- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Expression, Purification, and in vitro Enzyme Activity Assay of a Recombinant Aldehyde Dehydrogenase from Thermus thermophilus, using an Escherichia coli host
Published: Vol 12, Iss 9, May 5, 2022 DOI: 10.21769/BioProtoc.4401 Views: 3371
Reviewed by: Alba BlesaAnna M. KöhlerAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Thermus thermophilus CRISPR Cas6 Heterologous Expression and Purification
Junwei Wei [...] Yingjun Li
Jul 20, 2025 2193 Views

Prokaryotic Expression and Purification of the hSox2-HMG Domain
Lijie Yang [...] Jingjun Hong
Aug 20, 2025 2420 Views

On-Column Dual-Gradient Refolding for Efficient Recovery of Insoluble Affinity-Tagged Recombinant Proteins
Anna Vlaskina [...] Maxim Patrushev
Feb 5, 2026 174 Views
Abstract
Based on previous in-depth characterisation, aldehyde dehydrogenases (ALDH) are a diverse superfamily of enzymes, in terms of both structure and function, present in all kingdoms of life. They catalyse the oxidation of an aldehyde to carboxylic acid using the cofactor nicotinamide adenine dinucleotide (phosphate) (NAD(P)+), and are often not substrate-specific, but rather have a broad range of associated biological functions, including detoxification and biosynthesis. We studied the structure of ALDHTt from Thermus thermophilus, as well as performed its biochemical characterisation. This allowed for insight into its potential substrates and biological roles.
In this protocol, we describe ALDHTt heterologous expression in E. coli, purification, and activity assay (based on Shortall et al., 2021). ALDHTt was first copurified as a contaminant during caa3-type cytochrome oxidase isolation from T. thermophilus. This recombinant production system was employed for structural and biochemical analysis of wild-type and mutants, and proved efficient, yielding approximately 15–20 mg/L ALDHTt. For purification of the thermophilic his-tagged ALDHTt, heat treatment, immobilized metal affinity chromatography (IMAC), and gel filtration chromatography were used. The enzyme activity assay was performed via UV-Vis spectrophotometry, monitoring the production of reduced nicotinamide adenine dinucleotide (NADH).
Graphical abstract:
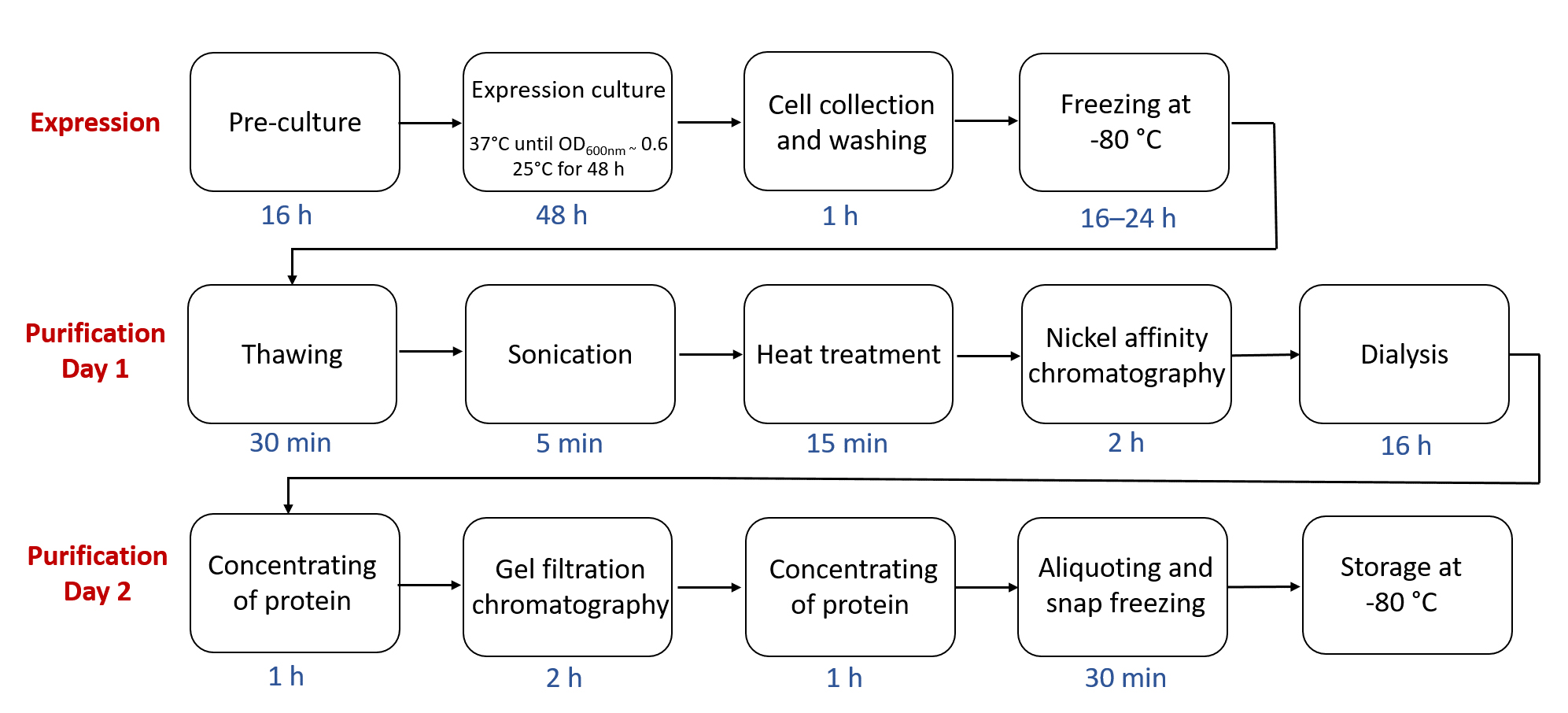
Flow chart outlining the steps in ALDHTt expression and purification, highlighting the approximate time required for each step.
Background
The aldehyde dehydrogenase ALDHTt was first identified and isolated as a contaminant, during cytochrome oxidase caa3-type native purification (Lyons et al., 2012) from Thermus thermophilus, which led to its further investigation. One of the common roles of ALDH family members in mammals is their inherent biosynthesis of retinoic acid assisted by cytochrome oxidases, which shows the possibility of such a role for this enzyme in T. thermophilus, due to the close relation between these two enzymes. Native purification of enzymes from T. thermophilus and other bacterial sources can be cumbersome, difficult, and lengthy, resulting in low protein yields (Soulimane, 2010; Robin et al., 2011). Therefore, recombinant protein production may be an attractive alternative route for the production of proteins for structural characterisation and functional analysis, allowing for high protein purity and yields. In 2018, the ALDHTt was first recombinantly expressed, purified, and its crystal structure determined (Hayes et al., 2018), revealing a novel C-terminal extension in the form of a tail, which contributes to active site regulation, thermostability and the oligomerization mode, aspects not before seen in the ALDH superfamily. The production protocol, employing a 48-h expression culture, heat treatment, immobilized metal affinity chromatography (IMAC) (Figure 1), and gel filtration chromatography, allows for yields of 15–20 mg of highly pure ALDHTt per litre of culture (Figure 2) (Shortall et al., 2021).
ALDHs are often not substrate specific, and can be characterised by their activity for the oxidation of aldehydes using the cofactor nicotinamide adenine dinucleotide (phosphate) (NAD(P)+) to the corresponding carboxylic acid, and nicotinamide adenine dinucleotide (phosphate) hydrate (NAD(P)H). Like other dehydrogenase enzymes, their activity is monitored most commonly via UV-Vis spectrometry for the production of NAD(P)H at 340 nm (Figure 3). ALDHTt can oxidise a range of aldehydes, including aliphatic, aromatic, and cyclics, at elevated temperatures, with the highest catalytic activity achieved with hexanal at 50°C (Shortall et al., 2021).
Conditions detailed here allowed for the production of a pure, soluble, and active form of ALDHTt. The protocol described can also serve as a starting strategy to express and purify similar proteins.
Materials and Reagents
Note: All reagents were stored at room temperature, unless otherwise stated here.
Ice
MilliQ water
Protein Expression
1.5 mL centrifuge tubes (Eppendorf, Sigma-Aldrich, catalog number: EP0030120086-1PAK)
50 mL sterile centrifuge tubes (Corning, Sigma-Aldrich, catalog number: CLS430290)
100 mm Petri dishes (Sigma-Aldrich, catalog number: P5731-500EA)
Cell spreader, sterile (Sigma-Aldrich, catalog number: HS8171A-500EA)
2 L cell culture Erlenmeyer flasks
10 mL serological pipettes, sterile (Sigma-Aldrich, catalog number: CLS4488-50EA)
25 mL serological pipettes, sterile (Sigma-Aldrich, catalog number: CLS4251-200EA)
Aluminium foil
pET22b(+)-ALDHTt (constructed in the lab by Hayes et al. (2018)), store at -20°C
BL21(DE3) chemically competent cells (prepared in the lab), store at -80°C
LB agar (Fisher Scientific, catalog number: BP9724-500)
LB broth (Sigma-Aldrich, catalog number: L3022)
Ampicillin sodium salt (Fisher Scientific, catalog number: A0166), store at 4°C
Tryptone (Fisher Scientific, catalog number: 1278-7099)
Yeast extract (Fisher Scientific, catalog number: 10225203)
Ammonium sulfate ((NH4)2SO4) (Sigma-Aldrich, catalog number: A4418)
Potassium phosphate monobasic (KH2PO4) (Sigma-Aldrich, catalog number: P0662)
Sodium phosphate dibasic (Na2HPO4) (Sigma-Aldrich, catalog number: S9763)
Glycerol (Fisher Scientific, catalog number: BP229-4)
(D)-(+)-Glucose (Sigma-Aldrich, catalog number: G7528)
α-Lactose monohydrate (Sigma-Aldrich, catalog number: L2643)
Magnesium sulfate heptahydrate (MgSO4·7H2O) (Sigma-Aldrich, catalog number: 230391)
ZY Media (see Recipes)
20× NPS (see Recipes)
50× 5052 (see Recipes)
1 M MgSO4 (see Recipes)
ZYP-5052 Auto-induction Media (see Recipes)
Protein Purification
1 L Super-Speed Centrifuge Bottles with Sealing Closure, Nalgene (ThermoFisher, Fisher Scientific, catalog number: 3140-1006)
50 mL Oak Ridge High-Speed Polycarbonate Centrifuge Tubes w/Sealing Cap (ThermoFisher, Fisher Scientific, catalog number: 3138-0050PK)
10 mL serological pipettes, sterile (Sigma-Aldrich, catalog number: CLS4488-50EA)
50 mL sterile centrifuge tubes (Corning, Sigma-Aldrich, catalog number: CLS430290)
0.45 μm syringe filters, nylon (Fisher Scientific, catalog number: 15131499)
20 mL plastic syringes (Fisher Scientific, catalog number: 15889152)
500 mL glass beaker
4 L glass beaker
Magnetic stir bars
Dialysis clips
Dialysis tubing, BiodesignTM Cellulose Dialysis Tubing Roll, 8000 Da MWCO (Fisher Scientific, catalog number: 12707486), store at 4°C
Amicon Ultra-15 centrifugal filters, 50 kDa MWCO (Merck Millipore, Sigma-Aldrich, catalog number: UFC9050)
200 μL PCR tubes (Sigma-Aldrich, catalog number: BR781301)
Liquid nitrogen (LN2)
Lysozyme from chicken egg white (~ 70,000 U/mg) (Sigma-Aldrich, catalog number: 62971), store at 4°C
Deoxyribonuclease I from bovine pancreas (Sigma-Aldrich, catalog number: DN25), store at -20°C
Magnesium chloride (MgCl2) (Sigma-Aldrich, catalog number: M8266)
Trizma base (Sigma-Aldrich, catalog number: T6066)
Hydrochloric acid (HCl) (Fisher Scientific, catalog number: 10053023)
β-mercaptoethanol (Sigma-Aldrich, catalog number: M3148)
Imidazole (Sigma-Aldrich, catalog number: I2399)
Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S7653)
Chelating Sepharose fast flow (Sigma-Aldrich, catalog number: GE17-0575-01), store at 4–30°C
Ethylenediamine tetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: E9884)
Nickel sulfate hexahydrate (NiSO4·6H2O) (Sigma-Aldrich, catalog number: 227676)
Sodium acetate (C2H3NaO2) (Sigma-Aldrich, catalog number: S2889)
Acetic acid (CH3COOH) (Sigma-Aldrich, catalog number: 695092)
Lysis Buffer (see Recipes)
Buffer A (see Recipes)
Buffer B (see Recipes)
Dialysis Buffer (see Recipes)
Gel Filtration Buffer (see Recipes)
ALDHTt Enzyme Assay
Plastic cuvettes, Fisherbrand Macrocuvettes (Fisher Scientific, catalog number: FB55923)
Hexanal (Sigma-Aldrich, catalog number: 115606)
β-nicotinamide adenine dinucleotide sodium salt (NAD+) (Sigma-Aldrich, catalog number: N0632), store at -20°C
Potassium phosphate monobasic (KH2PO4) (Sigma-Aldrich, catalog number: P0662)
Potassium phosphate dibasic (K2HPO4) (Sigma-Aldrich, catalog number: P3786)
Equipment
Biological safety cabinet
Autoclave
Lab balances
Water bath
Corning® 5 × 7 Inch Top PC-420D Stirring Hot Plate with Digital Displays, 120V/60Hz (Corning, catalog number: 6795-420D)
Shaking incubator (Eppendorf, New Brunswick Scientific Inova 40, Sigma-Aldrich, catalog number: EPM1299-0094)
-80°C freezer, New Brunswisk Scientific Ultra Low Temperature Freezer
Large scale centrifuge (ThermoFisher Scientific Sorvall RC6+ Centrifuge, Fisher Scientific, catalog number: 12121680)
ThermoFisher Scientific, Heraeus Megafuge 16R Centrifuge (ThermoFisher Scientific, Fisher Scientific, catalog number: 75004230)
VWR Microstar12 microcentrifuge (VWR, catalog number: 521-1651)
Probe sonicator (Bandelin Sonoplus HD 2200, catalog number: 2531)
Peristaltic Pump P-1 (Cytivia Life Sciences, catalog number: 18111091)
ÄKTAprime plus (Cytiva Life Sciences)
XK16 chromatography column (Sigma-Aldrich, catalog number: GE28-9889-37)
HiLoad 16/60 superdex 200 pg gel filtration column (Cytiva, Sigma-Aldrich, catalog number: GE28-9893-36)
NanoDropTM ND-1000 (ThermoFisher Scientific, catalog number: ND-ONE-W)
SDS-PAGE apparatus (Biorad Mini-PROTEAN Tetra Cell, catalog number: 1658005EDU)
Cary 60 UV-vis spectrophotometer (Agilent) equipped with a temperature controller (TC-1 temperature controller) (Quantum Northwest, catalog number: TC 1-MAN-2.2)
pH meter, Thermo Scientific Orion 2-star benchtop pH meter (Fisher Scientific, catalog number: Meter Kit 1111001)
Procedure
Protein Expression
Transformation: Transform the construct DNA pET-22b(+)-ALDHTt (Hayes et al., 2018) into E. coli BL21(DE3) chemically competent cells, via a standard heat shock in a water bath at 42°C for 30 s. Plate the resultant transformation on an agar plate supplemented with 100 μg/mL ampicillin, and grow at 37°C overnight.
Preculture: Innoculate 10 mL of LB broth supplemented with 100 μg/mL ampicillin with one colony from the transformation plate, and incubate with agitation at 250 rpm and 37°C for 16 h.
Main culture: Innoculate 1 L of ZYP-5052 auto-induction media (see Recipes below) supplemented with 50 μg/mL ampicillin with 10 mL of preculture (1% v/v), and incubate with agitation at 200 rpm and 25°C for 48 h.
Note: The main culture for expression is split, so as to have no more than 500 mL in a 2- L shake flask, for aeration purposes.
Protein Purification
Transfer E. coli cells to 1 L centrifuge bottles, and harvest the cells via centrifugation at 6,000 × g and 4°C for 15 min.
Note: Prior to cell collection, weigh the empty centrifuge bottles, to allow for determination of the cell mass after centrifuging.
Discard the supernatant, and autoclave the waste.
Weigh the cell pellet (typically ~35 g/L culture).
Supplement the lysis buffer with 0.25 mg/mL lysozyme, 20 μg/mL DNase I, and 200 mM MgCl2. Resuspend the pellet in lysis buffer (see Recipes below), adding 5 mL of lysis buffer for every 1 g of cells present.
Note: Resuspension of cells is carried out on ice using a 10-mL serological pipette for drawing the buffer-cell suspension up and down. If carried out at room temperature, this should be performed in a timely manner, as native cell proteases may degrade the desired protein.
Store the cell lysate in aliquots of 50 mL in Falcon tubes at -80°C overnight.
Note: Storage of cell lysate at -80°C overnight can be extended for up to 1 week.
Thaw the cell lysate in a water bath at 37°C.
Sonicate the cells using a probe sonicator with the power set between 60–70% (ultrasonic nominal output maximum of 200 W). Sonicate for 30 s, rest for 30 s, and repeat this process three times.
Note: Sonication was carried out in a cold room at 4°C, with the cell lysate solution surrounded by ice.
Heat the cell lysate in a water bath to 65°C for 15 min. Omission of this step results in an approximate 40% decrease in protein yield.
Collect the cell debris via centrifugation at 34,000 × g and 4°C for 30 min. Remove the supernatant that contains the ALDHTt, and store on ice. Autoclave the collected cells prior to disposal.
Note: Following the 30-min centrifugation for collection of the soluble fraction containing ALDHTt, the sample should be collected immediately after the centrifuge stops, to ensure the cells don’t re-enter solution.
Filter the collected soluble fraction through a 0.45 μm nylon syringe filter, and store in a 50 mL Falcon tube on ice. Keep 1 mL of sample for SDS-PAGE and Western blot analysis.
Pack an XK 16/20 column with approximately 10 mL of chelating Sepharose fast flow resin, and pre-equilibrate as follows. Using a peristaltic pump, deliver the following quantities of buffer to the column, at a rate of 1–3 mL/min: 0.5 column volume (CV) of 0.2 M EDTA, 0.5 M NaCl, pH 7, 2 CV of 0.5 M NaCl, 2 CV of MilliQ water, 0.2 CV of 0.2 M NiSO4, 5 CV of MilliQ water, 5 CV of 20 mM sodium acetate, 0.5 M NaCl pH 4, and finally 2 CV of buffer A (see Recipes below).
Load the entire filtered soluble fraction (approximately 80–100 mL from 1 L culture) onto the Ni-affinity column. Keep the solution on ice during loading. Collect the flow through for SDS-PAGE and Western blot analysis.
Connect the column to the Akta Prime system, and start the IMAC purification.
Wash the column with buffer A at 3 mL/min, until a stable (close to zero) Abs280nm is achieved.
Elute proteins at 3 mL/min, via an imidazole step gradient of 50, 100, 200, and 500 mM (Figure 1), using a combination of buffer A and buffer B (see Recipes below). Only increase the concentration of buffer B, and thus imidazole, when the Abs280nm obtained from the previous concentration is stable. Collect a fraction from each elution for SDS-PAGE and Western blot.
Note: ALDHTt should elute from Ni-affinity chromatography in the 200 mM imidazole fraction.
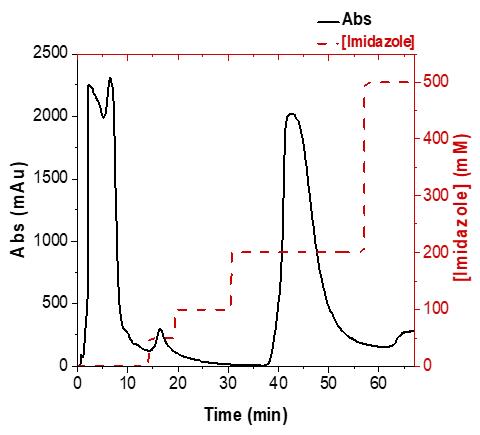
Figure 1. Elution profile of ALDHTt via Ni-affinity chromatography. Chromatogram displaying purification of ALDHTt via Ni-affinity chromatography using a step gradient of imidazole from 50–500 mM. E. coli host proteins are eluted from 10–50 mM imidazole in the first two peaks, while ALDHTt is eluted at 200 mM imidazole in the third peak.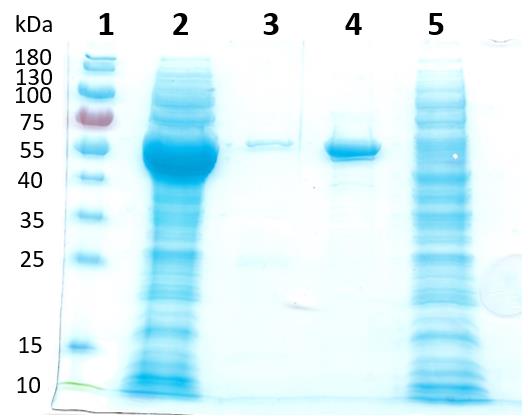
Figure 2. SDS-PAGE of ALDHTt expression and purification samples. Lane 1: PageRuler Pre-stained protein ladder (ThermoFisher Scientific), lane 2: E. coli BL21(DE3) cell lysate expressing ALDHTt, lane 3: Ni-affinity chromatography 200 mM imidazole elution, lane 4: purified ALDHTt following gel filtration chromatography, lane 5: Ni-affinity chromatography flow through (adapted from Shortall et al., 2021).Take the fractions containing the ALDHTt (typically 30–40 mL), and dialyse against 4 L of dialysis buffer (see Recipes below). Dialyse with gentle magnetic stirring at 4°C overnight.
Concentrate the protein sample using Amicon Ultra-15 centrifugal filters, 50 kDa MWCO (Merck Millipore), at 3,500 × g using 5 min spins, until a volume of approximately 1 mL (containing approximately 15–20 mg/mL) is obtained.
Gel filtration chromatography: Pre-equilibrate the HiLoad 16/60 Superdex 200 pg column with 2 CV (240 mL) of gel filtration buffer (see Recipes below) at 1 mL/min. Load approximately 1 mL of concentrated ALDHTt protein sample onto the column using the injection loop on the Akta Prime system, and run at 1 mL/min for 120 min, with collection of the protein peak.
Note: ALDHTt should begin to elute from gel filtration chromatography at approximately 65 mL. There should only be one peak on the gel filtration chromatogram. A slight shoulder at the start of the peak (on the left) can occur, but should not be collected.
Pool the fractions from gel filtration chromatography (approximately 20 mL), and concentrate to 25–30 mg/mL (or as desired) using Amicon Ultra-15 centrifugal filters, 50 kDa MWCO (Merck Millipore), at 3,500 × g using 5 min spins, until the desired concentration is obtained.
Monitor protein concentration by UV absorbance at 280 nm on a NanoDropTM 1000 spectrophotometer, using a sequence-derived extinction coefficient of 1671 M-1 cm-1. Use gel filtration buffer as the blank.
Prepare 30-μL aliquots, snap freeze in liquid nitrogen (LN2), and store at -80°C until further use. If the protein is to be used immediately, aliquot as desired, and store on ice until further use.
ALDHTt Enzyme Assay
Assay the ALDHTt, using hexanal as the substrate (2 mM) and NAD+ as the cofactor (2 mM), in 10 mM potassium phosphate pH 8.
Note: Hexanal is only slightly soluble in water, so stock solutions at low concentrations should be prepared. Solubility in water is ~6 g/L (60 mM). Shelf life of the hexanal solution prepared in buffer is one week.
Program the spectrophotometer equipped with a temperature controller to analyse at 340 nm and 50°C for 2 min. Add a pre-equilibration step of 1 min, to ensure all solutions are kept at 50°C. A short analysis time of 2 min is utilised to avoid loss of volatile aldehyde substrates due to evaporation.
Heat solutions of 5 mM hexanal, 10 mM NAD+, and 10 mM potassium phosphate pH 8 to 50°C, in a water bath.
Note: The enzyme activity of ALDHTt can be analysed at 20–50°C. However, with decreased temperature a decrease in enzymatic activity is observed.
Thaw a 30-μL aliquot of ALDHTt on ice.
Dilute the ALDHTt to obtain a concentration of 0.38 mg/mL using gel filtration buffer, and centrifuge at 4,050 × g for 5 min.
Note: Diluting the ALDHTt to a concentration of 0.38 mg/mL for the enzyme assay should be carried out using gel filtration buffer. For example, a 1:70 dilution of ALDHTt at a stock concentration of 26.65 mg/mL can be performed, by adding 10 μL of ALDHTt to 690 μL of gel filtration buffer.
To a plastic cuvette, add 360 μL of 10 mM NAD+, 680 μL of 10 mM potassium phosphate pH 8, and 40 μL of ALDHTt at 0.38 mg/mL. Insert the cuvette into the spectrophotometer, to undergo equilibration at 50°C for 1 min.
Add 720 μL of 5 mM hexanal to the cuvette and mix gently. Monitor the absorbance at 340 nm for 2 min, for the production of NADH.
Note: Enzymatic assaying of ALDHTt should result in an increase in absorbance at 340 nm (a positive slope) when monitored at 50°C (Figure 3).
Calculate enzyme activity employing Beer-Lambert’s Law, using the 6,220 M-1 cm-1 extinction coefficient of NADH at 340 nm (Equation 1).
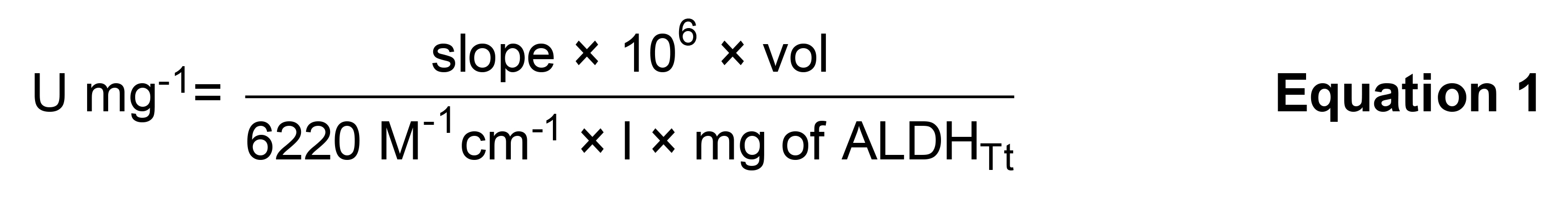
Note: One enzyme unit is equal to the production of 1 µmol/min NADH. l is equal to the path length of the cuvette, in this case l = 1 cm. Vol is equal to the volume of the cuvette, in this case vol = 1.8 mL.
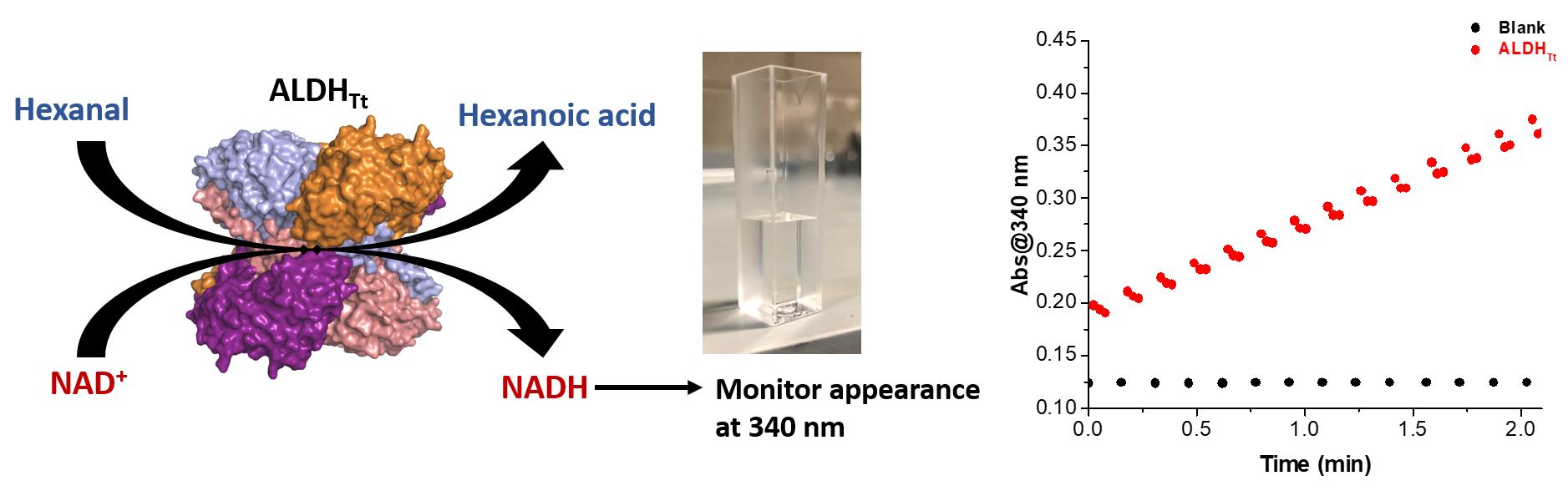
Figure 3. Principle for ALDHTt assaying using hexanal and NAD+ with activity monitored spectrophotometrically via the appearance of NADH. An example of the cuvette used and results obtained are demonstrated.
Data analysis
The original research article for the above expression, purification, and enzyme activity analysis was published in Shortall et al. (2021), https://doi.org/10.3390/cells10123535.
Recipes
Note: All solutions were prepared using MilliQ water (18.2 MΩ cm).
ZY Media
10% typtone
5% yeast extract
20× NPS (NPS = 100 mM PO4, 25 mM SO4, 50 mM NH4, 100 mM Na, 50 mM K)
0.5 M (NH4)2SO4
1 M KH2PO4
1 M Na2HPO4
50× 5052 (5052 = 0.5% glycerol, 0.05% glucose, 0.2% α-lactose)
25% glycerol
2.5% glucose
10% α-lactose
Note: Preparation of 50× 5052 can be assisted by gentle heating with magnetic stirring. 50× 5052 can be difficult to dissolve, and should not be autoclaved until all components are dissolved. If the solution is visibly displaying a brown tinge after autoclaving, it should be re-prepared.
1 M MgSO4
ZYP-5052 Auto-induction Media
~928 mL of ZY media
1 mL of 1 M MgSO4
20 mL of 50× 5052
50 mL of 20× NPS
50 μg/mL ampicillin
Note: Solutions for ZYP-5052 auto-induction media are prepared and autoclaved separately. These are combined to make the media on the day of the expression culture.
Lysis Buffer
20 mM Tris-HCl pH 7.5
5 mM β-mercaptoethanol
10 mM imidazole
500 mM NaCl
Buffer A
20 mM Tris-HCl pH 7.5
5 mM β-mercaptoethanol
10 mM imidazole
200 mM NaCl
Buffer B
20 mM Tris-HCl pH 7.5
5 mM β-mercaptoethanol
1 M imidazole
200 mM NaCl
Dialysis Buffer
50 mM Tris-HCl pH 7.5
5 mM β-mercaptoethanol
250 mM NaCl
Gel Filtration Buffer
50 mM Tris-HCl pH 7.5
5 mM β-mercaptoethanol
150 mM NaCl
Acknowledgments
This protocol is an extension of that described in Shortall et al. (2021) and adapted from Hayes et al. (2018). This research was funded by European Union’s Horizon 2020 Research and Innovation programme, Oyster (Open characterisation and modelling environment to drive innovation in advanced nano-architectured and bio-inspired hard/soft interfaces) under grant agreement No. 760827. Funding is also acknowledged from the Department of Chemical Sciences, University of Limerick and The Higher Education Authority, Ireland.
Competing interests
There are no conflicts of interest or competing interests.
References
- Hayes, K., Noor, M., Djeghader, A., Armshaw, P., Pembroke, T., Tofail, S. and Soulimane, T. (2018). The quaternary structure of Thermus thermophilus aldehyde dehydrogenase is stabilized by an evolutionary distinct C-terminal arm extension. Sci Rep 8(1): 1-14.
- Lyons, J. A., Aragão, D., Slattery, O., Pisliakov, A. V., Soulimane, T. and Caffrey, M. (2012). Structural insights into electron transfer in caa 3-type cytochrome oxidase. Nature 487(7408): 514-518.
- Robin, S., Arese, M., Forte, E., Sarti, P., Giuffre, A. and Soulimane, T. (2011). A sulfite respiration pathway from Thermus thermophilus and the key role of newly identified cytochrome c550. J Bacteriol 193(15): 3988-3997.
- Shortall, K., Durack, E., Magner, E. and Soulimane, T. (2021). Study of ALDH from Thermus thermophilus-Expression, Purification and Characterisation of the Non-Substrate Specific, Thermophilic Enzyme Displaying Both Dehydrogenase and Esterase Activity. Cells 10(12): 3535.
- Soulimane, T. (2010). Thermus thermophilus encodes an archaeal-like fructose-1,6-bisphosphatase: purification of native and recombinant protein for structural studies. Protein Expr Purif 74(2): 175-180.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Shortall, K., Magner, E. and Soulimane, T. (2022). Expression, Purification, and in vitro Enzyme Activity Assay of a Recombinant Aldehyde Dehydrogenase from Thermus thermophilus, using an Escherichia coli host. Bio-protocol 12(9): e4401. DOI: 10.21769/BioProtoc.4401.
Category
Biochemistry > Protein > Expression
Microbiology > Heterologous expression system > Escherichia coli
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








