- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
High-speed Atomic Force Microscopy Observation of Internal Structure Movements in Living Mycoplasma
(*contributed equally to this work) Published: Vol 12, Iss 5, Mar 5, 2022 DOI: 10.21769/BioProtoc.4344 Views: 3000
Reviewed by: Kristin L. ShinglerRon Saar DoverAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Assessing Different Ways of Bacillus subtilis Spreading over Abiotic Surfaces
Marco Bartolini and Roberto Grau
Nov 20, 2019 5482 Views

Assessing Swarming of Aerobic Bacteria from Human Fecal Matter
Arjun S. Byju [...] Sridhar Mani
May 5, 2021 3898 Views

Multiplexed Microfluidic Platform for Parallel Bacterial Chemotaxis Assays
Michael R. Stehnach [...] Jeffrey S. Guasto
Sep 5, 2024 2042 Views
Abstract
Dozens of Mycoplasma species belonging to the class Mollicutes bind to solid surfaces through the organelle formed at a cell pole and glide in its direction by a unique mechanism. In Mycoplasma mobile, the fastest gliding species in Mycoplasma, the force for gliding is generated by ATP hydrolysis on an internal structure. However, the spatial and temporal behaviors of the internal structures in living cells were unclear. High-speed atomic force microscopy (HS-AFM) is a powerful method to monitor the dynamic behaviors of biomolecules and cells that can be captured while maintaining their active state in aqueous solution. In this protocol, we describe a method to detect their movements using HS-AFM. This protocol should be useful for the studies of many kinds of microorganisms.
Graphic abstract:
Scannnig Mycoplasma cell
Background
Class Mollicutes, a small group of bacteria featuring a single-layered cell membrane, has as many as three unique motility mechanisms different from other organisms. Recently, the isolated internal structure of M. mobile was shown to hydrolyze ATP with conformational changes, suggesting that the internal structure functions as a force generator for gliding (Miyata and Hamaguchi, 2016; Nishikawa et al., 2019). However, the spatial and temporal behaviors and movements of internal motors in living cells have not been examined.
High-speed atomic force microscopy (HS-AFM) is a powerful method to monitor the dynamic behaviors of biomolecules and cells that can be captured while maintaining their active state in aqueous solution (Ando, 2018). In recent years, this approach has been dramatically improved, and the behaviors of more and more proteins have been elucidated (Kodera et al., 2010 and 2021; Ando, 2018; Kodera and Ando, 2020). In addition, HS-AFM has been applied to understanding the structures in the cell wall surface (Yamashita et al., 2012) and below the cell membrane (Zhang et al., 2017).
Here, we provide a protocol detailing how to detect the movements of the internal structure in a living M. mobile cell, based on our recent study (Kobayashi et al., 2021).
Materials and Reagents
1.5 mL microtube (Greiner Bio-One, catalog number: 616201)
Glass substrate stage for HS-AFM (custom-made glass rod with 2 mm in diameter and 2 mm in height, Japan Cell)
Cantilever (Olympus, BLAC10DS-A2)
#3000 sand paper (Riken Corundum, Precision Polishing Film Sheet #3000, catalog number: 3-9013-06)
100 mL beaker
Kimwipes (Nippon Paper Crecia, catalog number: 62015)
Filter paper for liquid replacement on glass stage (ADVANTEC, catalog number: 01531090) (Cut into triangles as shown in Figure 2)
25 cm2 tissue culture flask (AS ONE, catalog number: 2-8589-01)
Micro pipette tip (Greiner Bio-One, catalog numbers: 740290 and 739285)
Acetone (FUJIFILM Wako Pure Chemical Corporation, catalog number: 012-00343)
Potassium hydroxide (KOH) (Nacalai Tesque, catalog number: 28616-45)
Ethanol (FUJIFILM Wako Pure Chemical Corporation, catalog number: 057-00456)
Fluorosurf (FluoroTechnology, catalog number: FG-3020C-20)
Colorless nail polish (Shiseido, MAQuillAGE (Top base coat))
Deionized water
3-aminopropyldiethoxymethylsilane (APTES) (Shin-Etsu Chemical, catalog number: KBE-903)
Glutaraldehyde (Sigma-Aldrich, catalog number: G5882)
Heart infusion broth (BD, catalog number: 238400)
Yeast extract (BD, catalog number: 212750)
10 N NaOH (Nacalai Tesque, catalog number: 31511-05)
Horse serum (Thermo Fisher Scientific, GibcoTM, catalog number: 16050122)
Amphotericin B (Sigma-Aldrich, catalog number: A2942)
Ampicillin Na (Nacalai Tesque, catalog number: 02739-32)
di-Sodium hydrogenphosphate (Nacalai Tesque, catalog number: 31801-05)
Sodium dihydrogenphosphate dihydrate (FUJIFILM Wako Pure Chemical Corporation, catalog number: 192-02815)
NaCl (Nacalai Tesque, catalog number: 31320-05)
Glucose (Nacalai Tesque, catalog number: 16806-25
Phosphate-buffered saline containing glucose (PBS/G) (see Recipes)
Aluotto medium (see Recipes)
A mutant strain (gli521[P476R]) isolated from M. mobile 163K strain (ATCC, catalog number: 43663) (Uenoyama and Miyata, 2005)
Equipment
Micro pipette (Gilson, catalog numbers: F123600, F123601, and F123602)
Centrifuge (Sigma Laborzentrifugen, model: Sigma 1-14)
25°C incubator (Tokyo Rikakikai, model: LTI-400E)
Heater (TAITEC CORPORATION, CTU-Neo)
Magnetic stirrer (AS ONE, model: CT-1AT)
HS-AFM (An apparatus with the same performance is commercially available from Research Institute of Biomolecule Metrology Co., Ltd., model: SS-NEX)
Autoclave (TOMY SEIKO, model: LSX-700)
Procedure
Cell preparation
Inoculate 1 mL of frozen M. mobile stock into 10 mL of Aluotto medium in a 25 cm2 tissue culture flask and statically cultivate it at 25°C for a few days, to reach an optical density at 600 nm of 0.06-0.08.
Collect 2 mL of cultured cells by centrifugation at 12,000 × g and room temperature (RT) for 4 min, discard the supernatant, resuspend the cell pellet in 400 µL of PBS/G, and centrifuge it again at 12,000 × g and RT for 4 min (Note 1).
Repeat this process twice.
Resuspend the cells in 100 µL of PBS/G to 20-fold density of the original culture.
Glass stage treatment
Lightly press the substrate surface against the #3000 sandpaper and scrub 200 times back and forth to scratch the surface (The rough surface provides a more stable observation. This step is optional). Note that this process should not be applied to the other side.
Put the glass stage into the 1.5 mL microtube filled with acetone and shake it to remove glass debris. (Optional: as mentioned above)
To prepare saturated KOH-ethanol solution, add 10 g of KOH into 50 mL of ethanol, stir using a magnetic stirrer for 5 min, and transfer the supernatant of the saturated KOH-ethanol solution into a new beaker.
Put several glass stages into the prepared saturated KOH-ethanol solution.
Leave for 15 min to hydrophilize the glass surface.
Discard the solution, add some deionized water, and shake the beaker gently.
Repeat this process ten times to remove the chemicals.
Move the glass stages onto Kimwipes and remove the water.
Keep the glass stages with some slant in a heater, for 20 min at 40–45°C to dry.
Cell immobilization on the glass surface.
Treat the sides of the glass stage with Fluorosurf for hydrophobization.
Note: If not treated, the liquid placed on the substrate surface will spill over to the sides.
Fix the glass stage onto the Z-piezo of the HS-AFM scanner with nail polish (Figure 1).
Just before use, dilute 1 µL of APTES with 999 µL of water.
For silanization on the glass stage surface, load 5 µL of APTES solution onto the treated glass substrate immediately after the dilution and leave it for 10 min.
Rinse the substrate surface with 20 µL of water dropwise. Repeat this four times (Figure 2).
Replace the water on the glass substrate with 0.1% glutaraldehyde solution and leave for 5 min.
Rinse the substrate surface with 20 µL of water dropwise. Repeat this four times.
Replace the glutaraldehyde solution on the substrate with PBS/G.
Replace the PBS/G on the substrate to cell suspension and leave for 10 min at 25–28°C.
Rinse the substrate surface with 20 µL of PBS/G dropwise. Repeat this four times.
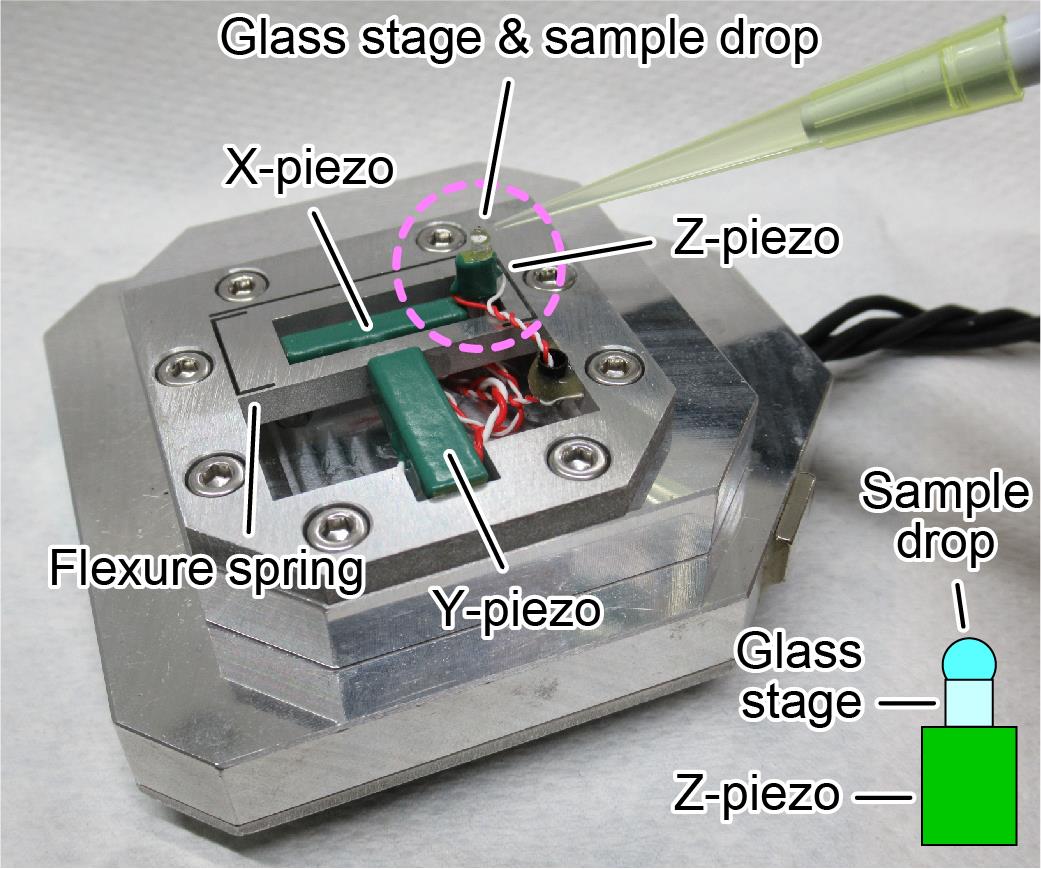
Figure 1. HS-AFM scanner with glass stage and sample drop. Piezoelectric elements are used to drive each axis in the XYZ directions. Z piezo, glass stage, and sample solution are seen in a pink dashed circle, as illustrated in the lower right. The glass stage is fixed onto the Z-piezo using nail polish.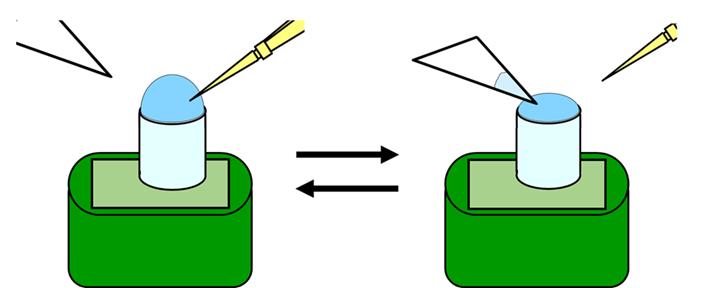
Figure 2. Procedure for liquid replacement on the glass stage.
Observation by HS-AFM.
Set the HS-AFM scanner equipped with the glass stage and sample solution upside down to the HS-AFM mechanical unit and start imaging (Figure 3).
Set the cantilever’s free oscillation amplitude (A0) and set-point amplitude (Asp) at ~2.5 nm and ~0.8 × A0, respectively. Under these conditions, the average tapping force <F> can be approximated as ~40 pN using the following equation:
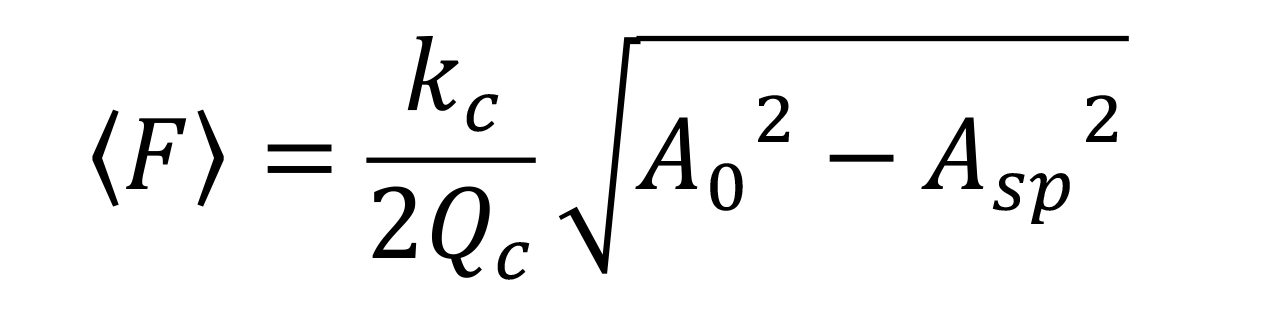
where kc and Qc are the spring constant and the quality factor of the cantilever, respectively. The nominal values of kc and Qc in liquid are ~0.08 N/m and 1.5, respectively. The detail conditions were described previously (Uchihashi et al., 2012).For searching cells (Figure 4a), scan the sample at 150 × 150 pixels with an imaging rate of 1,000 ms per frame in an area of 3,000 × 3,000 nm2.
To observe particle structure like Figure 4b, scan the cell surface with an imaging rate of 330 or 200 ms per frame in an area of 200 × 200 nm2, with 100 × 100 pixels.
Other details are common with general observation of samples in solution using HS-AFM (Uchihashi et al., 2012).
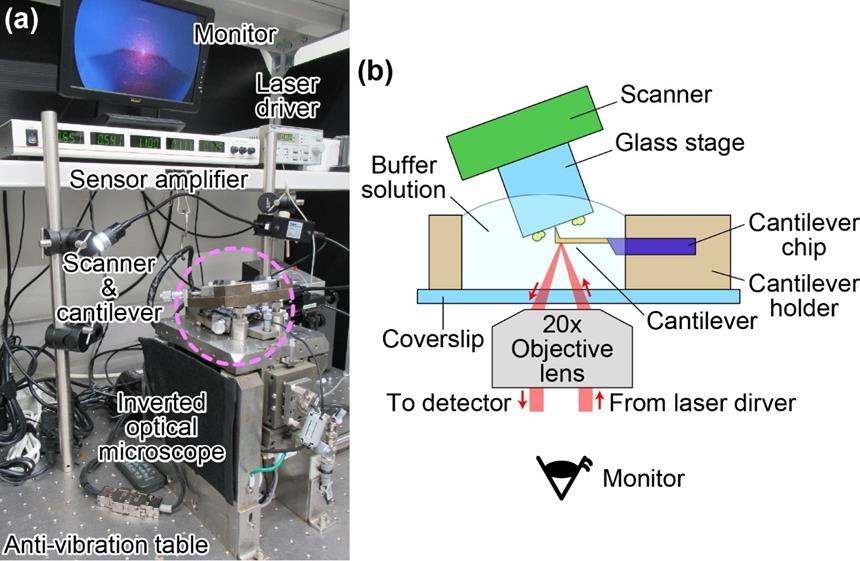
Figure 3. HS-AFM. (a) Picture showing the mechanical parts and some of the electrical devices of HS-AFM. The AFM unit (scanner and cantilever) is mounted on an inverted optical microscope which is placed on an anti-vibration table. (b) Schematic of experimental setup. Cantilever and M. mobile cells are not in scale. The optical view around the cantilever and the glass substrate can be observed through the monitor.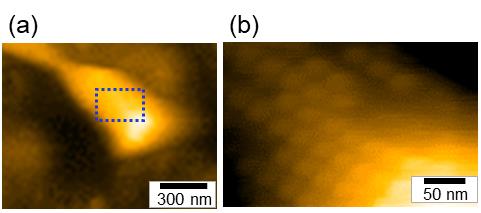
Figure 4. Representative images taken by HS-AFM. (a) Whole cell image (b) Particle structure image [Magnified image of the boxed area of the panel (a)]. This figure was modified a previous paper (Kobayashi et al., 2021).
Data analyses
All procedures were published in our previous paper (Kobayashi et al., 2021, https://journals.asm.org/doi/10.1128/mBio.00040-21). Briefly, videos were processed by three steps (i) to (iii) and analyzed for particle movements. (i) The image contrast was improved by a bandpass filter. (ii) Image drifts were corrected by a plugin for ImageJ, “align slices in stack” (Tseng et al., 2012). (iii) Image noises were reduced by averaging three consecutive slices. Then, each particle image was cropped, binarized, and traced for the mass center. All analyses were performed with ImageJ version 1.52A.
Notes
Suspension of cell pellet
More than 100 up-and-down movements with the pipette are recommended to separate cells completely.
Heat treatment of horse serum
For M. mobile growth and gliding, the horse serum should be inactivated by heat treatment at 56°C for 30 min.
Troubleshooting
Unable to obtain even a flat substrate image.
Cells or other suspended particles may be sticking to the cantilever. To shake them off, keep the cantilever away from the substrate surface, set the cantilever's amplitude at free oscillation for 1 min, and try imaging again. If this is not improved after several attempts, remove the scanner and wash the cantilever, using a pipetting water stream in the chamber.
No cells are found on the field.
Small molecules, including amino groups, may be bound to the glass stage and block cell binding. Prepare the cell suspension and the glass stage again carefully. For the cell preparation, increase the number of pipetting times to remove small molecules. Note that too many washes may also reduce cell number. Check the cell conditions by optical density at 600 nm and an optical microscope, because old cultures tend to include particles other than cells.
Cells easily detach from the substrate during observation.
The glass stage with a rough surface may be helpful [refer to Glass stage treatment option (Procedure B)].
The cell images look strange.
The cells may have dried out. Replace the cell suspension and the glass stage. Place a wet tissue at the edge, to humidify the area around the chamber.
The particle structures are not found on the cells.
This may be caused by damage on the cantilever tip. Replace the cantilever.
Recipes
Aluotto medium
1% heart infusion broth
0.56% yeast extract
0.035% 10 N NaOH
Note: Dissolve the above three reagents as a mixture, autoclave, cool to lower than 37°C, and add below three reagents in clean bench.
10% horse serum (Note 2)
0.025% amphotericin B
0.005% ampicillin Na
Phosphate-buffered saline containing glucose (PBS/G)
75 mM sodium phosphate (pH 7.3)
68 mM NaCl
20mM Glucose
Sterilize by using filtering or autoclaving
Acknowledgments
This work was supported by Grants-in-Aid for Scientific Research (B) and (A) (MEXT KAKENHI, Grant Numbers JP24390107, JP17H01544), and JST CREST (Grant Number JPMJCR19S5) to MM.
This protocol was adapted from a previously published paper (Kobayashi et al., 2021).
Competing interests
There are no conflicts of interest or competing interests.
References
- Ando, T., (2018). High-speed atomic force microscopy and its future prospects. Biophys Rev 10(2): 285-292.
- Kobayashi, K., Kodera, N., Kasai, T., Tahara, Y. O., Toyonaga, T., Mizutani, M., Fujiwara, I., Ando, T. and Miyata, M. (2021). Movements of Mycoplasma mobile gliding machinery detected by high-speed atomic force microscopy. mBio 12(3): e0004021.
- Kodera, N. and Ando, T. (2020). High-speed atomic force microscopy to study myosin motility. Adv Exp Med Biol 1239: 127-152.
- Kodera, N., Noshiro, D., Dora, S. K., Mori, T., Habchi, J., Blocquel, D., Gruet, A., Dosnon, M., Salladini, E. and Bignon, C. (2021). Structural and dynamics analysis of intrinsically disordered proteins by high-speed atomic force microscopy. Nat Nanotechnol 16(2): 181-189.
- Kodera, N., Yamamoto, D., Ishikawa, R. and Ando, T. (2010). Video imaging of walking myosin V by high-speed atomic force microscopy. Nature 468(7320): 72-76.
- Miyata, M. and Hamaguchi, T. (2016). Prospects for the gliding mechanism of Mycoplasma mobile. Curr Opin Microbiol 29: 15-21.
- Nishikawa, M. S., Nakane, D., Toyonaga, T., Kawamoto, A., Kato, T., Namba, K. and Miyata, M. (2019). Refined mechanism of Mycoplasma mobile gliding based on structure, ATPase activity, and sialic acid binding of machinery. mBio 10(6): e02846-19.
- Tseng, Q., Duchemin-Pelletier, E., Deshiere, A., Balland, M., Guillou, H., Filhol, O. and Thery, M. (2012). Spatial organization of the extracellular matrix regulates cell-cell junction positioning. Proc Natl Acad Sci U S A 109(5): 1506-1511.
- Uchihashi, T., Kodera, N. and Ando, T. (2012). Guide to video recording of structure dynamics and dynamic processes of proteins by high-speed atomic force microscopy. Nat Protoc 7(6): 1193-1206.
- Uenoyama, A. and Miyata, M. (2005). Gliding ghosts of Mycoplasma mobile. Proc Natl Acad Sci U S A 102(36): 12754-12758.
- Yamashita, H., Taoka, A., Uchihashi, T., Asano, T., Ando, T. and Fukumori, Y. (2012). Single-molecule imaging on living bacterial cell surface by high-speed AFM. J Mol Biol 422(2): 300-309.
- Zhang, Y., Yoshida, A., Sakai, N., Uekusa, Y., Kumeta, M. and Yoshimura, S. H. (2017). In vivo dynamics of the cortical actin network revealed by fast-scanning atomic force microscopy. Microscopy(Oxf) 66(4): 272-282.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Kobayashi, K., Kodera, N. and Miyata, M. (2022). High-speed Atomic Force Microscopy Observation of Internal Structure Movements in Living Mycoplasma . Bio-protocol 12(5): e4344. DOI: 10.21769/BioProtoc.4344.
Category
Microbiology > Microbial cell biology > Cell motility
Cell Biology > Cell movement > Cell motility
Cell Biology > Cell imaging > Atomic force microscopy
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link