- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Listeria innocua Biofilm Assay Using NanoLuc Luciferase
Published: Vol 12, Iss 3, Feb 5, 2022 DOI: 10.21769/BioProtoc.4308 Views: 3366
Reviewed by: Alba BlesaVinai Chittezham ThomasTimo A Lehti

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Mobilization of Plasmids from Bacteria into Diatoms by Conjugation Technique
Federico Berdun [...] Eduardo Zabaleta
Mar 5, 2024 1917 Views

Campylobacter jejuni Biofilm Assessment by NanoLuc Luciferase Assay
Tjaša Čukajne [...] Anja Klančnik
Feb 20, 2025 1636 Views

Artificial Metalloenzymes in Artificial Sanctuaries Through Liquid–Liquid Phase Separation
Kaixin Wang [...] Tong Wu
Oct 5, 2025 1566 Views
Abstract
Biofilms serve as a bacterial survival strategy, allowing bacteria to persist under adverse environmental conditions. The non-pathogenic Listeria innocua is used as a surrogate organism for the foodborne pathogen Listeria monocytogenes, because they share genetic and physiological similarities and can be used in a Biosafety Level 1 laboratory. Several methods are used to evaluate biofilms, including different approaches to determine biofilm biomass or culturability, viability, metabolic activity, or other microbial community properties. Routinely used methods for biofilm assay include the classical culture-based plate counting method, biomass staining methods (e.g., crystal violet and safranin red), DNA staining methods (e.g., Syto 9), methods that use metabolic substrates to detect live bacteria (e.g., tetrazolium salts or resazurin), and PCR-based methods to quantify bacterial DNA. The NanoLuc (Nluc) luciferase biofilm assay is a viable alternative or complement to existing methods. Functional Nluc was expressed in L. innocua using the nisin-inducible expression system and bacterial detection was performed using furimazine as substrate. Concentration dependent bioluminescence signals were obtained over a concentration range greater than three log units. The Nluc bioluminescence method allows absolute quantification of bacterial cells, has high sensitivity, broad range, good day-to-day repeatability, and good precision with acceptable accuracy. The advantages of Nluc bioluminescence also include direct detection, absolute cell quantification, and rapid execution.
Graphic abstract:
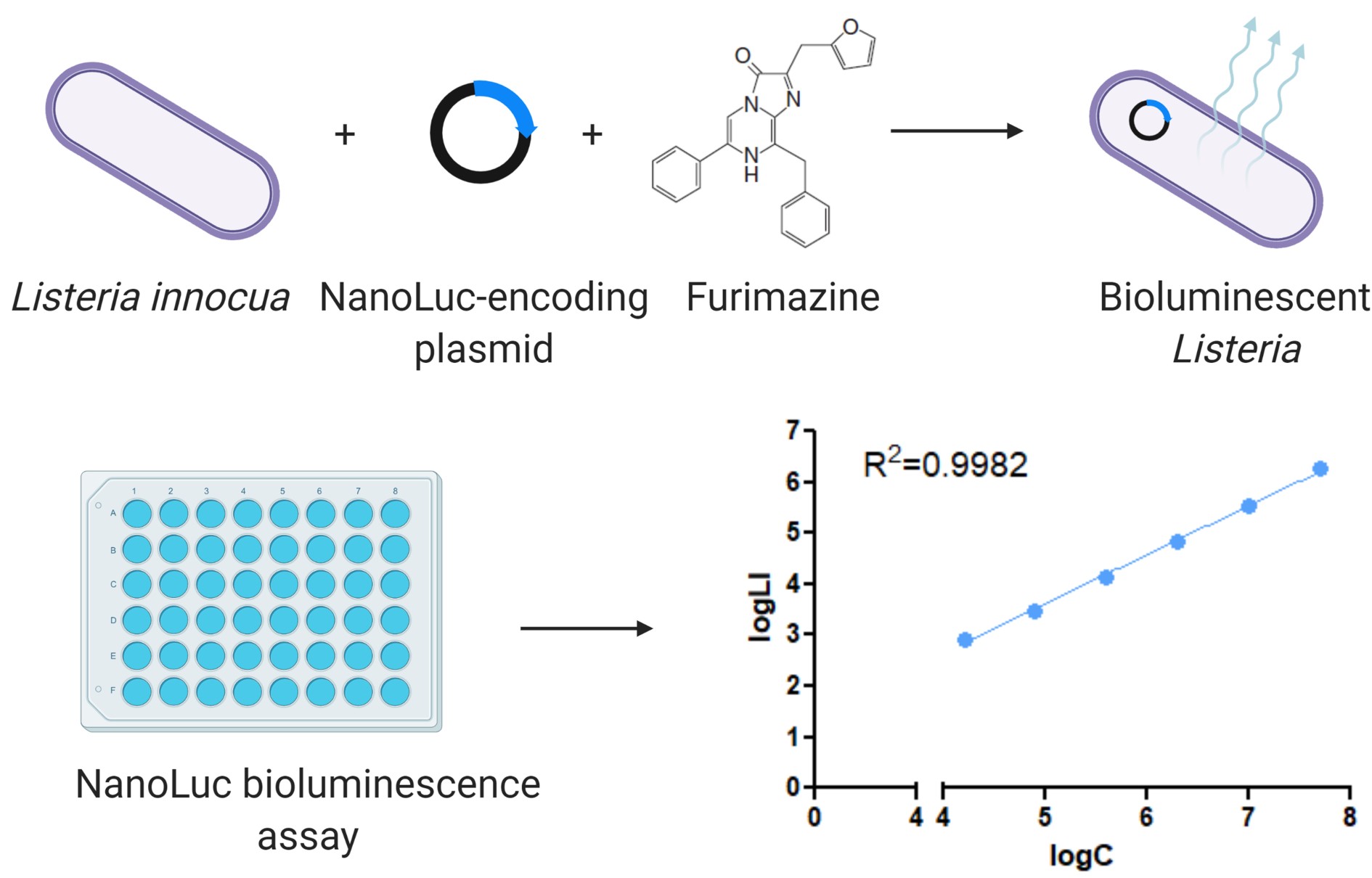
Engineering Listeria innocua to express NanoLuc and its application in bioluminescence assay.
Background
Bacteria form biofilms as a survival strategy, as they enable bacteria to survive and persist in adverse environmental conditions. Within biofilms, bacteria are protected from the human immune system and a variety of environmental factors (e.g., UV, acids, desiccation, salinity, antimicrobials, disinfectants). Opportunistic pathogenic bacteria commonly associated with food poisoning are linked to biofilms formed on surfaces in the food supply chain, and are a significant public health concern. These biofilms can cause persistent contamination and recurrent infections (Donlan, 2002; Chlebicz and Slizewska, 2018; Flemming and Wuertz, 2019). Listeria monocytogenes, the causative agent of listeriosis, is transmitted through the consumption of contaminated food and is associated with high hospitalization and mortality rates compared to other foodborne infectious diseases (EFSA and ECDC, 2019). L. monocytogenes can successfully colonize food processing environments, where it can persist in biofilms and cause outbreaks over several years (Buchanan et al., 2017). The non-pathogenic Listeria innocua has been proposed as a surrogate organism for L. monocytogenes (Costa et al., 2018). Their genomes are conserved (more than 2,500 orthologous genes) and co-linear (Glaser et al., 2001), and they share physiological similarities. However, L. innocua lacks the virulence factors of L. monocytogenes (Fairchild and Foegeding, 1993; Friedly et al., 2008; Silva-Angulo et al., 2015; Costa et al., 2018), so it can be used in Biosafety Level 1 laboratories. Biofilms formed by different strains of L. monocytogenes and L. innocua have often been described as thin and structurally variable (Costa et al., 2018; Lee et al., 2019). Currently available methods target thick and multilayer biofilms, far from the thin biofilms of Listeria.
Several methods have been developed for the quantification of cells and evaluation of listerial biofilms, including different approaches to determine biofilm biomass or culturability, viability, metabolic activity, or other cell properties. These include the classical culture-based plate counting method, biomass staining methods (e.g., crystal violet and safranin red), DNA staining methods (e.g., Syto 9), the use of chromogenic or fluorogenic metabolic substrates to detect live bacteria (e.g., tetrazolium salts or resazurin), and qPCR and digital droplet PCR to quantify bacterial DNA (Stiefel et al., 2016; Klančnik et al., 2017; Ripolles-Avila et al., 2019; Grudlewska-Buda et al., 2020).
The luciferase from Oplophorus gracilirostris, known as NanoLuc (Nluc), has been reported to have excellent properties, including small size (19 kDa), high stability, and high bioluminescence efficiency (150-fold higher compared to other luciferases) (England et al., 2016). The NanoLuc luciferase biofilm assay was compared with the other commonly used methods for detecting biofilms in microtiter plates, i.e., conventional plating and CFU counting, crystal violet staining assay, and resazurin fluorescence assay (Berlec et al., 2021). These methods differ in terms of research equipment and chemicals required, time needed to perform the assay, sensitivity and suitability for observing dynamic processes, and biofilm components detected. The Nluc bioluminescence assay detects all phenotypic subpopulations within the growing biofilm, culturable, viable-but-not culturable forms, and also damaged but non-lysed cells. This can be beneficial in assessing the overall biofilm population, which includes slow growing tolerant and persistent cells that are critical for biofilm persistence (Matereke and Okoh, 2020). Crystal violet staining binds to negatively charged molecules in the biofilm biomass, detecting live and dead cells, as well as the exopolymeric matrix (Costa et al., 2018). The major problem with the crystal violet assay is its variability and, thus, variations in the data due to the user performing the assay (Lourenco et al., 2012) and the high detection limit. Better reproducibility of data can be achieved with live cell methods. Since transition of Listeria cells to a viable-but-non-culturable state is also possible under stress conditions (Highmore et al., 2018), the disadvantage of the CFU counting assay was that it only determined culturable live cells. In contrast, all live cells were determined using the resazurin fluorescence assay, which is based on the detection of metabolically active cells (Van den Driessche et al., 2014). This method was less sensitive, requiring at least 106 CFU/mL to detect a change in fluorescence.
The NanoLuc luciferase biofilm assay is a viable complement or addition to existing methods. Limitations of this method include the requirement of genetic modification of the bacteria and the high cost, even though the cost is lower than that for the molecular biology methods (e.g., qPCR). On the other hand, the Nluc bioluminescence assay has several important advantages over other methods, including direct detection, absolute cell quantification, broad dynamic range (1.0 × 104 CFU/mL – 5.0 × 107 CFU/mL), low time requirement, and high sensitivity (1.0 × 104 CFU/mL).
The protocol can be adapted to other bacteria by appropriate modification of the genetic construct (plasmid or promoter). Due to its high sensitivity, it is particularly useful for studies on early steps of biofilm development and for the bacteria that do not form thick biofilms. It can be easily applied to the screening of substances for antibiofilm activity.
Materials and Reagents
White flat-bottomed 96-well plates (Corning, catalog number: 3990)
15 mL centrifuge tube, PP, screw cap (Brand, catalog number: 114818)
1.5 mL microcentrifuge tubes (Eppendorf Safe-Lock, catalog number: EP0030123611)
Filtermax filter top (TPP, catalog number: 99505)
Presterile 50 mL disposable reservoirs (Biotix, catalog number: SR-0050-1SC)
Listeria innocua ŽM39 transformed with pMSP::Nluc (Jožef Stefan Institute collection) (Figure 1)
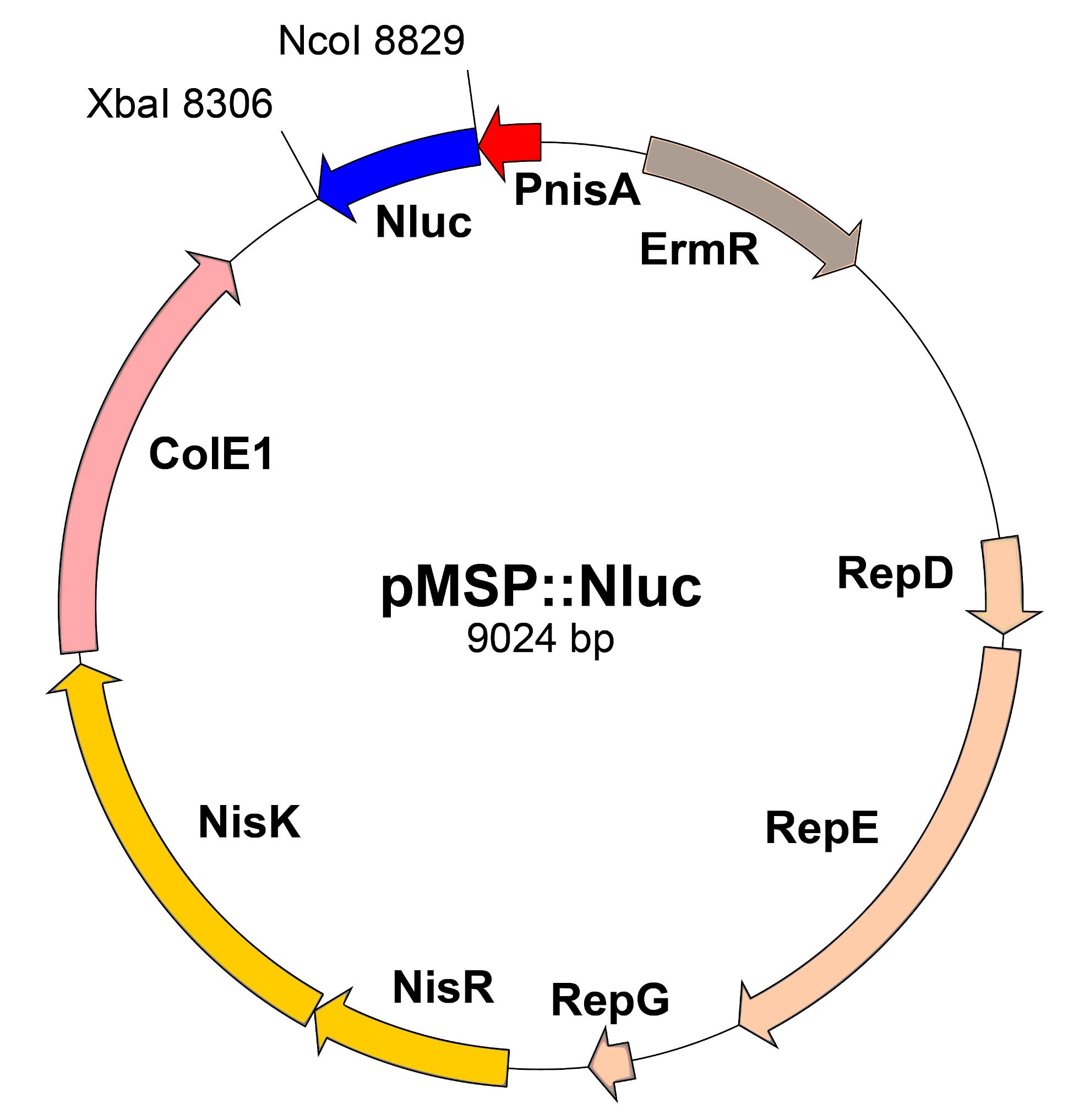
Figure 1. Scheme of pMSP::Nluc plasmid. PnisA: nisin promoter. ErmR: erythromycin resistance marker. RepD-G: replication genes. NisRK: nisin response genes. ColE1: E. coli replication gene. Nluc: Nanoluc.Tryptic soy broth (Merck, catalog number: 1.05459.0500)
Agar (Formedium, catalog number: AGA03)
Erythromycin (Merck, Sigma-Aldrich, catalog number: E5380)
Nisin (Merck, Sigma-Aldrich, catalog number: 72535)
Nano-Glo Luciferase Assay System (Promega, catalog number: N1120)
NaCl
KCl
Na2HPO4·2H2O
KH2PO4
Phosphate-buffered saline (PBS) (see Recipes)
TSA (see Recipes)
Equipment
Incubator (Binder)
Shaker/Incubator (Braun Certomat H)
Luminescence plate reader (Tecan, model: Infinite M-1000)
Pipette set (Eppendorf, catalog number: Z683892)
Multichannel pipette (Eppendorf, catalog number: 2231300048; model: Research Plus)
Biosafety cabinet (Labcaire)
Deep freezer (Sanyo, model: MDF-U53V)
Spectrophotometer (Perkin Elmer, catalog number: L7110187; model: Lambda Bio+)
Centrifuge (Hettich, catalog number: 1706; model: Rotina 380R)
Procedure
Reviving glycerol stock of L. innocua
Remove vial of permanent culture of Listeria innocua ŽM39 transformed with pMSP::Nluc from deep freezer and place it on ice.
Aseptically transfer culture to TSA plates containing 10 µg/mL erythromycin with sterile loop.
Incubate at 37°C for 24 h.
Overnight culture for biofilm inoculum or calibration curve culture preparation
Prepare 5 mL of TSB medium with 10 μg/mL erythromycin in a 15 mL tube.
Transfer biomass equivalent to three medium sized colonies from the TSA agar plate and inoculate it into the medium prepared above.
Incubate with shaking (190 RPM) at 37°C for 18-24 h.
Setting up biofilm in microtiter plate
Centrifuge overnight culture at 3,000 × g and 4°C for 10 min.
Resuspend the pellet in 5 mL of TSB medium with 10 μg/mL erythromycin and 5 μL of nisin for Nluc induction (stock solution concentration: 1 mg/mL).
Dilute the suspension until its OD600 value corresponds to 1 × 107 CFU/mL (see Note 1).
Dilute bacterial suspension from Step C3 as required [e.g., 100-fold in TSB with 10 μg/mL erythromycin and 5 μL of nisin (stock solution concentration: 1 mg/mL) to obtain dilution corresponding to 1 × 105 CFU/mL].
Pipette 50 μL of the bacterial suspensions per well onto the microtiter plate in triplicate.
Incubate at 30°C (see Note 2) for a selected time period (e.g., 4 h, 8 h, 24 h, 48 h, or 72 h).
Wash biofilm three time using 80 µL of PBS. After last wash, add 50 µL of PBS to each well. Proceed with bioluminescence measurement.
Preparation of calibration curve (see Note 3)
Before the measurement (8 h), prepare 5 mL of TSB medium with 10 μg/mL erythromycin and 5 μL of nisin (stock solution concentration: 1 mg/mL) in a 15 mL tube.
Transfer 100 µL of the overnight culture (prepared in section B) to the medium and incubate with shaking (190 RPM) at 37°C for 8 h.
Immediately before the measurement, calibrate the OD600 of the suspension to correspond to 5 × 107 CFU/mL. This is the bacterial output suspension for the calibration curve.
In 1.5 mL tubes, prepare five consecutive five-fold dilutions that will serve as a calibration curve.
Transfer 50 μL of dilutions to the biofilm plate in duplicate.
Bioluminescence measurement
Prepare the appropriate amount of substrate by mixing one volume of Nano-Glo Luciferase Assay Substrate with 50 volumes of Nano-Glo Luciferase Assay Buffer (e.g., 200 μL of Substrate in 10 mL of Buffer). Prior to use, incubate the substrate at 37°C to equal the temperature of the samples.
Add 50 µL of substrate per well quickly with a multichannel pipette. Wait 3 min and then measure the bioluminescence for the first time using plate reader (e.g., Infinite M-1000, settings: Luminescence mode; Integration time, 1 s; Settle time, 150 ms; Figure 2).
Measure bioluminescence at 10 min intervals three more times (see Note 4).
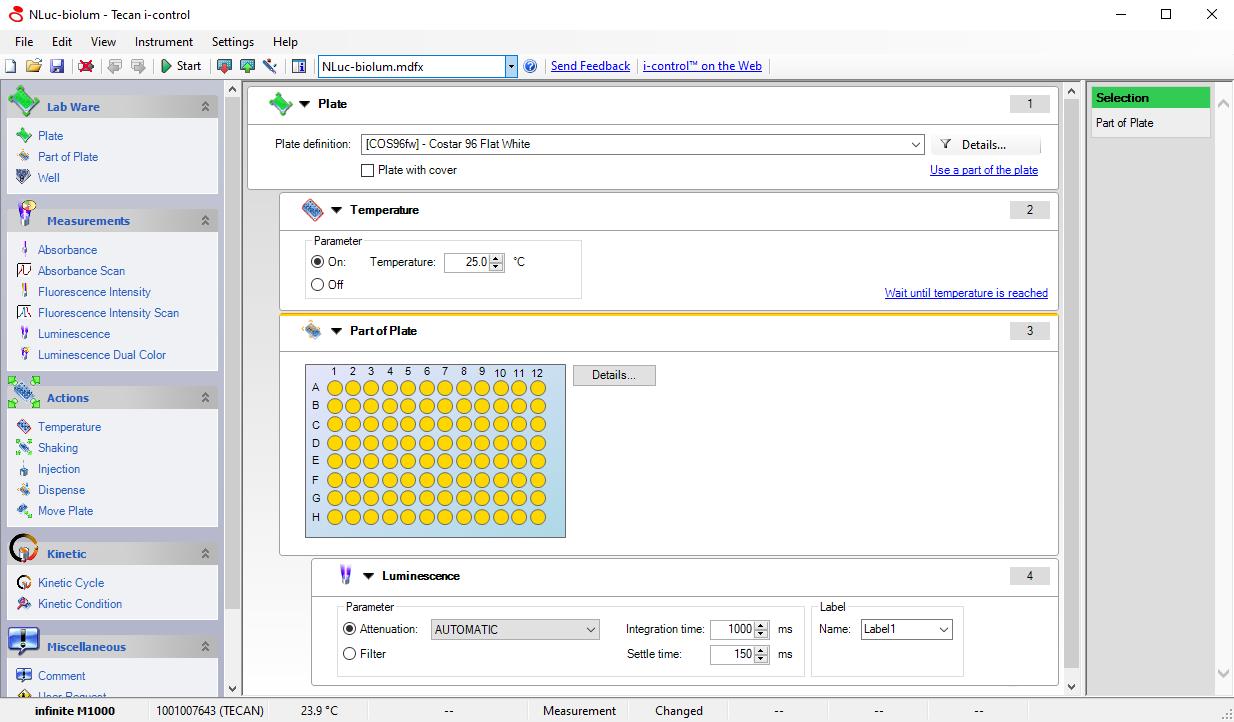
Figure 2. Screenshot of microplate reader settings for measuring bioluminescence of Nluc.Calculating concentration of bacteria in biofilms
Use Excel to calculate logarithms of bacterial concentrations and logarithms of corresponding average bioluminescence and use them to plot the calibration curve. A representative calibration curve is shown in Figure 3.
Use linear regression (Excel functions: Slope and Intercept) to calculate biofilm cell concentrations from measured bioluminescence.
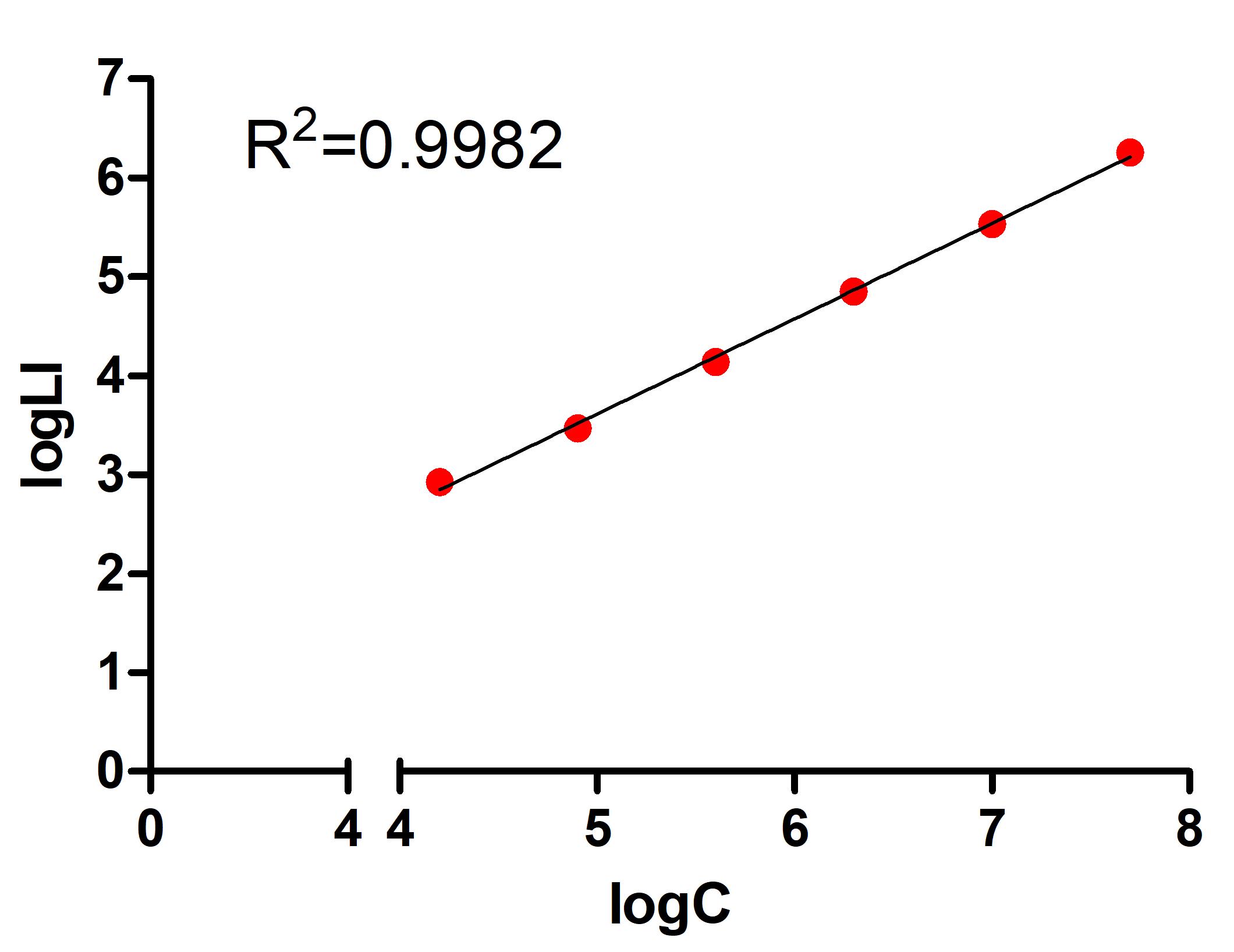
Figure 3. Representative calibration curve of Listeria innocua biofilm assay. LogC: logarithm of bacterial concentration. LogLI: logarithm of luminescence intensity.
Data analysis
Bioluminescence measurements are saved in Excel format, and Excel can be used to analyze the data. The coefficient of determination (R2) of the calibration curve is determined, and data are considered for analysis only if R2 is greater than 0.95. All measurements are repeated in two biological replicates, each of which is repeated in three technical replicates.
Notes
Relationship between OD600 and CFU number should be established in advance, by plating bacterial suspensions with different OD600 (serial dilutions) and counting the numbers of colonies (CFU). Measured OD600 values depend on the spectrophotometer used and are therefore not specified.
Listeria is cultured at 37°C for optimal cell growth. However, culturing at 30°C was suggested for the studies of biofilm formation (Lemon et al., 2007), due to higher motility of the cells.
In time-course experiments of biofilm growth, each time point (or each plate measured) requires a new calibration curve, prepared from the 8 h culture that is inoculated from the overnight culture. Measurement after the first 10 min is usually selected for calculation; however, strength of the signal and reproducibility should be taken into account, and measurements after 20 min and 30 min can also be considered.
Careful pipetting is essential for reproducible results.
Recipes
PBS
PBS is prepared by 10× dilution and sterile filtration of 10× PBS.
10× PBS:
80 g NaCl
2 g KCl
17.8 g Na2HPO4·2H2O
2.7 g KH2PO4
Fill to 1 L with dH2O.
TSA
30 g TSB
15 g agar
Fill to 1 L with dH2O and autoclave 15 min at 121°C. Pour the agar into Petri dishes in sterile conditions and leave the agar to solidify.
Acknowledgments
This study was funded by the Slovenian Research Agency (Grant Numbers P4-0127, J4-9327, J4-1771). This protocol was derived from Berlec et al. (2021).
Competing interests
The authors declare no competing interests.
References
- Buchanan, R. L., Gorris, L. G. M., Hayman, M. M., Jackson, T. C. and Whiting, R. C. (2017). A review of Listeria monocytogenes: An update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control 75: 1-13.
- Berlec, A., Janež, N., Sterniša, M., Klančnik, A. and Sabotič, J. (2021). Expression of NanoLuc Luciferase in Listeria innocua for Development of Biofilm Assay. Front Microbiol 12: 636421.
- Chlebicz, A. and Slizewska, K. (2018). Campylobacteriosis, Salmonellosis, Yersiniosis, and Listeriosis as Zoonotic Foodborne Diseases: A Review. Int J Environ Res Public Health 15(5): 863.
- Costa, A., Lourenco, A., Civera, T., and Brito, L. (2018). Listeria innocua and Listeria monocytogenes strains from dairy plants behave similarly in biofilm sanitizer testing. LWT 92: 477-483.
- Donlan, R. M. (2002). Biofilms: microbial life on surfaces. Emerg Infect Dis 8(9): 881-890.
- England, C. G., Ehlerding, E. B. and Cai, W. (2016). NanoLuc: A Small Luciferase Is Brightening Up the Field of Bioluminescence. Bioconjug Chem 27(5): 1175-1187.
- European Food Safety Authority and European Centre for Disease Prevention and Control (EFSA and ECDC). (2019). The European Union One Health 2018 Zoonoses Report. EFSA J 17(12): e05926.
- Fairchild, T. M. and Foegeding, P. M. (1993). A proposed nonpathogenic biological indicator for thermal inactivation of Listeria monocytogenes. Appl Environ Microbiol 59(4): 1247-1250.
- Friedly, E. C., Crandall, P. G., Ricke, S., O'Bryan, C. A., Martin, E. M. and Boyd, L. M. (2008). Identification of Listeria innocua surrogates for Listeria monocytogenes in hamburger patties. J Food Sci 73(4): M174-178.
- Flemming, H. C., and Wuertz, S. (2019). Bacteria and archaea on Earth and their abundance in biofilms. Nat Rev Microbiol 17(4): 247-260.
- Glaser, P., Frangeul, L., Buchrieser, C., Rusniok, C., Amend, A., Baquero, F., Berche, P., Bloecker, H., Brandt, P., Chakraborty, T., et al. (2001). Comparative genomics of Listeria species. Science 294(5543): 849-852.
- Grudlewska-Buda, K., Skowron, K. and Gospodarek-Komkowska, E. (2020). Comparison of the intensity of biofilm formation by Listeria monocytogenes using classical culture-based method and digital droplet PCR. AMB Express 10(1): 75.
- Highmore, C. J., Warner, J. C., Rothwell, S. D., Wilks, S. A. and Keevil, C. W. (2018). Viable-but-Nonculturable Listeria monocytogenes and Salmonella enterica Serovar Thompson Induced by Chlorine Stress Remain Infectious. mBio 9(2): e00540-18.
- Klančnik, A., Šikić Pogačar, M., Trošt, K., Tušek Žnidarič, M., Mozetič Vodopivec, B. and Smole Možina, S. (2017). Anti-Campylobacter activity of resveratrol and an extract from waste Pinot noir grape skins and seeds, and resistance of Camp. jejuni planktonic and biofilm cells, mediated via the CmeABC efflux pump. J Appl Microbiol 122(1): 65-77.
- Lourenco, A., Rego, F., Brito, L. and Frank, J. F. (2012). Evaluation of methods to assess the biofilm-forming ability of Listeria monocytogenes. J Food Prot 75(8): 1411-1417.
- Lee, B. H., Cole, S., Badel-Berchoux, S., Guillier, L., Felix, B., Krezdorn, N., Hebraud, M., Bernardi, T., Sultan, I. and Piveteau, P. (2019). Biofilm Formation of Listeria monocytogenes Strains Under Food Processing Environments and Pan-Genome-Wide Association Study. Front Microbiol 10: 2698.
- Lemon, K. P., Higgins, D. E. and Kolter, R. (2007). Flagellar motility is critical for Listeria monocytogenes biofilm formation. J Bacteriol 189(12): 4418-4424.
- Matereke, L. T. and Okoh, A. I. (2020). Listeria monocytogenes Virulence, Antimicrobial Resistance and Environmental Persistence: A Review. Pathogens 9(7): 528.
- Ripolles-Avila, C., Cervantes-Huaman, B. H., Hascoet, A. S., Yuste, J. and Rodriguez-Jerez, J. J. (2019). Quantification of mature Listeria monocytogenes biofilm cells formed by an in vitro model: A comparison of different methods. Int J Food Microbiol 289: 209-214.
- Silva-Angulo, A. B., Zanini, S. F., Rosenthal, A., Rodrigo, D., Klein, G., and Martinez, A. (2015). Comparative study of the effects of citral on the growth and injury of Listeria innocua and Listeria monocytogenes cells. PLoS One 10: e0114026.
- Stiefel, P., Rosenberg, U., Schneider, J., Mauerhofer, S., Maniura-Weber, K., and Ren, Q. (2016). Is biofilm removal properly assessed? Comparison of different quantification methods in a 96-well plate system. Appl Microbiol Biotechnol 100(9): 4135-4145.
- Van den Driessche, F., Rigole, P., Brackman, G. and Coenye, T. (2014). Optimization of resazurin-based viability staining for quantification of microbial biofilms. J Microbiol Methods 98: 31-34.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Berlec, A., Janež, N., Sterniša, M., Klančnik, A. and Sabotič, J. (2022). Listeria innocua Biofilm Assay Using NanoLuc Luciferase. Bio-protocol 12(3): e4308. DOI: 10.21769/BioProtoc.4308.
Category
Microbiology > Microbial biofilm
Microbiology > Pathogen detection > Bioluminescence
Biological Sciences > Microbiology
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









