expressed as means of five individual experiments. C. Phase contrast micrograph showing the LMSC colony (i), and T75 flask showing crystal violet stained colonies of LMSC (ii). The graph represents the colony forming efficiency of LMSC over the passages. Percentage of colonies expressed as means ± standard deviation (n = 5). D. Immunostaining analysis showing the expression of fatty acid binding protein 4 (FABP4), osteocalcin, and aggrecan in adipogenic, osteogenic, and chondrogenic induced cells, respectively. No staining has been seen for FABP4 in undifferentiated (UD) controls, but weak staining was observed for osteocalcin and aggrecan in UD controls. Nuclear counterstaining with DAPI (blue).
Reprinted from Polisetti et al. (2022), licensed under a CC BY 4.0.
Recipes
Collagenase solution (2 mg/mL)
Reagent Collagenase A Components and Preparation 500 mg Collagenase A 220 mL of DMEM High Glucose 25 mL of fetal calf serum 5 mL of Penicillin-Streptomycin Mix well by inverting Method of Sterilization Sterile Filter (0.2 µm) Note Prepare 10 mL aliquots Storage -20°C MesenCult Medium
Medium MesenCultTM medium complete Components and Preparation 450 mL of MesenCultTM MSC Basal Medium 0 mL of MesenCultTM MSC Stimulatory Supplement 5 mL of Penicillin-Streptomycin Mix well by inverting Method of Sterilization None Note Prepare aliquots if needed Storage 1 month at 2–8°C FACS buffer (2% FBS and 0.5mM EDTA in DPBS)
Components and Preparation 1 mL of FBS 25 µL of 0.5 M EDTA 24 mL of DPBS Mix well by inverting Method of Sterilizaton None Note Always use fresh
Acknowledgments
This protocol was adapted from previous work (Polisetti et al., 2022).
Competing interests
The authors declare that they have no competing interests.
References
- Al-Jaibaji, O., Swioklo, S. and Connon, C. J. (2019). Mesenchymal stromal cells for ocular surface repair. Expert Opin Biol Ther 19(7): 643-653.
- Chen, S. Y., Hayashida, Y., Chen, M. Y., Xie, H. T. and Tseng, S. C. (2011). A new isolation method of human limbal progenitor cells by maintaining close association with their niche cells. Tissue Eng Part C Methods 17(5): 537-548.
- Chen, S. Y., Han, B., Zhu, Y. T., Mahabole, M., Huang, J., Beebe, D. C. and Tseng, S. C. (2015). HC-HA/PTX3 Purified From Amniotic Membrane Promotes BMP Signaling in Limbal Niche Cells to Maintain Quiescence of Limbal Epithelial Progenitor/Stem Cells. Stem Cells 33(11): 3341-3355.
- Dziasko, M. A., Armer, H. E., Levis, H. J., Shortt, A. J., Tuft, S. and Daniels, J. T. (2014). Localisation of epithelial cells capable of holoclone formation in vitro and direct interaction with stromal cells in the native human limbal crypt. PLoS One 9(4): e94283.
- Funderburgh, J. L., Funderburgh, M. L. and Du, Y. (2016). Stem Cells in the Limbal Stroma. Ocul Surf 14(2): 113-120.
- Ghareeb, A. E., Lako, M. and Figueiredo, F. C. (2020). Recent Advances in Stem Cell Therapy for Limbal Stem Cell Deficiency: A Narrative Review. Ophthalmol Ther 9(4): 809-831.
- Gonzalez, G., Sasamoto, Y., Ksander, B. R., Frank, M. H. and Frank, N. Y. (2018). Limbal stem cells: identity, developmental origin, and therapeutic potential. Wiley Interdiscip Rev Dev Biol 7(2).
- González, S. and Deng, S. X. (2013). Presence of native limbal stromal cells increases the expansion efficiency of limbal stem/progenitor cells in culture. Exp Eye Res 116: 169-176.
- Li, G. G., Zhu, Y. T., Xie, H. T., Chen, S. Y. and Tseng, S. C. (2012). Mesenchymal stem cells derived from human limbal niche cells. Invest Ophthalmol Vis Sci 53(9): 5686-5697.
- Li, G., Zhang, Y., Cai, S., Sun, M., Wang, J., Li, S., Li, X., Tighe, S., Chen, S., Xie, H. and Zhu, Y. (2018). Human limbal niche cells are a powerful regenerative source for the prevention of limbal stem cell deficiency in a rabbit model. Sci Rep 8(1): 6566.
- Li, Y., Inoue, T., Takamatsu, F., Kobayashi, T., Shiraishi, A., Maeda, N., Ohashi, Y. and Nishida, K. (2014). Differences between niche cells and limbal stromal cells in maintenance of corneal limbal stem cells. Invest Ophthalmol Vis Sci 55(3): 1453-1462.
- Nakatsu, M. N., Gonzalez, S., Mei, H. and Deng, S. X. (2014). Human limbal mesenchymal cells support the growth of human corneal epithelial stem/progenitor cells. Invest Ophthalmol Vis Sci 55(10): 6953-6959.
- Ordonez, P. and Di Girolamo, N. (2012). Limbal epithelial stem cells: role of the niche microenvironment. Stem Cells 30(2): 100-107.
- Polisetti, N., Giessl, A., Zenkel, M., Heger, L., Dudziak, D., Naschberger, E., Stich, L., Steinkasserer, A., Kruse, F. E. and Schlotzer-Schrehardt, U. (2021). Melanocytes as emerging key players in niche regulation of limbal epithelial stem cells. Ocul Surf 22: 172-189.
- Polisetti, N., Schlunck, G., Reinhard, T., Kruse, F. E. and Schlotzer-Schrehardt, U. (2020). Isolation and ex vivo Expansion of Human Limbal Epithelial Progenitor Cells. Bio-protocol 10(18): e3754.
- Polisetti, N., Sharaf, L., Schlotzer-Schrehardt, U., Schlunck, G. and Reinhard, T. (2022). Efficient Isolation and Functional Characterization of Niche Cells from Human Corneal Limbus. Int J Mol Sci 23(5).
- Polisetti, N., Zenkel, M., Menzel-Severing, J., Kruse, F. E. and Schlotzer-Schrehardt, U. (2016). Cell Adhesion Molecules and Stem Cell-Niche-Interactions in the Limbal Stem Cell Niche. Stem Cells 34(1): 203-219.
- Polisetty, N., Fatima, A., Madhira, S. L., Sangwan, V. S. and Vemuganti, G. K. (2008). Mesenchymal cells from limbal stroma of human eye. Mol Vis 14: 431-442.
- Rama, P., Ferrari, G. and Pellegrini, G. (2017). Cultivated limbal epithelial transplantation. Curr Opin Ophthalmol 28(4): 387-389.
- Shortt, A. J., Secker, G. A., Munro, P. M., Khaw, P. T., Tuft, S. J. and Daniels, J. T. (2007). Characterization of the limbal epithelial stem cell niche: novel imaging techniques permit in vivo observation and targeted biopsy of limbal epithelial stem cells. Stem Cells 25(6): 1402-1409.
- Veréb, Z., Póliska, S., Albert, R., Olstad, O. K., Boratkó, A., Csortos, C., Moe, M. C., Facskó, A. and Petrovski, G. (2016). Role of Human Corneal Stroma-Derived Mesenchymal-Like Stem Cells in Corneal Immunity and Wound Healing. Sci Rep 6: 26227.
- Xiao, Y. T., Qu, J. Y., Xie, H. T., Zhang, M. C. and Zhao, X. Y. (2020). A Comparison of Methods for Isolation of Limbal Niche Cells: Maintenance of Limbal Epithelial Stem/Progenitor Cells. Invest Ophthalmol Vis Sci 61(14): 16.
- Xie, H. T., Chen, S. Y., Li, G. G. and Tseng, S. C. (2012). Isolation and expansion of human limbal stromal niche cells. Invest Ophthalmol Vis Sci 53(1): 279-286.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Polisetti, N., Sharaf, L., Reinhard, T. and Schlunck, G. (2022). Isolation and ex vivo Expansion of Limbal Mesenchymal Stromal Cells. Bio-protocol 12(14): e4471. DOI: 10.21769/BioProtoc.4471.
Category
Stem Cell > Adult stem cell > Mesenchymal stem cell
Biological Engineering > Biomedical engineering
Cell Biology > Cell isolation and culture > Cell isolation > Flow cytometry
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










