- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Simple Technique for Direct Immobilization of Target Enzymes from Cell Lysates Based on the SpyTag/SpyCatcher Spontaneous Reaction
Published: Vol 12, Iss 1, Jan 5, 2022 DOI: 10.21769/BioProtoc.4282 Views: 3841
Reviewed by: Alessandro DidonnaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Endo-1,4-β-D-xylanase Assay Using Azo-Xylan and Variants Thereof
Luca Bombardi [...] Salvatore Fusco
Apr 20, 2025 1917 Views

Activation of X-Succinate Synthases for Fumarate Hydroalkylation Using an In Vitro Activation Method
Anshika Vats [...] Mary C. Andorfer
Jun 20, 2025 2511 Views

An Optimized Enzyme-Coupled Spectrophotometric Method for Measuring Pyruvate Kinase Kinetics
Saurabh Upadhyay
Aug 20, 2025 2444 Views
Abstract
Many of the current methods for enzyme purification and immobilization suffer from several drawbacks, such as requiring tedious multistep procedures or long preparation, and being environmentally unfriendly, due to the chemicals and conditions involved. Thus, a simple technique for direct purification and immobilization of target enzymes from cell lysates was proposed. The elastin-like polypeptides (ELPs)-SpyCatcher chimera could mediate the formation of silica carriers within seconds and the target enzymes were then covalently immobilized on silica carriers via SpyCatcher/SpyTag spontaneous reaction. These tailor-made carriers were easily prepared, with precisely controlled morphology and size, as well as none-consuming surface modification needed, which could specifically immobilize the SpyTag-fused target enzymes from the cell lysate without pre-purification.
Background
Enzymes are green biocatalysts with high activity in industrial manufacture. However, enzymes suffer from some problems which may hinder their industrial applications. Firstly, the process of enzyme purification is long and tedious, while in other cases it just includes one chromatographic step (Lin et al., 2020). Meanwhile, enzymes are soluble and thus need to be immobilized, for further reutilization. Hence, we propose a novel and simple technique that could directly purify and immobilize target enzymes from cell lysates (Figure 1). Briefly, new ELPs [K5V4F-40] were fused to the N-terminal of SpyCatcher (K5-C), and the K5-C chimera was purified by the inverse transition cycling (ITC) method. Then, the purified K5-C was self-encapsulated to form the K5-C modified silica NPs (K5-C@SiO2), via ELPs-mediated biomimetic silicification. On the other hand, the target enzyme was fused to the N-terminal of SpyTag and the SpyTag-fused enzymes could be directly purified and immobilized from cell lysate via the covalent bonds between the SpyCatcher and SpyTag. To verify the feasibility of this technique, we immobilized β-1,3-xylanase on K5-C@SiO2, with high activity recovery, good immobilization efficiency, and excellent reusability.
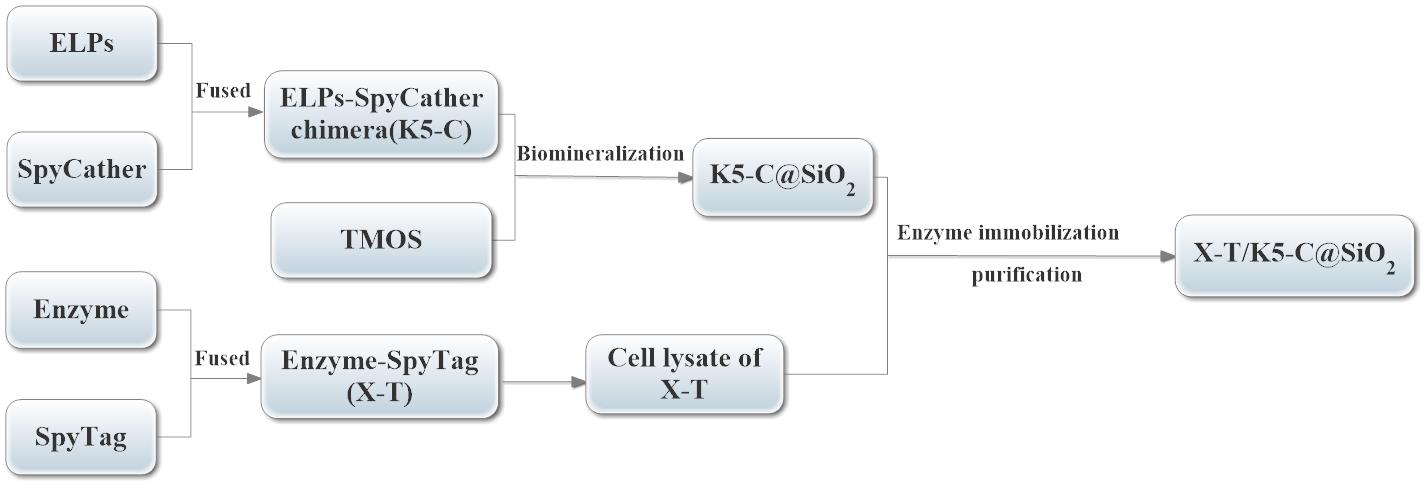
Figure 1. The flow chart of the simple technique for direct immobilization of target enzymes from cell lysates.
Materials and Reagents
1.5 mL microtubes (Axygen, catalog number: MCT150LC)
500 mL erlenmeyer flask (Shuniu, catalog number: GG17)
0.45 μm sterilized filter (Millipore, catalog number: SLHV033)
Dialysis bags (Solarbio, 8000-14000, catalog number: YA1072)
Caulerpa lentillifera (Nha trang, Vietnam,Vmax)
Escherichia coli BL21(DE3) cell (Klang, catalog number: KL9050510)
Plasmid: pET-22b(+), ambenzyl resistant, provided by Suzhou Jinweizhi Biotechnology Company, and preserved by our laboratory.
Yeast extract (Oxoid, catalog number: LP0021)
Tryptone (Oxoid, catalog number: LP0042)
Ampicillin (Amresco, catalog number: LP0339)
Isopropyl-β-D-Thiogalactoside:IPTG (Solarbio, catalog number: I8070)
NaCl (Xilong scientific, catalog number: 7647145)
PageRuler prestained protein ladder (Therom, catalog number: 142546)
Tetramethoxysilane:TMOS (Macklin, catalog number: T819504)
SiO2 standard solution (Macklin, catalog number: I821744)
NaOH (Xilong Scientific, catalog number: 100154)
Sulfuric acid solution (Xilong Scientific, catalog number: 7664939)
NaClO4 (Xilong Scientific, catalog number: 7791073)
Anhydrous ethanol (Xilong Scientific, catalog number: 64175)
Acetic acid (Xilong Scientific, catalog number: 64197)
Dichloroethanol (Merck, catalog number: 107073)
BCA protein assay kit (Yanxi, catalog number: PDBCA500)
SDS-PAGE gel preparation kit (Beyotime,catalog number: P0012A)
Nickel affinity column (Smart-Lifesciences, catalog number: SA003025)
Elastin-like polypeptides (ELPs, K5V4F means the ratio of K:V:F=5:4:1)
Phosphate buffer solution (PBS, 100 mmol/L, pH 7.0) (see Recipes)
Citrate phosphate buffer (CPB, 20 mmol/L, pH 6.6) (see Recipes)
TB medium (see Recipes)
Equipment
Eppendorf mixer (Eppendorf, catalog number: 5382000074)
-80°C freezer (Therom, catalog number: 905)
Beaker (Shuniu, catalog number: 056245)
Gel imaging analysis system (Tanon, catalog number: GIS-2008)
Ultrasonic cell disruption system (Scientz, catalog number: TY92-II)
Temperature controlled ultraviolet spectrophotometer (Analyticjena, catalog number: SPECORD40)
High speed refrigerated centrifuge (Eppendorf, catalog number: 5418RL)
Constant temperature shaker (HerryTech, catalog number: GG-100C)
Weigh scale (Sartorius, catalog number: BSA2202S)
Pipette (Eppendorf Research plus)
Scanning electron microscopy (Hitachi, catalog number: S-4800)
Transmission electron microscopy (Hitachi, catalog number: H7650)
Grinder (Leimai, catalog number: FS-100)
Mesh (Shuyin, catalog number:JD-12)
Dryer (Jinchen, catalog number: JC-9023AE)
Flasks (Shuniu, catalog number: GG-17)
Spectrophotometer (Mapada, catalog number: GDJ355)
Software
NCBI (https://www.ncbi.nlm.nih.gov)
ImageJ (NIH; https://imagej.nih.gov/ij/download.html)
ProtParam (http://web.expasy.org/protparam/)
Procedure
Extraction of β-1,3-Xylan from Caulerpa lentillifera
Wash the fresh Caulerpa lentillifera three times, and dry the clean Caulerpa lentillifera in a dryer for 24 h. Then, grind the Caulerpa lentillifera into a powder through a 200-mesh screen.
Add 20 g powder of Caulerpa lentillifera into 1 L of NaOH solution (300 mmol/L), heat the mixture at 100°C for 30 min, then centrifuge the mixture at 5,500 × g at room temperature for 20 min. Finally, discard the supernatant and then wash the precipitate with 1 L ddH2O under room temperature twice (Iriki et al., 1960).
Move the precipitate to a beaker containing a sulfuric acid solution (250 mmol/L, 1 L), and repeat the previous step.
Resuspend the precipitate in 2 L of NaClO4 solution (1%), stir the mixture at 150 rpm at 25°C for 2 h, and then wash the precipitate with ddH2O twice.
Prepare the precipitate in 800 mL of NaOH solution (2.5 mol/L) with stirring at 100 rpm in an ice bath for 2 h, discard the precipitate, and add 3.2 L of anhydrous ethanol to the supernatant at 4°C for 12 h.
Centrifuge at 13,500 × g for 20 min at 4°C to collect the precipitate. Wash the precipitate with 100 mL of anhydrous acetic acid (5.7 mol/L) and 200 mL of ddH2O, respectively, then freeze-dry the precipitate at 4°C for 36 h.
Preparation of glycol β-1,3-xylan
Mix β-1,3 xylan (3 g), NaOH solution (14%, 150 mL), and 18 mL of dichloroethanol, stirring the mixture in an ice bath at 100 rpm for 1 h. Then, keep the mixture at room temperature for 24 h.
After 24 h, use acetic acid to neutralize the mixture to pH7 with pH test strips, and then put it into a dialysis bag to remove impurities by ddH2O dialysis.
After dialysis, heat the sugar solution at 80°C to concentrate to 10 mL, and then freeze-dry the sugar solution at 4°C for 72 h (Cai et al., 2020).
Protein expression and purification
Express Xyl3088 (GenBank accession No. MK253053), K5-C (No. MN136291), and Xyl3088-Tag (X-T, No. MN136290) in Escherichia coli BL21(DE3) cells, respectively.
Inoculate 2 mL of Xyl3088 (or K5-C or X-T) and 0.2 mL of ampicillin (100 mg/mL) into 200 mL of TB medium in a 500 mL Erlenmeyer flask, and incubate at 37°C for 4 h with agitation at 200 rpm.
Add 1 mL of isopropyl-β-thiogalactopyranoside (IPTG, 0.5 mmol/L) in TB medium, and incubate at 25°C for 16 h at 180 rpm.
Harvest the cells at 5,500 × g for 20 min at 4°C, and disrupt cells by sonication (Power: 300 W; ultrasonic cycle: 150 runs; pulse: 4 s; stop: 2 s) on ice.
Centrifuge at 13,500 × g for 20 min at 4°C, to remove the insoluble cell debris; the supernatant (cytoplasmic fraction) is the crude enzyme extract.
Protein purification
Purify the recombinant proteins of Xyl3088 and X-T by a nickel affinity column.
Filter the cell lysate of Xyl3088 and X-T through a 0.45 μm sterilized filter.
Equilibrate the nickel affinity column with 15 mL of equilibrium solution (50 mmol/L Tris-Cl, 500 mmol/L NaCl, pH 7.4).
Add the washing buffer (50 mmol/L Tris-Cl, 40 mmol/L imidazole, 500 mmol/L NaCl, pH 7.4) to the column to wash the crude protein.
Elute the Xyl3088 and X-T proteins adsorbing on the column by the elution buffer (50 mmol/L Tris-Cl buffer, 250 mmol/L imidazole, 500 mmol/L NaCl, pH 7.0).
Dialyze the purified Xyl3088 and X-T proteins extensively with 50 mmol/L Tris−Cl and then store at 4°C.
Purify the supernatant containing soluble crude protein (K5-C) by the method of inverse transition cycling (ITC).
Prepare 29.25 g NaCl in 20 mL of K5-C solution and incubate the mixture at 37°C for 15 min.
Centrifuge at 13,500 × g for 20 min at 37°C to collect the aggregated K5-C.
Add 10 mL of ice-cool phosphate buffer solution (7.0) into the tube to resolubilize the aggregated K5-C.
Centrifuge at 13,500 × g for 20 min at 4°C to collect the supernatant.
Repeat the ITC process twice, to achieve high purity of the fusion K5-C proteins (Lim et al., 2007).
Analyze the purity of recombinant proteins by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE,12%).
Calculate the purity of the Xyl3088, X-T, and K5-C by ImageJ.
For the purity of Xyl3088, SDS-PAGE yielded a clear thick band of 49 kDa (Figure 2), it was consistent with the theoretical molecular weight values of 48853 Da calculated by ProtParam (http://web.expasy.org/protparam/).
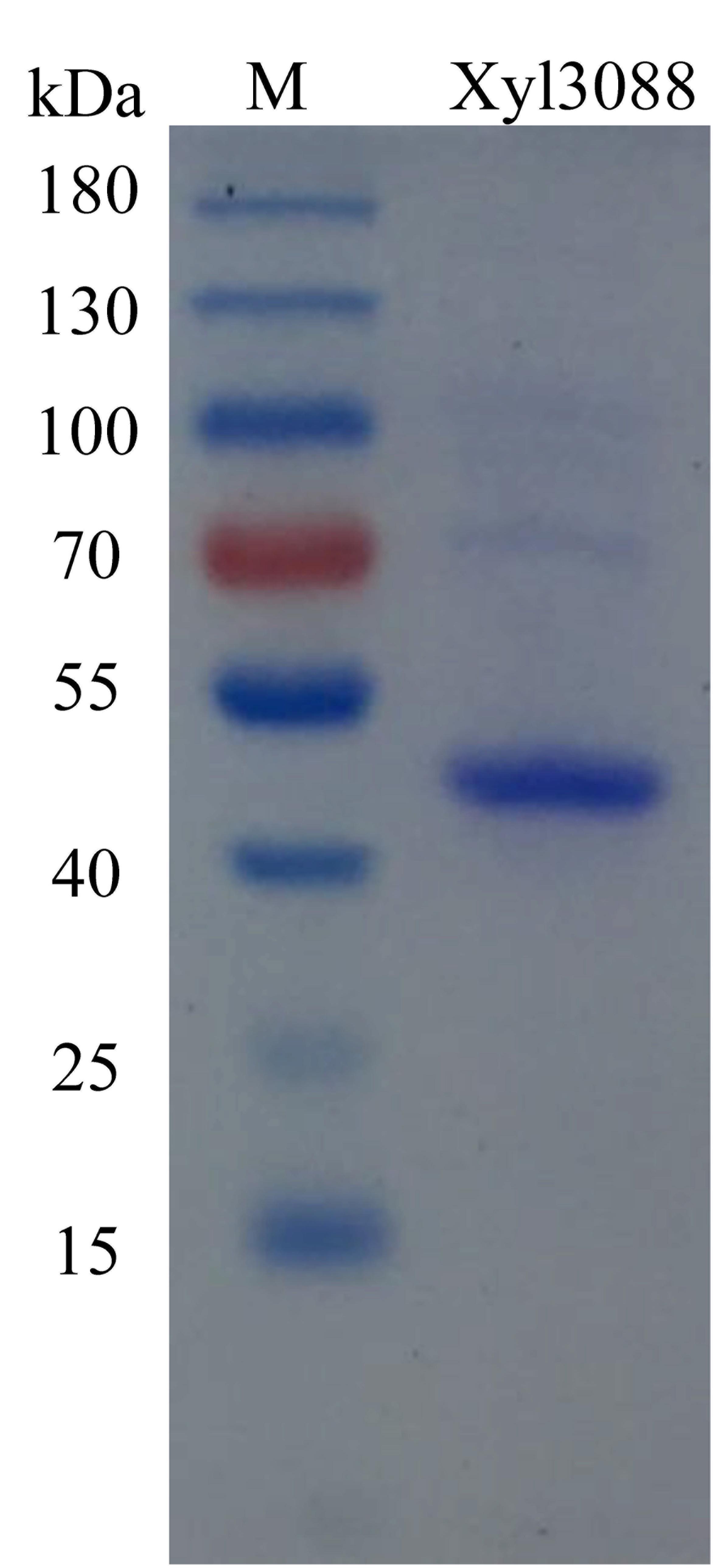
Figure 2. The SDS-PAGE of purifired recombination Xyl3088.
Preparation of silica nanoparticles
Prepare 1.522 g tetramethyl orthosillicate (TMOS) in 7 mL of HCl (1 mmol/L), set the volume to 10 mL, and keep the fresh TMOS at room temperature for 10 min (Cai et al., 2021).
Blend fresh TMOS with K5-C (300 μmol/L) at the ratio of 1:9 (v:v), and keep the reaction at 4°C for 10 min.
Centrifuge at 5,500 × g for 3 min at 4°C to collect the silica precipitation.
Wash the silica precipitation three times with 1.5 mL of precooled PBS buffer, to collect the new formed silica nanoparticles (NPs) containing K5-C proteins (K5-C@silica).
K5-C leakage test
Resuspend the silica NPs in 3 mL of CPB buffer (20 mmol/L, pH 6.6). Store the suspension at 4°C.
At given times, centrifuge at 13,500 × g for 5 min at 4°C to collect the silica NPs and the supernatants, respectively.
Measure the supernatants by spectrophotometer at 280 nm to analyse the K5-C protein.
Add the supernatants back to tubes to resuspend K5-C@silica for further testing.
Characterization of K5-C@silica
Disperse the synthesized K5-C@silica in 1 mL of ddH2O and air-dry overnight. Analyze the silica morphology and size of K5-C@silica using scanning electron microscopy (SEM) and transmission electron microscopy (TEM). Calculate the elementary composition of K5-C@silica by energy-dispersive X-ray spectroscopy (EDS). The SEM photos of the white precipitation formed by K5-C displayed that they were spherical, rough, and their diameters ranged from 200 to 600 nm (Figure 3).

Figure 3. The SEM micrograph of K5-C@silica. Scale bar is 1 μm.Enzyme self-immobilization
As SpyCatcher and SpyTag can spontaneous form the isopeptide bond in vitro, we mix the crude enzyme of X-T (300 mL) with K5-C@silica (60 mg), and then incubate the tubes at 30°C for 1 h to formulate XTC-K5 immobilized enzymes (immobilized Xyl3088).
Centrifuge at 5,500 × g for 3 min at 4°C to collect the immobilized Xyl3088.
Wash the immobilized Xyl3088 with 1.5 mL of CPB buffer (20 mmol/L, pH 6.6) three times and keep the immobilized enzymes at 4°C for later use.
Enzyme Activity Assays
Assay the enzymatic activities of the free and immobilized Xyl3088, using β-1,3-xylan as a substrate.
Estimate the enzyme activity of the free and immobilized Xyl3088, by using the modified 3,5-dinitrosalicylic acid (DNS) assay.
Prepare 50 μL of diluted enzyme and 350 μL of β-1,3-xylan (1%) together, and incubate the mixture at 45°C (pH6.6) for 10 min.
After 10 min, add 400 μL of DNS reagent to stop the reaction, and quickly transfer the tube to a 100°C bath for 5 min.
Define one international unit (IU) as the amount of enzyme that released 1 μmol of reducing sugar per minute.
Assessing the optimum temperature and pH.
Determine the effects of temperature on β-1,3-xylan hydrolytic activity of the free and immobilized Xyl3088, by assessing enzyme activity in a temperature range of 25-75°C. Define the highest activities as 100%, and calculate the others as a relative percentage.
Meanwhile, determine the effects of pH on β-1,3-xylan hydrolytic activity of the free and immobilized Xyl3088, by assessing enzyme activity in a pH range of 4.0-9.0.
Calculate the immobilization efficiency, immobilization yield and activity recovery of the immobilized enzymes as following (Sheldon, 2014; Boudrant et al., 2019).
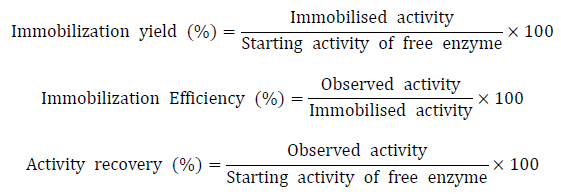
At last, the immobilization yield of Xyl3088 was up to 96%. Immobilized Xyl3088 showed 88.6% of the activity recovery and had 85.6% of the immobilization efficiency.
Enzyme thermal stability and reusability
Enzyme reusability
Briefly, define the first round of specific activity of immobilized Xyl3088 as 100% (Schoene et al., 2014).
After each cycle, centrifuge at 13,500 × g for 1 min at 4°C, to collect the silica NPs and the supernatant.
Measure the activity of the supernatant on glycol β-1,3-xylan hydrolytic ability, and caculate the reusability activity of immobilized Xyl3088 in this cycle.
Wash the silica NPs with CPB buffer (20 mmol/L, pH 6.6), to remove the reducing sugar.
Add the new substrate solution into reaction system containing immobilized Xyl3088, to start a new enzymatic reaction cycle.
Thermal stability
Incubate the free and immobilized Xyl3088 in Tris-Cl buffer (50 mmol/L, pH 7.0) without substrate at the given temperature (40°C, 45°C, 50°C).
Calculate the residual activities at each temperature by taking the activity at 45°C as 100%.
Kinetic Studies
Estimate the kinetic parameters of the free or immobilized Xyl3088, with increasing β-1,3-xylan concentrations ranging from 1 to 10 mg/mL.
Monitor the concentration of reducing sugar at 540 nm by spectrophotometer.
Calculate the maximum rate (Vmax) and Michaelis constant (Km) of the free or immobilized Xyl3088 by using the Michaelis-Menten model.
Recipes
Phosphate buffer solution (PBS, 100 mmol/L, pH 7.0)
A solution: 200 mmol/L Na2HPO4
B solution: 200 mmol/L NaH2PO4
Adjust pH to 7.0 with A solution and B solution.
Citrate phosphate buffer (CPB, 20 mmol/L, pH 6.6)
A solution: 20 mmol/L Na2HPO4
B solution: 10 mmol/L Citric acid
Adjust pH to 6.6 with A solution and B solution.
TB medium
Yeast extract 4.8 g
Tryptone 2.4 g
Glycerol 0.9 mL
ddH2O 180 mL
Acknowledgments
We thank Dr. Li Xialan and Dr. Chen Mingxia for experimental support and helpful comments on the manuscript. This protocol was supported by Foundation of Fujian Educational Committee for Young and middle-aged Teachers (JAT200496) and Natural Science Foundation of Fujian Province of China (2021J011110).
The technique for direct immobilization of target enzymes from cell lysates was as described in Cai et al. (2021). "A novel all-in-one strategy for purification and immobilization of β-1,3-xylanase directly from cell lysate as active and recyclable nanobiocatalyst". Microb Cell Fact 20(1): 37.
Competing interests
There are no conflicts of interest or competing interests.
References
- Boudrant, J., Woodley, J. M., Fernandez-Lafuente, R. (2019). Parameters necessary to define an immobilized enzyme preparation. Process Biochem 90: 66-80.
- Cai, L. X., Chu, Y. M., Liu, X., Qiu, Y., Zhang, G. Y. (2021). A novel all-in-one strategy for purification and immobilization of β-1,3-xylanase directly from cell lysate as active and recyclable nanobiocatalyst.Microb Cell Fact 20(1): 37.
- Cai, L. X., Liu, X., Qiu, Y., Liu, M. Q., Zhang, G. Y. (2020). Enzymatic degradation of algal 1,3-xylan: from synergism of lytic polysaccharide monooxygenases with β-1,3-xylanases to their intelligent immobilization on biomimetic silica nanoparticles. Appl Microbiol Biot 104(12): 5347-5360.
- Iriki, Y., Suzuki, T., Nisizawa, K., Miwa, T. (1960). Xylan of siphonaceous green algae. Nature 187: 82-83.
- Lim, D. W., Trabbic-Carlson, K., MacKay, J. A., Chilkoti, A. (2007). Improved non-chromatographic purification of a recombinant protein by cationic elastin-like polypeptides. Biomacromolecules 8: 1417-1424.
- Lin, Y. Q., Qiu, Y., Cai, L. X., Zhang, and G. Y. (2020). Investigation of the ELP-mediated silicification-based protein self-Immobilization using an acidic target enzyme. Ind Eng Chem Res 59(44): 19829-19837.
- Schoene, C., Fierer, J., Bennett, DSP., Howarth, P. M. (2014). SpyTag/SpyCatcher cyclization confers resilience to boiling on a mesophilic enzyme. Protein Eng 53: 6215-6218.
- Sheldon, R. A. (2014). Green and sustainable manufacture of chemicals from biomass: state of the art. ChemInform 2014, 45(19):950-963.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Cai, L., Lin, Y., Chu, Y., Chen, X., Liu, L., Zhang, M. and Zhang, G. (2022). A Simple Technique for Direct Immobilization of Target Enzymes from Cell Lysates Based on the SpyTag/SpyCatcher Spontaneous Reaction. Bio-protocol 12(1): e4282. DOI: 10.21769/BioProtoc.4282.
Category
Biochemistry > Protein > Activity
Biological Engineering > Biomedical engineering
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









