- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Large-scale Analysis of Sleep in Zebrafish
Published: Vol 12, Iss 3, Feb 5, 2022 DOI: 10.21769/BioProtoc.4313 Views: 4914
Reviewed by: Andrea Herrera-SolisJesús Hernández Falcón Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In vivo Imaging of Calcium Activities from Pancreatic β-cells in Zebrafish Embryos Using Spinning-disc Confocal and Two-photon Light-sheet Microscopy
Jia Zhao [...] Yanmei Liu
Dec 5, 2021 4631 Views

Long-term in toto Imaging of Cellular Behavior during Nerve Injury and Regeneration
Weili Tian [...] Hernán López-Schier
May 5, 2023 2424 Views

Generation of Zebrafish Maternal Mutants via Oocyte-Specific Knockout System
Chong Zhang [...] Ming Shao
Nov 5, 2024 2248 Views
Abstract
Over the past decade, zebrafish have emerged as a powerful model for the study of vertebrate sleep and wake behaviors. Experimental evidence has demonstrated behavioral, anatomical, genetic, and pharmacological conservation of sleep between zebrafish and mammals, suggesting that discoveries in zebrafish can inform our understanding of mammalian sleep. Here, we describe a protocol for performing sleep behavioral experiments in larval zebrafish, using a high-throughput video tracking system. We explain how to set up a sleep behavioral experiment and provide guidelines on how to analyze the data. Using this protocol, a typical experiment can be completed in less than five days, and this method provides a scalable platform to perform genetic and pharmacological screens in a simple and cost-effective vertebrate model. By combining high-throughput behavioral assays with several advantageous features of zebrafish, this model system provides new opportunities to make discoveries that clarify the genetic and neurological mechanisms that regulate sleep.
Background
Sleep is an evolutionarily conserved behavior that is operationally defined by the presentation of three broadly-accepted behavioral criteria: (i) a period of behavioral quiescence that is rapidly reversible; (ii) an increased arousal threshold during these periods of quiescence, and (iii) homeostatic control, such that when deprived of sleep, an animal’s need to sleep is increased (Campbell and Tobler, 1984). While mammalian sleep assays have traditionally relied on measurements of cortical and muscle electrical activity using electroencephalogram (EEG) and electromyogram (EMG), recently experiments using both mammalian and non-mammalian model organisms have shifted to monitoring sleep based on the above behavioral criteria (Hendricks et al., 2000; Shaw et al., 2000; Zhdanova et al., 2001; Prober et al., 2006; Pack et al., 2007; Yokogawa et al., 2007; Raizen et al., 2008; Zimmerman et al., 2008; Churgin et al., 2017; Nath et al., 2017). Doing so allows for simpler, higher-throughput, and less expensive sleep studies that enable large-scale screens using some model systems (Cirelli et al., 2005; Koh et al., 2008; Chiu et al., 2016; Huang et al., 2017; Toda et al., 2019), and show broad agreement with sleep states defined using EEG and EMG in mammals (Pack et al., 2007; Fisher et al., 2012).
Studies using zebrafish, a diurnal vertebrate, have employed automated video recording and analysis of locomotor behaviors, along with measures of arousal threshold and sleep rebound following sleep deprivation, to define sleep and wake states (Zhdanova et al., 2001; Prober et al., 2006; Yokogawa et al., 2007; Elbaz et al., 2012). These and other studies have demonstrated behavioral, anatomical, genetic, and pharmacological conservation of sleep between zebrafish and mammals (Zhdanova et al., 2001; Prober et al., 2006; Renier et al., 2007; Yokogawa et al., 2007; Rihel et al., 2010a; Lee et al., 2019; Oikonomou et al., 2019). Most of these studies use larval zebrafish at 5-7 days post-fertilization (dpf), due to several advantageous features of this stage of development. These features include a small size that permits large-scale experiments (Prober et al., 2006), a relatively simple behavioral repertoire that facilitates analysis of sleep that is not confounded by other complex behaviors, amenability to pharmacological treatment due to a lack of a mature blood brain barrier (Jeong et al., 2008), and optical transparency which enables non-invasive monitoring of neuronal activity (Ahrens et al., 2012; Leung et al., 2019; Marques et al., 2020) and optogenetic manipulation of specific neuronal circuits (Singh et al., 2015) using intact and behaving animals. Inactive larval zebrafish exhibit an increased arousal threshold in response to mechanical (Zhdanova et al., 2001), acoustic (Elbaz et al., 2012; Gandhi et al., 2015) and visual (Prober et al., 2006) stimuli. These studies found that larval zebrafish which show no locomotor activity for one or more minutes, have a dramatic increase in arousal threshold, suggesting that one or more minutes of inactivity corresponds to a sleep state. This sleep is apparently controlled by a homeostatic mechanism, since larval zebrafish exhibit sleep rebound following sleep deprivation (Zhdanova et al., 2001; Lee et al., 2019; Oikonomou et al., 2019). These observations provide a basis to study the regulation of sleep and wake states using larval zebrafish, which has enabled large-scale genetic (Chiu et al., 2016), and pharmacological (Rihel et al., 2010a) screens, which have identified novel mechanisms that regulate vertebrate sleep, as well as targeted mechanistic sleep studies (Appelbaum et al., 2010; Singh et al., 2015 and 2017; Yelin-Bekerman et al., 2015; Chen et al., 2017; Lee et al., 2017, 2019, 2020; Oikonomou et al., 2019; Reichert et al., 2019; Zada et al., 2019; Ly et al., 2020; Özcan et al., 2020). Zebrafish studies have also identified mechanisms that underlie circadian (Gandhi et al., 2015) and homeostatic (Lee et al., 2019; Oikonomou et al., 2019; Reichert et al., 2019) regulation of sleep, thus addressing longstanding and important questions in the sleep field.
In most larval zebrafish sleep studies, individual fish are placed in each well of a 96-well plate at 4-dpf and their behavior can be monitored until 7- or 8-dpf (Figure 1). During this time, the animals obtain nutrients from their yolk sac, and thus food does not need to be provided. This allows sleep to be studied in sealed plates, which prevents evaporation of the medium from each well and generates more robust and uniform sleep data. However, in some cases it is preferable to use plates that are not sealed, such as when testing the acute effect of a drug on sleep, which requires that the drug be added during an experiment after collecting baseline sleep data. During zebrafish sleep experiments, white lights are turned on during the day and off at night to simulate day/night cycles, the locomotor activity of each fish is continuously monitored using infrared lights and an infrared camera, which records the time and duration of movement of each fish. Wild-type zebrafish exhibit robust diurnal sleep/wake cycles starting at 5-dpf (Prober et al., 2006), and robust sleep data can be collected during the following 48-72 h. Here, we provide a detailed protocol on how to perform larval zebrafish sleep studies.
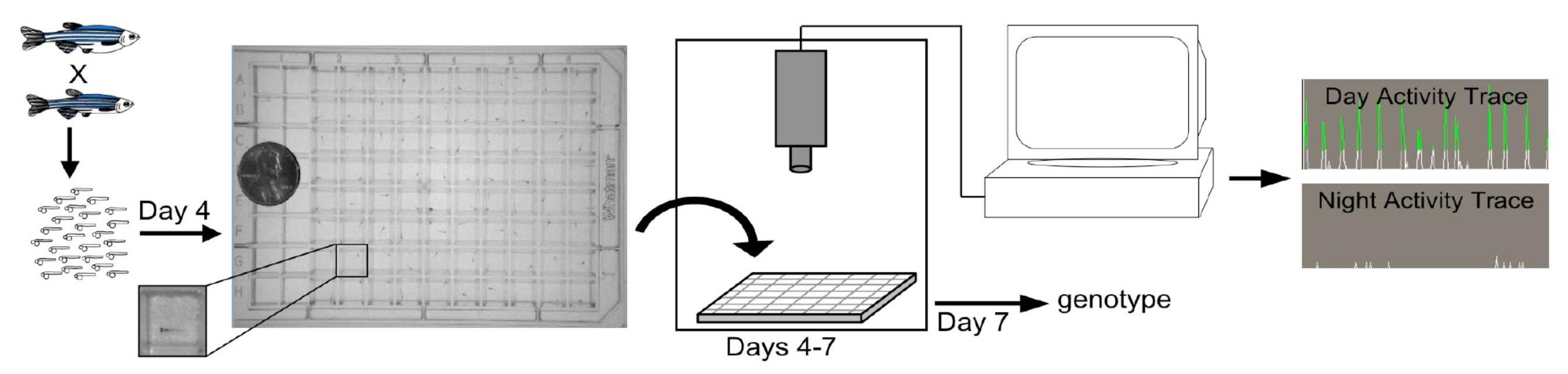
Figure 1. Zebrafish locomotor activity assay. Individual larvae are placed in each well of a 96-well plate on the fourth day of development. The plate is illuminated by white lights from 9am-11pm and is continuously illuminated by infrared lights. The locomotor activity of each larva is monitored with an infrared camera. Sample 40-second activity traces for a single larva are shown.
Materials and Reagents
All materials and reagents are kept at room temperature.
96-well plate, 650 µL, clear polystyrene, square well, flat bottom (Whatman, catalog number: 7701-1651)
MicroAmp Optical Adhesive Film (Applied Biosystems, catalog number: 4311971)
12-channel Pipette with variable volume adjustment from 20 to 300 µL (any vendor)
50 mL reagent reservoir (Genesee Scientific, catalog number: 28-125BX)
100 mm × 15 mm round Petri dishes (Biopioneer, catalog number: GS82-1473-001)
Plastic transfer pipets, 7 mL, non-graduated (VWR, catalog number: 490016614)
PCR adhesive seal applicator (Thermo Fisher Scientific, catalog number: AB1391)
Zebrafish larvae (4-dpf) of TLAB hybrid strain
NaCl (any vendor)
KCl (any vendor)
CaCl2·2H2O (any vendor)
MgCl2·7H2O (any vendor)
Methylene blue hydrate (Sigma, catalog number: 66720)
E3 medium for zebrafish embryos (see Recipes)
Methylene Blue solution (see Recipes)
Blue E3 medium solution (see Recipes)
Equipment
ZebraBox videotracking system (Viewpoint Life Sciences, Inc., model: ZebraBox, 4th generation) (Figure 2)
Stereo microscope with brightfield illumination (any vendor)
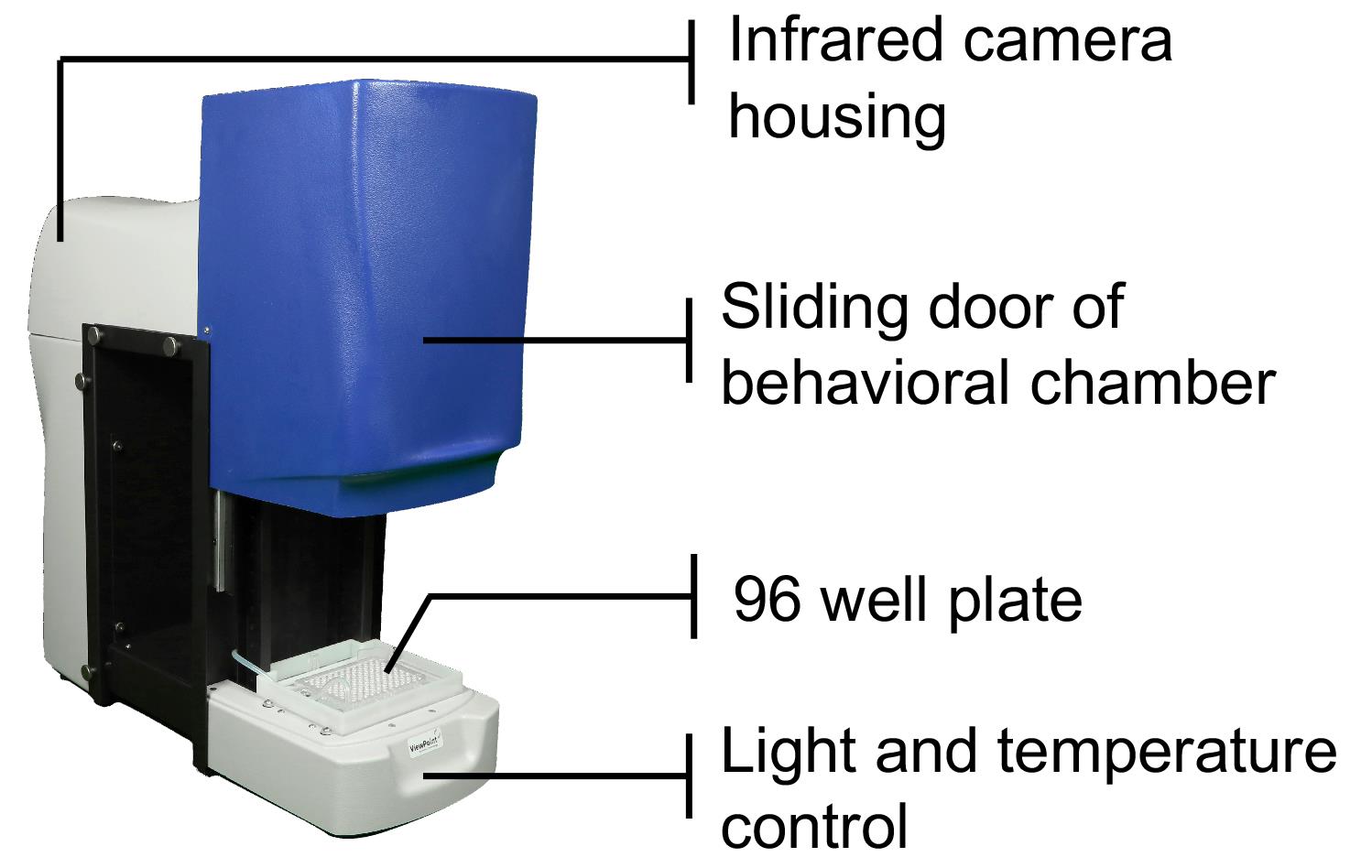
Figure 2. A high throughput video tracking system to monitor larval zebrafish behavior.
Software
ZebraLab (Viewpoint Life Sciences, Inc.)
MATLAB (Mathworks, https://mathwork.com)
Custom MATLAB scripts (Prober Lab, https://github.com/proberlab/videotracker-scripts)
Excel (Microsoft, https://office.microsoft.com/excel/)
Prism (GraphPad Software, https://graphpad.com/)
Procedure
Solution Preparation
Prepare E3, methylene blue, and blue E3 solutions ahead of time. Recipes are provided at the end of this protocol.
Animal handling
Raise animals in accordance with local institutional animal care and usage committee recommendations. Take care to minimize stress to ensure the health of animals and maximize reproducibility of behavioral experiments.
Five days before the planned start of a behavioral experiment, set up mating tanks of well-fed sexually mature adult zebrafish pairs in the afternoon or evening, with a divider separating the male and female fish. See Notes 1, 2, and 3 on experimental design.
The next morning, remove the dividers and tip the tanks to encourage zebrafish mating.
Collect zebrafish embryos in blue E3 embryo medium and rinse well with system water to remove debris.
House zebrafish embryos in 100 mm × 15 mm Petri dishes, immersed in blue E3, with no more than 50 embryos per Petri dish, and raise under 14 h light: 10 h dark conditions at 28.5°C. Use embryos from large healthy clutches of greater than 100 embryos, so that comparisons can be made between experimental and control siblings.
At 24 h post-fertilization (hpf), carefully check all Petri dishes and remove debris and unfertilized embryos, as these can cause undue stress and impact development and behavior.
At 48 hpf, carefully check all Petri dishes and remove debris.
At 72 hpf, carefully check all Petri dishes and remove chorion fragments from hatched embryos. Change blue E3 media to standard E3 media.
At 4 dpf (~100-110 hpf), set up animals and reagents for video tracking experiment. First, check on the health of the zebrafish under a stereo microscope. Zebrafish should be swimming upright with inflated swim bladders (Figures 3 and 4A). Do not use larvae with an uninflated swim bladder, or larvae that are resting on their sides. Clutches with large numbers of larvae with uninflated swim bladders should not be used.
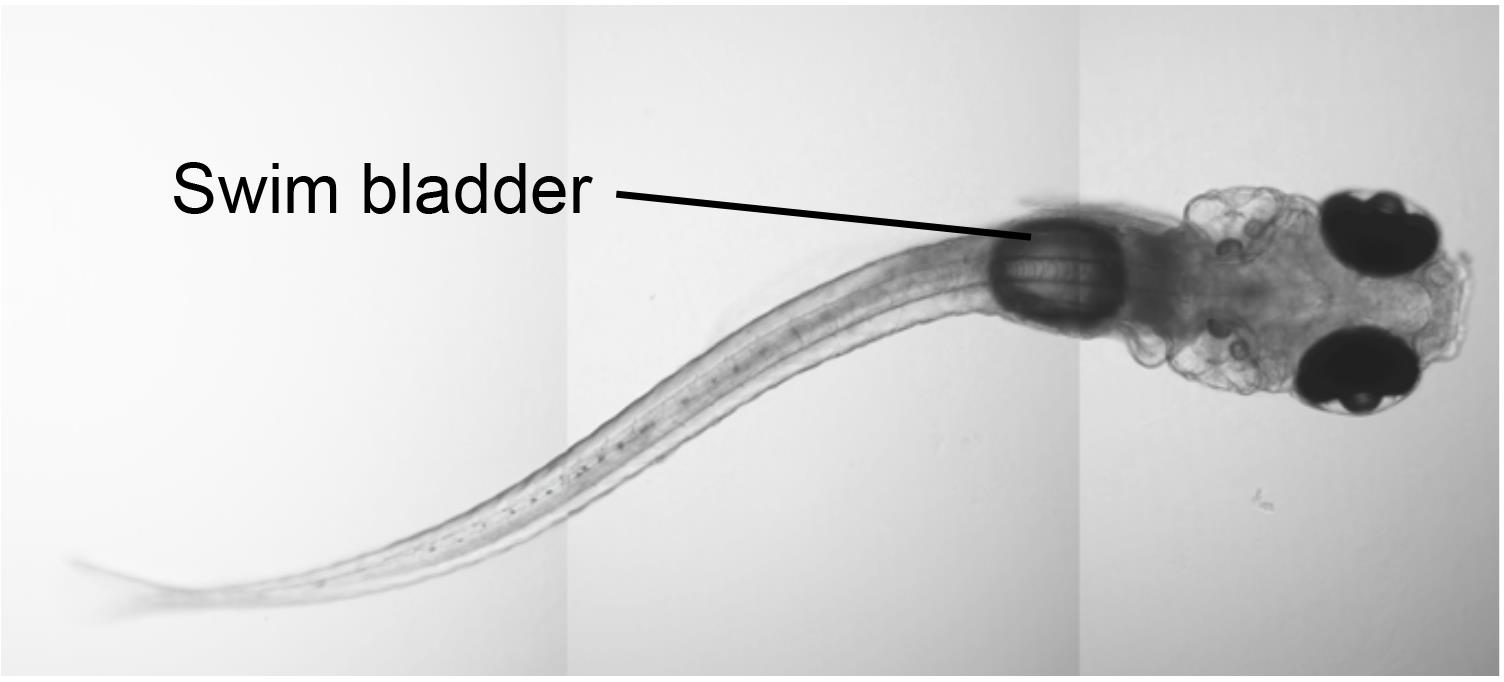
Figure 3. Example of a 5-dpf larval zebrafish with an inflated swim bladder.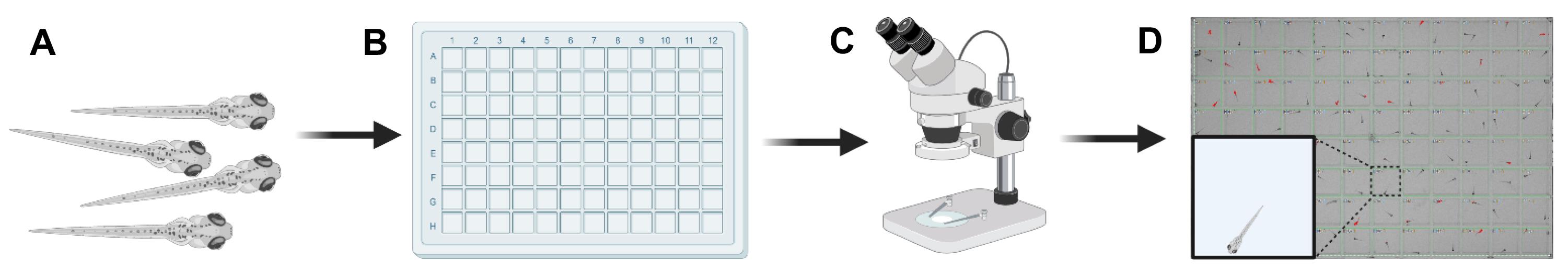
Figure 4. Workflow summary of animal handling. A. Step 8: Ensure that zebrafish larvae are healthy. B. Step 11: Gently transfer animals into 96-well plates. C. Step 12: Check that all animals are healthy under a stereo microscope, before placing the 96-well plate into the video tracking rig. D. Step 14: Seal the 96-well plate to prevent evaporation. A single zebrafish larva should be in each well. Red highlights indicate animals in which locomotor activity is detected by the video tracking system. Wells with air bubbles or more than one animal are excluded from analysis. Figure 4 was created with BioRender.com. White scale bar (inner well) ≈ 8.5 mm.Fill a 50 mL reagent reservoir with E3 medium heated to 28.5°C.
Gently fill a flat-bottom square-well 96-well plate (with a volume of 650 μL per well) with 600 μL of E3 medium per well using a 12-channel pipette. Be careful not to introduce air bubbles into the wells, as they can create video tracking artifacts. See Note 4 on using drugs in the experimental design.
Gently place a single healthy larva into each square well of the 96-well plate, using a disposable transfer pipette (Figure 4B).
Using a stereo microscope, check each well to make sure all animals look healthy, are upright, and have an inflated swim bladder (Figure 4C).
Add E3 medium to each well, so that it bulges on top of each well, but not so much that the surface tension breaks and pools with neighboring wells.
Seal the 96-well plate with an optical adhesive film using a seal applicator. The type of film used to seal the plate is very important, as some seals prevent O2 permeability or are toxic in some other way. The E3 medium will spill over the sides of the 96-well plate, but if done carefully, there should be no bubbles in most of the wells. An image of a well-sealed plate is shown below (Figure 5). See Note 5 on using seals in the experimental design.
Alternative steps: Steps 13 and 14 can be omitted, in which case E3 medium should be added to the top of each well, without creating bulges. This is important for the video tracker, to accurately track animal movements. Due to evaporation, fresh E3 medium should be added to each well each day. We usually perform this soon after the lights turn on in the morning. Working with open plates (Video 1) is useful when access to the fish during the experiment is required (e.g., for adding drugs to the wells during an experiment).
Video 1. Example of an un-sealed 96-well plate monitored using the Quantization mode with the following parameters: detection sensitivity, 25; burst (threshold for large movements), 900; freeze (threshold for no movement), 10; integration period (bin size), 60 s.Double-click to play video. Red highlights indicate animals in which locomotor activity is detected by the video tracking system.
Video tracking experiment
We use the ZebraBox, a commercial video tracking system from Viewpoint Life Sciences, Inc., to monitor zebrafish behavior from 4-8 dpf. Newer versions of the ZebraBox have been developed since the referenced article was published (Lee et al., 2017), and here we provide guidelines for using the fourth generation Zebrabox (latest version as of Spring 2021 of this writing). Like previous versions, the fourth generation Zebrabox contains an enclosed system, a high-resolution camera fitted with a fixed-angle lens, and a filter that transmits infrared light. White light illumination levels for standard behavioral experiments are set at ~1,200 lux during the day. We use a customized chamber that allows the 96-well plate to be immersed in recirculating water at 28.5°C, to maintain a constant and optimal temperature, which is essential for robust sleep/wake cycles (see Note 6). ZebraBoxes should be in a quiet and climate-controlled room, to ensure reproducibility of behavioral experiments. Video tracking data from two Zebraboxes can be recorded by a single computer, allowing for the simultaneous use of two 96-well plates in parallel.
Prior to placing a 96-well plate inside the ZebraBox, ensure that the distilled water within the chamber is clean and continuously recirculating at 28.5°C. This is important because floating debris in the water may produce movement artifacts in the video tracking data.
Place a 96-well plate inside the ZebraBox chamber and ensure that the plate is secure and not floating.
Open the video tracking software (ZebraLab; Viewpoint Life Sciences) on the computer accompanying the ZebraBox.
For most experiments, we use the Quantization mode with the following parameters: detection sensitivity: 25; burst (threshold for large movements): 900; freeze (threshold for no movement): 10; integration period (bin size): 60 s (Video 1). However, these values are only guidelines and must be empirically determined for each ZebraBox.
Determining the values of these parameters involves calibration of the signal-to-noise ratio with a focus on false positive signal reduction. To this end, one can either increase the detection sensitivity value (i.e., the change in pixel intensity that is required for that pixel to register as having changed its value) or increase the freezing threshold (i.e., the number of pixels per location that need to change from one frame to the next, for that location to be considered as registering a movement). The detection sensitivity and freezing threshold must be calibrated empirically for each ZebraBox. To determine these values, we use a 96-well plate containing both freely-swimming larvae and larvae that are anesthetized using 0.016% w/v tricaine, and record them for 1-3 h at each detection sensitivity and freezing threshold value (fish thus anesthetized will last for up to 8 hours without decomposition). Then, we analyze the data to identify the levels of false positive signals that are generated by the anesthetized fish with each of these parameters. In our hands, occasional values under 0.5 s of “movement” (for 60 s bins) from anesthetized fish has been an acceptable false positive signal. If higher levels are observed, increasing the sensitivity value or the freezing threshold can reduce the noise; however, this will also reduce the ability to detect real movements from the non-anesthetized fish. If a video tracker requires much higher detection sensitivity/freezing values than those mentioned here to ensure low levels of noise, we recommend servicing the machine.
Open the grid interface, which overlays each well with a green box. Align the grid interface with the square wells of the 96-well plate (Figure 5).
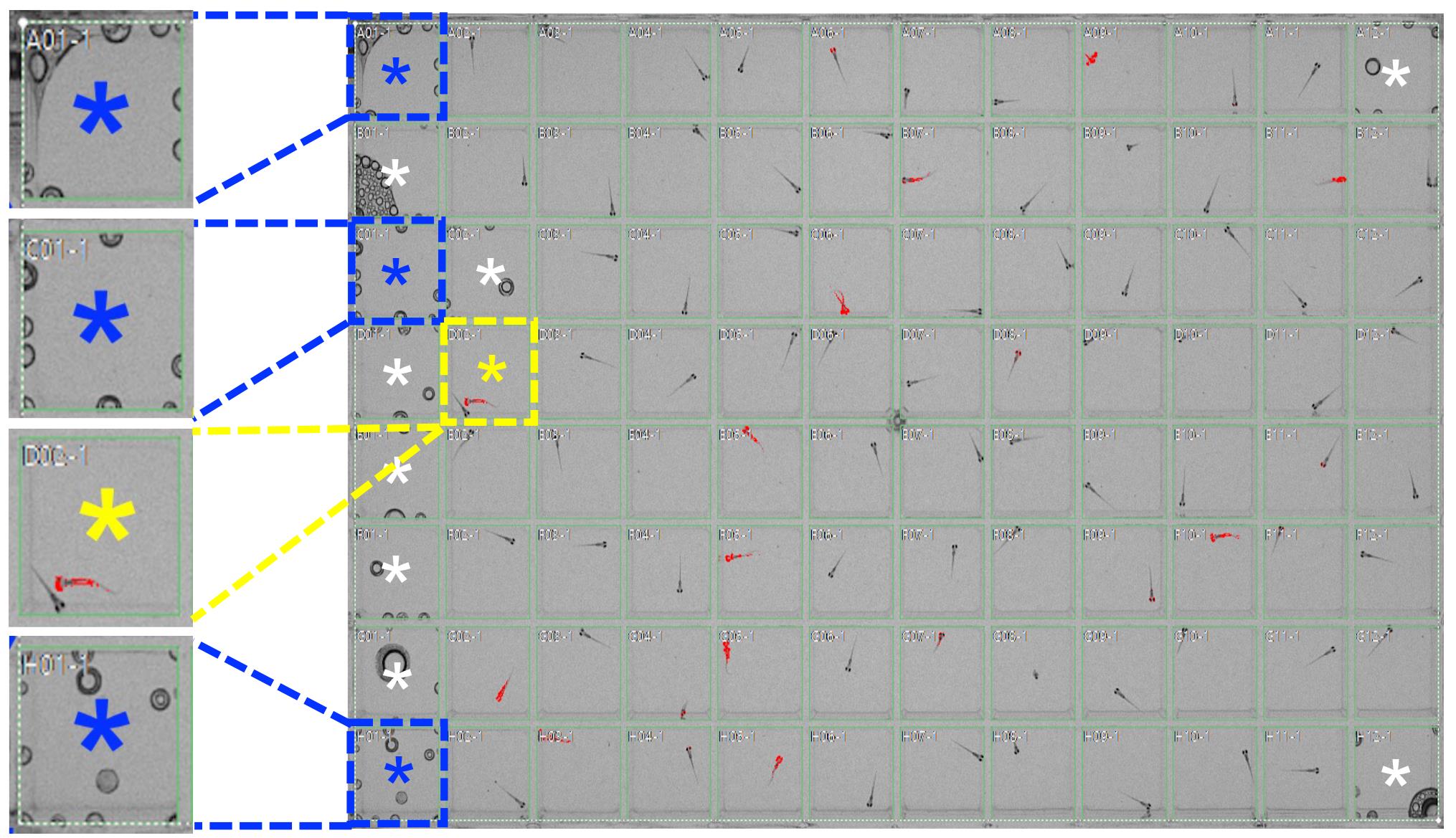
Figure 5. Example of the grid interface (green) that overlays the separate areas where behavior will be monitored (main figure). The PCR adhesive seal was applied from left to right on top of the plate. Most of the bubbles appear on the side where the seal was initially applied. Wells marked with an asterisk will be excluded from analysis due to the presence of air bubbles or multiple animals, respectively. Examples of such wells are enlarged and shown on the left. Red highlights indicate animals in which locomotor activity is detected by the video tracking system. White scale bar (inner well) ≈ 8.5 mm.Program the white lights to turn on and off at the desired times. We typically set them to turn on at 9 a.m. and off at 11 p.m., since these are the lighting conditions under which the animals have been raised. The chamber is continuously illuminated with infrared light to allow for video tracking regardless of whether the white lights are on or off.
Program the length of the experiment on the ZebraLab software interface. Behavioral experiments typically begin at 4 dpf and end at 7 dpf.
Save the ZebraLab software protocol for each experiment in a folder on your computer. Name and save the raw data files that will be generated.
Start the behavioral experiment using the ZebraLab software user interface.
Allow the behavioral experiment to run its planned time course (e.g., 48-72 h).
End the behavioral experiment on ZebraLab and allow the data to compile. A large .CSV or .XLS file is generated that contains locomotor activity and inactivity raw data. Take a screenshot of the green grid overlay on the 96-well plate as a quality control step, to ensure that the data collected for each well was confined to its assigned well and has not shifted to overlap with a neighboring well.
Remove the 96-well plate from the ZebraBox and inspect it using a stereo microscope. Note wells that contain bubbles or animals that appear to be unhealthy (e.g., uninflated swim bladder), and exclude these wells from analysis.
Genotype the animals using PCR or, if applicable, based on the presence or absence of a fluorescent marker using a fluorescent stereo microscope. See Notes 7 and 8 for relevant comments on experimental design.
Data analysis
To analyze sleep and wake behaviors for each animal, we use custom MATLAB scripts that build upon previous versions (Prober et al., 2006; Lee et al., 2017, 2020; Oikonomou et al., 2019). The VTs_to_DATA_middur.m script extracts the middur values from the .CVS/.XLS file generated by the Viewpoint software and organizes it in a matrix where each column is a fish and each row is a time bin. The last column of the matrix is the Zeitgeber time (ZT). The VT_analysis.m script extracts activity, waking activity, sleep, and sleep architecture (sleep bout number, sleep bout duration, and sleep latency) data for each fish, for each day and night, plots the results for each genotype (with or without SEM shading), and saves the results in a .mat file.
Custom Analysis Script Installation
Download the custom analysis script from Github (https://github.com/proberlab/videotracker-scripts).
Create a folder for each experiment that you wish to analyze (e.g., “/mutantX_20210821” for an experiment performed with mutant X fish on August 21, 2021). Within that folder set up the following folder structure (you can edit the script to change the folder scheme as you see fit):
/analysis_output
/data_original
/matalab_code
/matlab_data_processed
Place the "VT_analysis.m" and "VTs_to_DATA_middur.m" scripts in the “/matalab_code” folder. The other scripts are functions called by the two main scripts and should be placed in your MATLAB function folder.
Format Raw Data for Custom Analysis Script
Data from two Zebraboxes are generally recorded on a single computer, allowing for the simultaneous collection of data from two 96-well plates, or up to 192 animals. The output data is thus presented in a 1-192 well format.
Open the raw .CSV or .XLS data file in TextEdit or a similar text editor. Duplicate the file and save as a UTF-8.TXT file. We usually add the suffix “_UTF8” after the file name. We commonly use the format year-month-date e.g., YYYYMMDD_VT_experiment-name_UTF8.txt, where VT refers to the number of the video tracker. This step is necessary because MATLAB does not recognize the raw .CSV data file when it is saved in UTF-16 format. Place this file in the /matlab_data_processed folder.
Open the “VTs_to_DATA_middur.m” script in Matlab and change the filename (“input_file” variable) to match your case (Figure 6):

Figure 6. Screen capture of “VTs_to_DATA_middur.m” script in Matlab.Purple labeled filename in line 7 indicates the input file variable to be updated.
This script extracts the main variable (“middur”, seconds of movement per bin) for each fish. The data is then organized in a matrix with each column representing a fish, and each row a timepoint. The first two columns give the start and end times (in seconds) for each bin. The last column gives the Zeitgeber Time (ZT) with lights on at 9 a.m. (ZT 0) and lights off at 11 p.m. (ZT 14). The script will generate a .TXT file with the same name as the input file and the suffix '_DATA_middur.txt'. This is referred to as a “data file” in the following steps.
[Optional] Open the data file in Excel. At this point you can manually trim your data to remove partial days. For example, if the fish were loaded at the evening of 4 dpf, and the experiment was stopped at the morning of 7 dpf, the data leading up to 9 am of 5 dpf and the data collected after 9 am of 7 dpf can be removed and the file saved as “YYYYMMDD_VT_experiment-name_UTF8_TRIMMED.txt”. This new file will contain data from only the full days and nights 5 and 6 of development. This makes downstream analysis and plotting more streamlined.
Create a text (.txt) file in Excel that indicates the genotype, treatment, or condition for each well of the 96-well plate. For example, if your experiment contains mutant larvae obtained from an in-cross of heterozygous mutant fish, your genotyping list might look like the following (Figure 7):
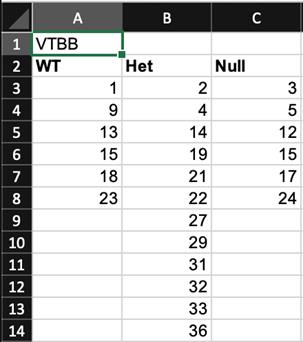
Figure 7. Screen capture of an example genotyping list text file in Excel.Green highlighted cell indicates the video tracker number that should be updated.
Cell A1: VTBB indicates which video tracker one is using. For example, video tracker number 1 in the lab should be updated to “VT01”.
Row 2: This indicates the genotype, treatment, or condition of each animal, which in this example consists of homozygous wild-type, heterozygous mutant (Het) and homozygous mutant (Null). The file can include up to 8 genotypes, treatments, or conditions.
Rows 3-14: This indicates which genotype, treatment, or condition is in each well. A minimum of three animals are required for each genotype, treatment, or condition.
We commonly name this file using the format YYYYMMDD_VT_genotype_N.txt where YYYYMMDD refers to year, month, and date, VT refers to the number of the video tracker, and N refers to the number of genotypes, treatments, or conditions. For the above example, an appropriate file name would be 20210821_10_genotype_3.txt. Place this file in the /matalab_data_processed folder.
Open the “VT_analysis.m” script within the /matlab_code folder. Find the following section and update the filenames of the data and genotype files (Figure 8):

Figure 8. Screen capture of “VT_analysis.m” script in Matlab.Purple labeled filenames in lines 9 and 10 indicate the data and genotype file names that need to be updated.
As mentioned in Section C, step 4, there will be some false positive movement signals from the video trackers (ideally not more than 0.5 s per 60 s time bin). Although this has a negligible effect on the amount of locomotor activity quantified, it can significantly distort sleep levels. To correct for this, we use a cut-off of 0.5 to 1 when converting activity levels to sleep (Figure 9). For example, a cut-off of 1 means that any 60 s bin that has movement of less than 1 s is considered sleep. This value might be different for different hardware set ups, and each ZebraBox needs to be calibrated for the parameters discussed in Section C, step 4.
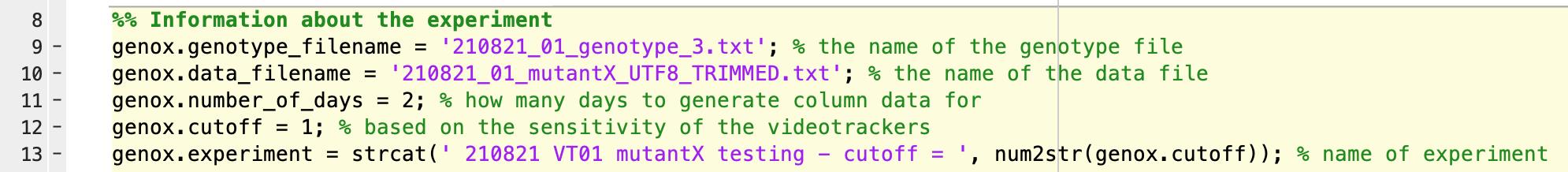
Figure 9. Screen capture of “VT_analysis.m” script in Matlab.The input in line 12 indicates the cut-off value that needs to be determined empirically for each ZebraBox.
Click “Run” to run the script.
The script extracts locomotor activity, waking activity (amount of locomotor activity while awake), sleep, and sleep architecture (sleep bout number, sleep bout duration, and sleep latency following light transitions) data for each fish and for each day and night period. It plots the results for each genotype (Figure 10 is an example), and saves the plots in different image file formats, as well as the results in a .mat file. These files are saved in the “/analysis_output” folder.
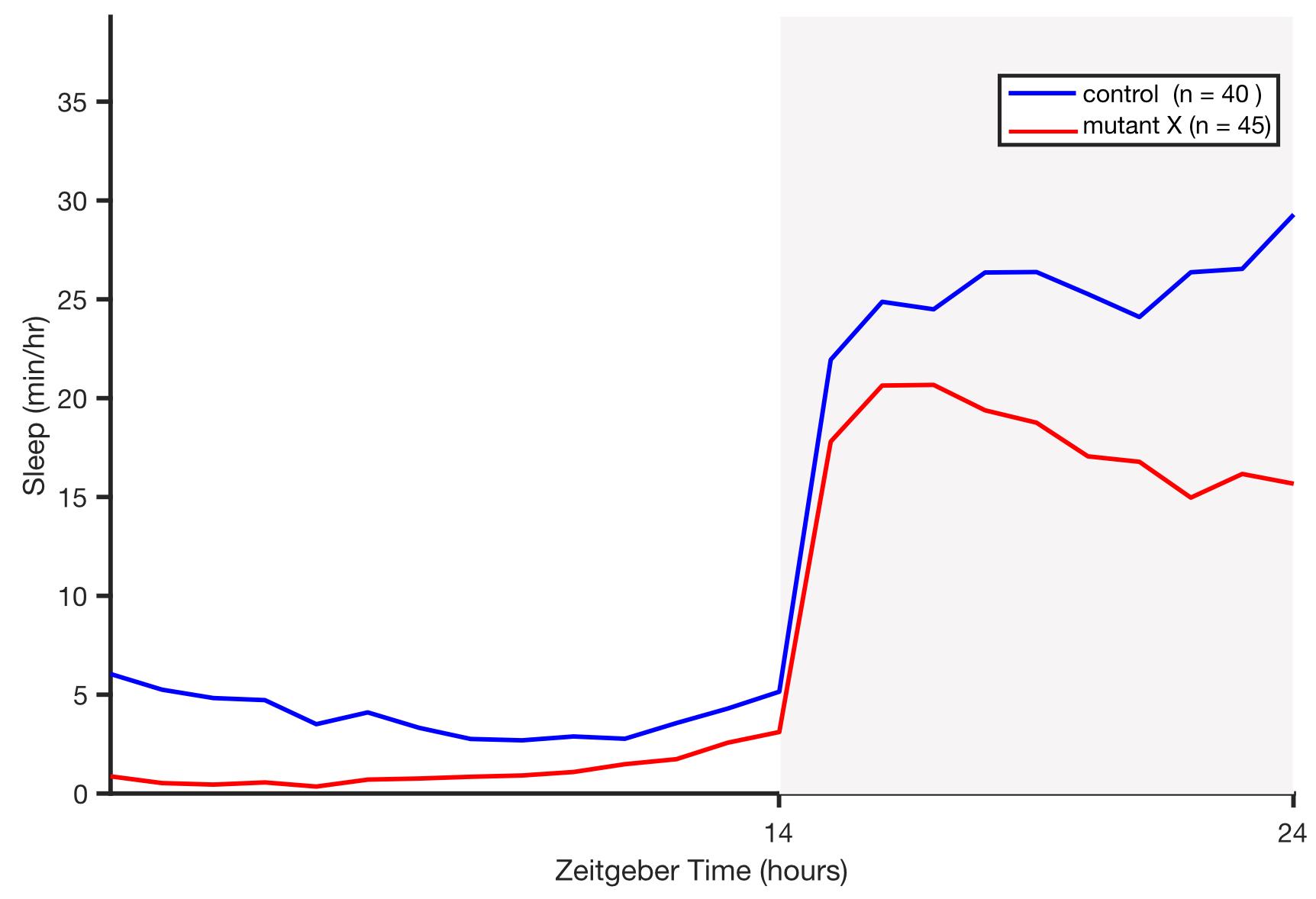
Figure 10. Example sleep plot output that compares sleep between mutant and wild-type siblings during a 24-hour experiment.Night is indicated by gray shading. n indicates the number of animals of each genotype. In this example, mutant animals sleep less than their wild-type sibling controls.
Statistical Analysis of Behavioral Data
To prepare the behavioral data for statistical analysis, open the. MAT summary data file in MATLAB.
In the MATLAB Workspace, an icon labeled “genox” will appear. Double-click this icon (Figure 11).
Figure 11. Screen capture of. MAT summary data file in Matlab and “genox” icon.The following information will appear (Figure 12):

Figure 12. Screen capture of Matlab data tables found within .MAT summary data file.Red arrow indicates the “summarytable” icon to open next.
Matlab Data as .MAT file
The dataset is organized as follows:
Name: Each [cell] contains the genotype name. Datasets are kept in the same order as listed in the genotype file.
Data: Each [cell] contains a n x m dataset, where n is the one-minute middur data for each timepoint (rows) and m is each well (columns).
fishID: Each [cell] contains an array with the well number for each genotype.
Zeitgeber: The time data for the experiment, with 0 = lights on, 14 = lights off.
Lightschedule: A map of the time data, marking the all night timepoints with a 1 and all day timepoints with a 0.
Time: Minutes from the start of the experiment.
Daynumber: A map of the time data, marking each day of the experiment (1, 2, 3, etc.) for each timepoint.
Lightboundries: Marks the night:day transitions and day:night transitions with a 1. All other datapoints are 0 s.
Tenminute: The same as Data, except the middur data is summed in 10 min bins.
Avewaking: Same as data, but all 0 bins are marked as NaNs.
Avewakechart1h: Same as data, but only active minutes are averaged into each 1 h measurement.
Avewakechart_10min: Same as data, but only active minutes are averaged into each 10-minute measurement.
Sleep: Each [cell] contains an n × m dataset, where n indicates the one-minute wake or sleep state for each timepoint (rows), and m is the well (columns). Each waking minute is denoted with a 0, and each sleep minute is denoted with a 1.
Sleepchart1h: The same as Sleep, except the data is summed in 1 h bins.
Sleepchart_10min: The same as Sleep, except the data is summed in 10 min bins.
Sleepcontinuity: The same as Sleep, except each sleep bout progressively increases in length, and resets to 0 when the fish moves. For example, if a fish is active, the sleep continuity value is a 0. The first sleep minute of a bout is 1, the 2nd min is 2, the 3rd min is 3, etc.
Sleepboutstart: The same as Sleep, except only the first sleep minute for each bout is marked as a 1. All other points are 0 s.
Sleeplatency: At each day:night and night:day transition, it marks all of the active minutes with a 1, until the first sleep bout. All other data is a 0.
Summarytable: Has several parts separated by genotype and/or condition and recaptures the summary information found in the tabular format described above in detail below.
Click on “summarytable” icon (Figure 12). The following information will appear (Figure 13):
Figure 13. Screen capture of Matlab data tables found within genox.summarytable.These tables provide a comprehensive summary for each genotype and/or condition of total sleep, number of sleep bouts, average sleep bout length, sleep latency, waking activity, and total locomotor activity for each day and night period that was measured. Some parameters, like “sleep” for example, are described in total amount for each day or night period, or in the “hourly_sleep” table, in average minutes of sleep per hour.
If desired, copy and paste relevant data from the “summarytable” from MATLAB to Excel and organize as preferred.
Use statistical analyses to determine if data are normally or non-normally distributed, followed by appropriate statistical tests and corrections for multiple comparisons. We typically use Prism, a statistical software package, for this purpose. See Note 9 on experimental design.
Notes
Cautionary point: For experiments that use mutant animals, perform an in-cross of heterozygous mutant fish to compare the behavior of homozygous wild-type, heterozygous mutant, and homozygous mutant sibling larvae to each other, to minimize variation due to genetic background.
Cautionary point: For experiments that use transgenic animals, heterozygous transgenic animals should be outcrossed to non-transgenic animals of the same parental strain (typically TLAB) to compare the behavior of transgenic and non-transgenic sibling larvae.
Cautionary point: We have observed that different commonly used zebrafish strains exhibit large differences in sleep behaviors, so the same background strain should be used when comparing experimental and control groups of larvae.
For experiments that test the effects of specific drugs on sleep, drugs can be added directly to wells during an experiment using plates that are not sealed (Rihel et al., 2010a). Alternatively, drugs may be added at the beginning of an experiment if it is desirable to use a sealed plate, although this requires the inclusion of an empty row of wells separating different treatment conditions because drugs may transfer to neighboring wells during the sealing process. We have observed rare cases in which a drug has different effects on sleep in sealed plates compared to open plates. Most drugs are prepared as stock solutions dissolved in DMSO and are diluted to the final desired concentration in E3 medium. The effective concentration of most drugs is between 100 nM and 1 mM. The final concentration of DMSO should not exceed 0.1%, because DMSO itself can affect behavior when used at higher concentrations.
There are advantages and disadvantages to using sealed plates. When using sealed plates, it is less work to maintain an experiment because the medium in each well does not evaporate, and thus it is not necessary to add E3 medium to each well each day. As a result, drug and salt concentrations remain more stable over the course of an experiment in sealed plates compared to unsealed plates. The addition of medium each day to unsealed plates also induces a behavioral artifact, which is avoided when using sealed plates. In addition, sleep/wake cycles are more robust and are maintained for longer periods of time using sealed plates, which is especially helpful for experiments that extend into 7- or 8-dpf. However, as noted above, sealed plates cannot be used for acute drug perturbations and, in rare cases, drugs induce different behavioral effects in sealed plates compared to unsealed plates, likely due to reaction of the drug with the seal adhesive. Other drawbacks to sealed plates are that the fish are exposed to the seal adhesive, oxygen levels in the E3 medium may decrease during an experiment, and often some wells must be excluded from the analysis due to the presence of air bubbles.
Cautionary point: Zebrafish should be raised and behaviorally assayed at 28.5°C. A difference of as little as 2°C above or below this temperature can adversely affect sleep. A water bath that circulates 28.5°C water below and around the 96-well plate within the ZebraBox is recommended, to maintain a constant, uniform, and optimal temperature.
Due to the variable nature of behaviors such as sleep, we typically require at least fifteen animals per genotype or condition in each experiment, and we perform comparisons between identically raised siblings whenever possible. Meaningful inferences between groups can be difficult to make when less than fifteen animals per genotype or condition are used.
Cautionary point: To minimize bias, behavioral experiments should be performed blind to genotype, when possible, with animals genotyped after each experiment using PCR or fluorescent markers.
We generally perform at least three independent experiments using identical treatment conditions, before drawing any conclusions.
Recipes
E3 medium 60× stock solution (1 L)
17.29 g NaCl
0.76 g KCl
2.9 g CaCl2·2H2O
4.9 g MgCl2·7H2O
To prepare a 60× stock, dissolve the ingredients in a final volume of 1 L of nanopure water. Adjust the pH to 7.0 with NaOH. Autoclave. To prepare a 1× working solution, dilute 333 mL of the 60× stock to a final volume of 20 L with nanopure water. Leave it overnight to mix at room temperature. Adjust the pH to 6.8-7.0 with sodium bicarbonate or acetic acid the next day.
Methylene Blue solution (1 g/L stock)
To prepare a methylene blue stock solution, dissolve 1 g of methylene blue hydrate (Sigma) in 1 L of water.
Blue E3 medium solution
To the 20 L of 1× E3 medium described above, add 20 mL of 1% methylene blue (as antifungal agent) and mix well.
Acknowledgments
This protocol builds upon methods developed by Jason Rihel, David Prober and Alexander Schier (Prober et al., 2006; Rihel et al., 2010b), which were subsequently updated based on experimental and analytical advances (Lee et al., 2017, 2019 and 2020; Oikonomou et al., 2019). This work was supported by grants from the NIH (DAL: K99NS097683, F32NS084769; GO: F32NS082010; DAP: R35NS122172, R01MH121601), a NARSAD Young Investigator Grant (DAL: 25392) and a Caltech BBE Postdoctoral Fellowship (DAL).
Competing interests
The authors declare no conflicts of interest.
Ethics
Animal husbandry and experiments involving zebrafish were performed in accordance with the California Institute of Technology Institutional Animal Care and Use Committee (IACUC) guidelines and by the Office of Laboratory Animal Resources at the California Institute of Technology (animal protocol 1580).
References
- Appelbaum, L., Wang, G., Yokogawa, T., Skariah, G. M., Smith, S. J., Mourrain, P. and Mignot, E. (2010). Circadian and homeostatic regulation of structural synaptic plasticity in hypocretin neurons. Neuron 68(1): 87-98.
- Ahrens, M. B., Li, J. M., Orger, M. B., Robson, D. N., Schier, A. F., Engert, F. and Portugues, R. (2012). Brain-wide neuronal dynamics during motor adaptation in zebrafish. Nature 485(7399): 471-477.
- Campbell, S. S. and Tobler, I. (1984). Animal sleep: a review of sleep duration across phylogeny. Neurosci Biobehav Rev 8(3): 269-300.
- Cirelli, C., Bushey, D., Hill, S., Huber, R., Kreber, R., Ganetzky, B. and Tononi, G. (2005). Reduced sleep in Drosophila Shaker mutants. Nature 434(7037): 1087-1092.
- Chiu, C. N., Rihel, J., Lee, D. A., Singh, C., Mosser, E. A., Chen, S., Sapin, V., Pham, U., Engle, J., Niles, B. J., et al. (2016). A Zebrafish Genetic Screen Identifies Neuromedin U as a Regulator of Sleep/Wake States. Neuron 89(4): 842-856.
- Chen, S., Reichert, S., Singh, C., Oikonomou, G., Rihel, J. and Prober, D. A. (2017). Light-Dependent Regulation of Sleep and Wake States by Prokineticin 2 in Zebrafish. Neuron 95(1): 153-168.e156.
- Churgin, M. A., Jung, S. K., Yu, C. C., Chen, X., Raizen, D. M. and Fang-Yen, C. (2017). Longitudinal imaging of Caenorhabditis elegans in a microfabricated device reveals variation in behavioral decline during aging. Elife 6: e26652.
- Elbaz, I., Yelin-Bekerman, L., Nicenboim, J., Vatine, G. and Appelbaum, L. (2012). Genetic ablation of hypocretin neurons alters behavioral state transitions in zebrafish.J Neurosci 32(37): 12961-12972.
- Fisher, S. P., Godinho, S. I., Pothecary, C. A., Hankins, M. W., Foster, R. G. and Peirson, S. N. (2012). Rapid assessment of sleep-wake behavior in mice. J Biol Rhythms 27(1): 48-58.
- Gandhi, A. V., Mosser, E. A., Oikonomou, G. and Prober, D. A. (2015). Melatonin is required for the circadian regulation of sleep. Neuron 85(6): 1193-1199.
- Hendricks, J. C., Finn, S. M., Panckeri, K. A., Chavkin, J., Williams, J. A., Sehgal, A. and Pack, A. I. (2000). Rest in Drosophila is a sleep-like state. Neuron 25(1): 129-138.
- Huang, H., Zhu, C. T., Skuja, L. L., Hayden, D. J. and Hart, A. C. (2017). Genome-Wide Screen for Genes Involved in Caenorhabditis elegans Developmentally Timed Sleep. G3 (Bethesda) 7(9): 2907-2917.
- Jeong, J. Y., Kwon, H. B., Ahn, J. C., Kang, D., Kwon, S. H., Park, J. A. and Kim, K. W. (2008). Functional and developmental analysis of the blood-brain barrier in zebrafish. Brain Res Bull 75(5): 619-628.
- Koh, K., Joiner, W. J., Wu, M. N., Yue, Z., Smith, C. J. and Sehgal, A. (2008). Identification of SLEEPLESS, a sleep-promoting factor. Science 321(5887): 372-376.
- Lee, D. A., Andreev, A., Truong, T. V., Chen, A., Hill, A. J., Oikonomou, G., Pham, U., Hong, Y. K., Tran, S., Glass, L., et al. (2017). Genetic and neuronal regulation of sleep by neuropeptide VF. Elife 6: e25727.
- Lee, D. A., Liu, J., Hong, Y., Lane, J. M., Hill, A. J., Hou, S. L., Wang, H., Oikonomou, G., Pham, U., Engle, J., et al. (2019). Evolutionarily conserved regulation of sleep by epidermal growth factor receptor signaling.Sci Adv 5(11): eaax4249.
- Lee, D. A., Oikonomou, G., Cammidge, T., Andreev, A., Hong, Y., Hurley, H. and Prober, D. A. (2020). Neuropeptide VF neurons promote sleep via the serotonergic raphe. Elife 9: e54491.
- Leung, L. C., Wang, G. X., Madelaine, R., Skariah, G., Kawakami, K., Deisseroth, K., Urban, A. E. and Mourrain, P. (2019). Neural signatures of sleep in zebrafish. Nature 571(7764): 198-204.
- Ly, S., Lee, D. A., Strus, E., Prober, D. A. and Naidoo, N. (2020). Evolutionarily Conserved Regulation of Sleep by the Protein Translational Regulator PERK. Curr Biol 30(9): 1639-1648 e1633.
- Marques, J. C., Li, M., Schaak, D., Robson, D. N. and Li, J. M. (2020). Internal state dynamics shape brainwide activity and foraging behaviour. Nature 577(7789): 239-243.
- Nath, R. D., Bedbrook, C. N., Abrams, M. J., Basinger, T., Bois, J. S., Prober, D. A., Sternberg, P. W., Gradinaru, V. and Goentoro, L. (2017). The Jellyfish Cassiopea Exhibits a Sleep-like State. Curr Biol 27(19): 2984-2990 e2983.
- Oikonomou, G., Altermatt, M., Zhang, R. W., Coughlin, G. M., Montz, C., Gradinaru, V. and Prober, D. A. (2019). The Serotonergic Raphe Promote Sleep in Zebrafish and Mice. Neuron 103(4): 686-701 e688.
- Özcan, G. G., Lim, S., Leighton, P. and Allison, W. T. (2020). Sleep is bi-directionally modified by amyloid beta oligomers. Elife 9: e53995.
- Prober, D. A., Rihel, J., Onah, A. A., Sung, R. J. and Schier, A. F. (2006). Hypocretin/orexin overexpression induces an insomnia-like phenotype in zebrafish. J Neurosci 26(51): 13400-13410.
- Pack, A. I., Galante, R. J., Maislin, G., Cater, J., Metaxas, D., Lu, S., Zhang, L., Von Smith, R., Kay, T., Lian, J., et al. (2007). Novel method for high-throughput phenotyping of sleep in mice. Physiol Genomics 28(2): 232-238.
- Renier, C., Faraco, J. H., Bourgin, P., Motley, T., Bonaventure, P., Rosa, F. and Mignot, E. (2007). Genomic and functional conservation of sedative-hypnotic targets in the zebrafish. Pharmacogenet Genomics 17(4): 237-253.
- Raizen, D. M., Zimmerman, J. E., Maycock, M. H., Ta, U. D., You, Y. J., Sundaram, M. V. and Pack, A. I. (2008). Lethargus is a Caenorhabditis elegans sleep-like state. Nature 451(7178): 569-572.
- Reichert, S., Pavón Arocas, O. and Rihel, J. (2019). The Neuropeptide Galanin Is Required for Homeostatic Rebound Sleep following Increased Neuronal Activity. Neuron 104(2): 370-384.e375.
- Rihel, J., Prober, D. A., Arvanites, A., Lam, K., Zimmerman, S., Jang, S., Haggarty, S. J., Kokel, D., Rubin, L. L., Peterson, R. T., et al. (2010a). Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science 327(5963): 348-351.
- Rihel, J., Prober, D. A. and Schier, A. F. (2010b). Monitoring sleep and arousal in zebrafish. Methods Cell Biol 100: 281-294.
- Shaw, P. J., Cirelli, C., Greenspan, R. J. and Tononi, G. (2000). Correlates of sleep and waking in Drosophila melanogaster. Science 287(5459): 1834-1837.
- Singh, C., Oikonomou, G. and Prober, D. A. (2015). Norepinephrine is required to promote wakefulness and for hypocretin-induced arousal in zebrafish. Elife 4: e07000.
- Singh, C., Rihel, J. and Prober, D. A. (2017). Neuropeptide Y Regulates Sleep by Modulating Noradrenergic Signaling. Curr Biol 27(24): 3796-3811 e3795.
- Toda, H., Williams, J. A., Gulledge, M. and Sehgal, A. (2019). A sleep-inducing gene, nemuri, links sleep and immune function in Drosophila.Science 363(6426): 509-515.
- Yokogawa, T., Marin, W., Faraco, J., Pezeron, G., Appelbaum, L., Zhang, J., Rosa, F., Mourrain, P. and Mignot, E. (2007). Characterization of sleep in zebrafish and insomnia in hypocretin receptor mutants. PLoS Biol 5(10): e277.
- Yelin-Bekerman, L., Elbaz, I., Diber, A., Dahary, D., Gibbs-Bar, L., Alon, S., Lerer-Goldshtein, T. and Appelbaum, L. (2015). Hypocretin neuron-specific transcriptome profiling identifies the sleep modulator Kcnh4a. Elife 4: e08638.
- Zada, D., Bronshtein, I., Lerer-Goldshtein, T., Garini, Y. and Appelbaum, L. (2019). Sleep increases chromosome dynamics to enable reduction of accumulating DNA damage in single neurons. Nat Commun 10(1): 895.
- Zhdanova, I. V., Wang, S. Y., Leclair, O. U. and Danilova, N. P. (2001). Melatonin promotes sleep-like state in zebrafish. Brain Res 903(1-2): 263-268.
- Zimmerman, J. E., Raizen, D. M., Maycock, M. H., Maislin, G. and Pack, A. I. (2008). A video method to study Drosophila sleep. Sleep 31(11): 1587-1598.
Article Information
Copyright
Lee et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Lee, D. A., Oikonomou, G. and Prober, D. A. (2022). Large-scale Analysis of Sleep in Zebrafish. Bio-protocol 12(3): e4313. DOI: 10.21769/BioProtoc.4313.
- Lee, D. A., Andreev, A., Truong, T. V., Chen, A., Hill, A. J., Oikonomou, G., Pham, U., Hong, Y. K., Tran, S., Glass, L., et al. (2017). Genetic and neuronal regulation of sleep by neuropeptide VF. Elife 6: e25727.
Category
Neuroscience > Basic technology
Drug Discovery
Biological Sciences > Biological techniques
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









