- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantitative Determination of Primary Cilia Protein Distribution Using Immunofluorescence Staining and MATLAB Analysis
Published: Vol 11, Iss 23, Dec 5, 2021 DOI: 10.21769/BioProtoc.4248 Views: 4109
Reviewed by: Khyati Hitesh ShahFRANCESC GARCIA-GONZALOAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

An Automated Imaging Method for Quantification of Changes to the Endomembrane System in Mammalian Spheroid Models
Margaritha M. Mysior and Jeremy C. Simpson
Jun 5, 2025 1650 Views

Quantifying Intracellular Distributions of HaloTag-Labeled Proteins With SDS-PAGE and Epifluorescence Microscopy
Julia Shangguan and Ronald S. Rock
Jul 20, 2025 2501 Views

Fluorescence Lifetime-Based Separation of FAST-Labeled Cellular Compartment
Aidar R. Gilvanov [...] Yulia A. Bogdanova
Oct 5, 2025 1323 Views
Abstract
Primary cilia are microtubule-based sensory organelles surrounded by membrane. They can detect mechanical and chemical stimuli. The last few years have uncovered cilia as unique signaling hubs that host a number of receptors and effector molecules. Thus, defining how specific proteins localize and are distributed along the cilium is critical to understanding its function.
Quantitative immunofluorescence can be used to accurately assess the localization of receptors and signaling molecules within the primary cilia. However, image analysis can be time consuming, and there are limited programs that can accurately determine staining intensity along the cilia. To overcome these issues, we developed a series of MATLAB scripts to accurately measure staining intensity along the length of the cilia, in both a semi-automated and automated fashion. Here, we describe the scripts and include a protocol for image analysis for each. With these scripts, the protocols can be used to analyze the distribution of any ciliary protein using immunofluorescence images.
Background
Primary cilia are microtubule-based sensory organelles that function as antenna-like structures emerging from the cell body. They detect mechanical and chemical signals from the environment and are involved in a range of biological processes critical for homeostasis (Goetz and Anderson, 2010; Anvarian et al., 2019). Not surprisingly, genetic cilia defects result in various clinical manifestations, called ciliopathies (Goetz and Anderson, 2010). Interestingly, the ciliary membrane hosts unique receptors and effector molecules that control a number of signaling pathways and biological processes. These include oncogenic receptor tyrosine kinases (RTKs), such as insulin-like growth factor receptor (IGF-1R) or platelet-derived growth factor receptor alpha (PDGFR-α), and G-protein coupled receptors (GPCRs), such as smoothened (SMO) (Christensen et al., 2012; Schou et al., 2015).
We previously reported that primary cilia mediate resistance to several kinase inhibitors in cancer cells (Jenks et al., 2018). Drug resistant cells show more cilia, longer cilia, the appearance of cilia terminal fragments (CTF), and increased activation of the cilia-specific Hedgehog signaling pathway (Jenks et al., 2018). Significantly, targeting cilia or ciliary signaling can sensitize drug-resistant cells to their respective inhibitors (Jenks et al., 2018). Interestingly, drug resistant cells show changes in the distribution of cilia proteins, including Kif7 and IFT81. While control cells showed Kif7 and IFT81 distributed along the cilia and at the cilia tip, drug resistant cells showed a significant decrease in the distribution of these proteins along the cilia (Jenks et al., 2018). In addition, we observed changes in a number of proteins and ciliary receptors in drug resistant cells, highlighting the importance of developing accurate image analysis tools for cilia research.
These results presented us with the challenge of being able to measure the distribution of proteins along the cilium. The accurate quantification of fluorescence intensity in microscopy images is an important part of cilia research. For this purpose, two open source software packages have been described (Lauring et al., 2019; Hansen et al., 2021). To accurately quantify the distribution of proteins along the cilia, we developed a series of MATLAB scripts that support cilia analysis using the improfile function of MATLAB and adaptive threshold for binarization. Our protocols are versatile, intuitive, and start with a manual script that is then fully automated. As cilia and cilia staining can be heterogeneous, our automated protocol can be adapted to different conditions by changing the pixel intensity threshold, named “level” in the script. These scripts are outlined here, along with a detailed step-by-step guide to use them with examples. A supplementary section with each script is included.
Materials and Reagents
12 mm round coverslips (Fisher Scientific, catalog number: 11846933)
6-well plates (Corning, catalog number: 353046)
16% Formaldehyde (Thermo Fisher Scientific, catalog number: 28908)
NaCl (VWR International, catalog number: 10241)
KCl (VWR International, catalog number: 10198)
KH2PO4 (VWR International, catalog number: 10203)
Na2HPO4 (VWR International, catalog number: 10249)
HCl (VWR International, catalog number: 45002)
Triton-X-100 (Sigma-Aldrich, catalog number: T8787)
Bovine Serum Albumin (BSA) (Sigma-Aldrich, catalog number: A7906)
4′,6-Diamidine-2′-phenylindole dihydrochloride (DAPI) (Thermo Fisher Scientific, catalog number: D1306)
Methanol (VWR International, catalog number: 20847.318)
Prolong Gold Mounting Media (Thermo Fisher Scientific catalog number: P10144)
Superfrost adhesion slides (Thermo Fisher Scientific, catalog number: 10149870)
Poly-L-lysine (Sigma-Aldrich, catalog number: P6282)
PTEM Buffer (see Recipes)
1× Phosphate Buffered Saline (1× PBS) (see Recipes)
1× PBST (see Recipes)
1× PBSBT (see Recipes)
Primary antibodies: mouse anti-acetylated tubulin (1:2,000 dilution; Sigma-Aldrich, catalog number: T7451)
Mouse anti-γ-tubulin (1:500 dilution; Santa Cruz, catalog number: sc-51715)
Rabbit anti-IFT81 antibody (1:200 dilution; Proteintech, catalog number: 17744-1-AP)
Rabbit anti-Arl13B (1:500; Proteintech, catalog number: 17711-1-AP)
Secondary antibodies: Alexa Fluor 488 (Thermo Fisher Scientific, catalog number: A-21141), 594 (Thermo Fisher Scientific, catalog number: A11037), or 680 (Thermo Fisher Scientific, catalog number: A31562) (1:500 dilution)
Cell lines
A204 and RPE-1 cell lines were used as indicated. Cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) and Dulbecco’s Modified Eagle Medium: Nutrient Mixture F-12 (DMEM:F12) media with 10% FBS, respectively.
Equipment
Orbital shaker
Epifluorescence microscope: Upright microscope (Axio Imager M2; Zeiss) equipped with 100× oil objectives, 1.4 numerical aperture (N.A.), a camera (ORCA R2; Hamamatsu Photonics), and a computer with image-processing software (Zen)
Zeiss 710 confocal microscope with Zen software
Computer specifications: To run MATLAB – Intel or AMD x86-64 processor with four logical cores and AVX2, 8 GB RAM, Windows 7 or newer operating system
Software
Mathworks MATLAB (version R2020b) with the image processing toolbox
Excel (Microsoft)
Zen (Zeiss image acquisition software)
Procedure
Seeding cells and fixation for ciliogenesis assay
Wash 13 mm coverslips in 1 N HCl at 55°C overnight in a flask in an orbital shaker at 140 × g.
After extensive washing, coat coverslips with 0.1 mg/ml poly-L-lysine for 1 h at room temperature.
Wash coverslips with water and let dry overnight.
Before plating, place coverslips under UV light for 15 min.
Seed cells onto coverslips at a density of 4 × 105 per well in 6 well plates (Diameter = 3.5 cm). This is approximately a 60% density, so the cells will be around 80% the next day. This can vary between cell lines, and we suggest optimization.
Allow cells to attach for 24 h.
Wash cells 3x with serum free media (SFM) and keep in SFM for 48 h.
Wash 2x with 1× PBS (alternatively with PTEM buffer) and proceed to fix (4% formaldehyde diluted in 1× PBS or Methanol at -20°C, as desired).
Immunostaining protocol
Note: All steps to be carried out at room temperature.
Place coverslips in a humid chamber and wash 2x with 1× PBST to permeabilize cells. Leave in second wash for 5 min.
Block cells using 3% (w/v) bovine serum albumin in 1× PBST (PBSBT) for 5 min.
Add primary antibodies diluted in PBSBT and incubate for 1 h.
Wash coverslips 3x with 1× PBST for 5 min.
Add secondary antibodies diluted 1:500 in PBSBT and incubate for 1 h.
Wash coverslips 3x with 1× PBST for 5 min.
Add DAPI diluted in PBSBT and incubate for 2 min.
Wash 2x, apply drops of ProLong Gold to a slide, and mount. Allow to dry in the dark overnight.
After 24 h, seal with nail polish.
Image acquisition
Images were acquired using an 100× oil objective (1.4 NA). Exposure times were set equally for all images amongst different conditions. Images were acquired using Zeiss analysis software (Zen).
Description of MATLAB scripts and analysis
CILIA 1 (Four channel raw intensity): cilia_linesc_raw_int_v9
This script measures fluorescence intensity (channel 2) along a linescan drawn manually and outputs average intensity in 10 equal segments along the linescan. This allows the user to compare cilia of different length. Cilia are segmented in a user- interactive manner using the improfile function from MATLAB, which retrieves the intensity values of pixels along a multiline path defined by the user. This script also measures the length of the segmented cilium. The data is exported as an excel spreadsheet and saved using the same filename as the TIFF file. A cilia marker (i.e., acetylated tubulin or Arl13B) can be used to define the cilium path. Intensity profiles are then retrieved from the channels of interest. To reduce noise, the average fluorescence intensity of three pixels (above, on, and below the path) is measured for each position along the cilium (taking an average of multiple pixels reduces the impact of a single noisy pixel). A description of how to use this script is detailed below.
Four (4) fluorescent channels. For the example shown: channel 1 (C1) is the centriole marker (γ-tubulin), channel 2 (C2) is the protein of interest (IFT81), channel 3 (C3) is the cilia marker (acetylated tubulin), and channel 4 (C4) is the nuclear stain (DAPI). Cilia are manually defined, and then the script will divide cilia into 10 equal sections (bins). The raw pixel intensity of channel 3 will be measured along the defined cilia and presented as 10 cilia sections running from the base to the cilia tip. In the example below, this script is used to quantify IFT81 (Figures 1-8).
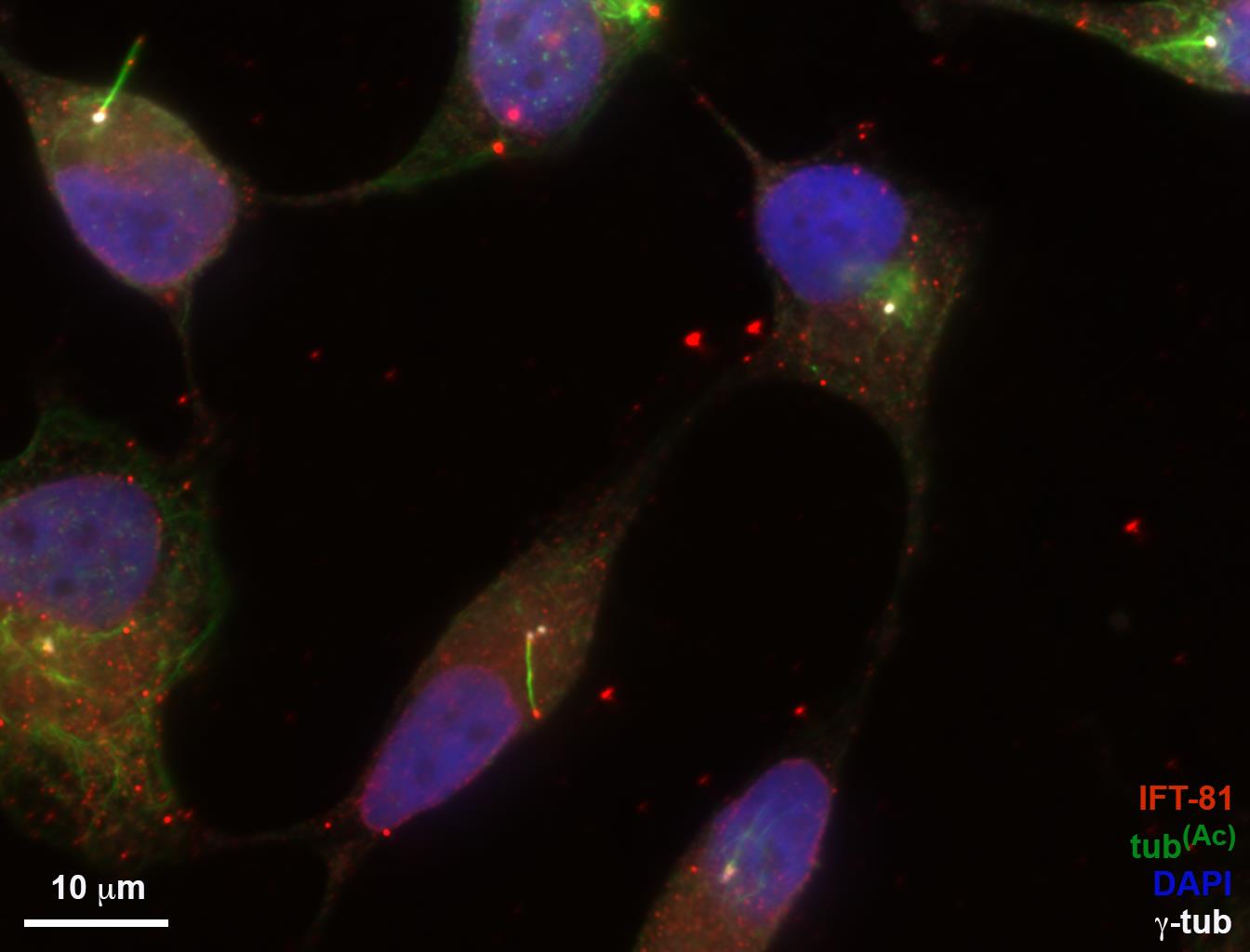
Figure 1. A204 IFT81 image used for CILIA 1 script MATLAB analysis. A204 cells were serum starved for 48 h to induce ciliogenesis, then fixed and stained with antibodies for IFT81 (red), acetylated tubulin (green), and γ-tubulin (white). The nuclear stain is DAPI (blue). Scale bar = 10 μm.
Opening MATLAB, importing data, and preparing script
Image exporting and saving
Export images as TIFF file format; an individual image is required for each fluorescent channel. The channels for analysis have been exported in the following order in the example shown: centriole marker, channel 1 (C1), signal to be analyzed, channel 2 (C2), acetylated tubulin or cilia marker of choice, channel 3 (C3) and DAPI or alternative nuclear stain, channel 4 (C4). An image of an overlay of all channels C1 + C2 + C3 + C4 is included in the file exported. In order for the script to work correctly, channel 2 (C2) must be the channel to be analyzed and channel 3 (C3) a cilia marker. The markers used for channel 1 and 4 can be changed if required.
Open MATLAB.
Add the four script files to the MATLAB directory (left sidebar) by dragging files.
Add the TIFF image files to be analyzed to the left sidebar (Figure 2).
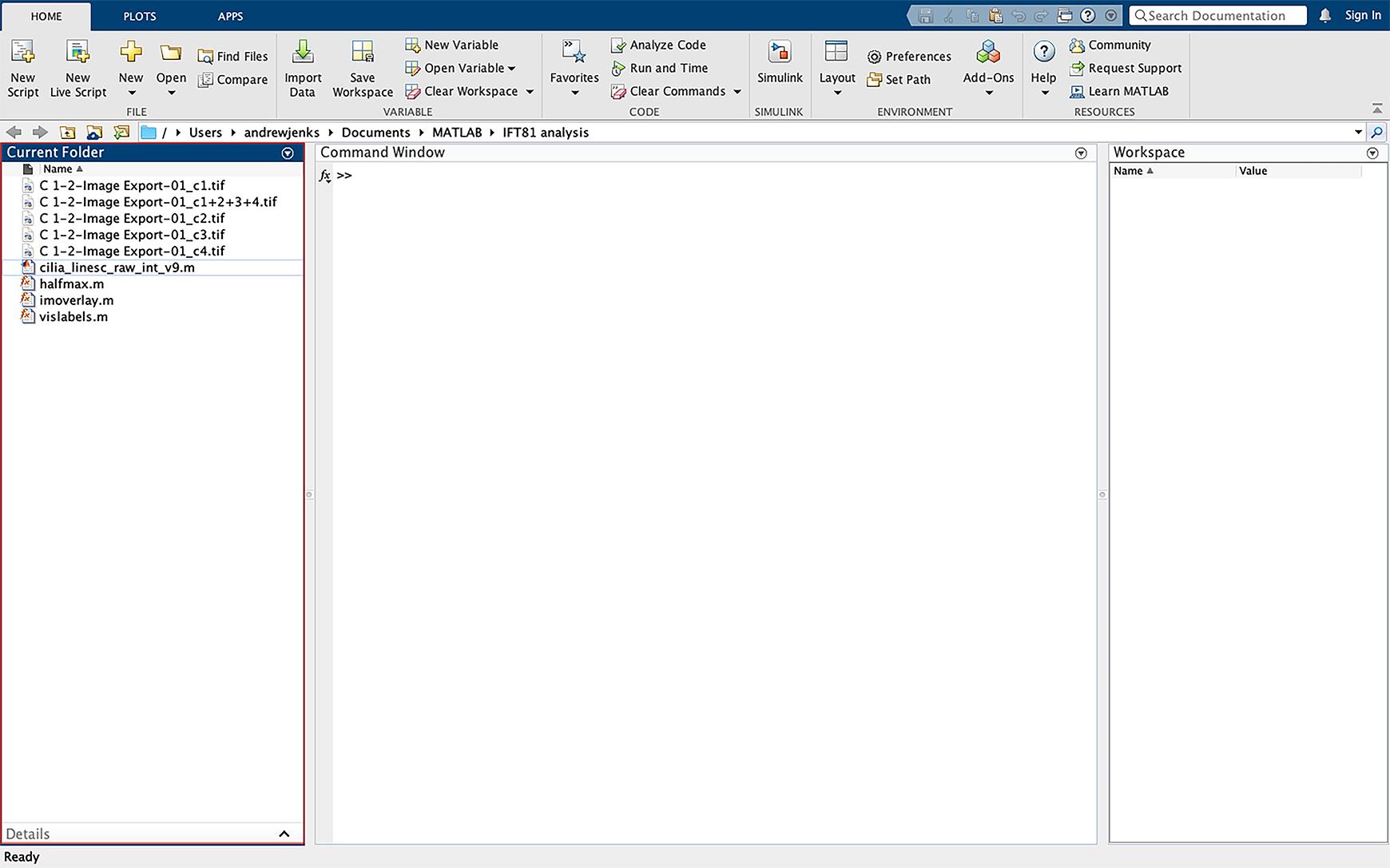
Figure 2. Importing data into MATLAB. Drag the script files and images to be analyzed into the MATLAB left sidebar of the operating interface. Images must be imported as the individual fluorescent channels as TIFF files.In line 12 of the script, change the name of the file to the same as the file to be analyzed. Exclude the TIFF file extension from the file name in the script (Figure 3).
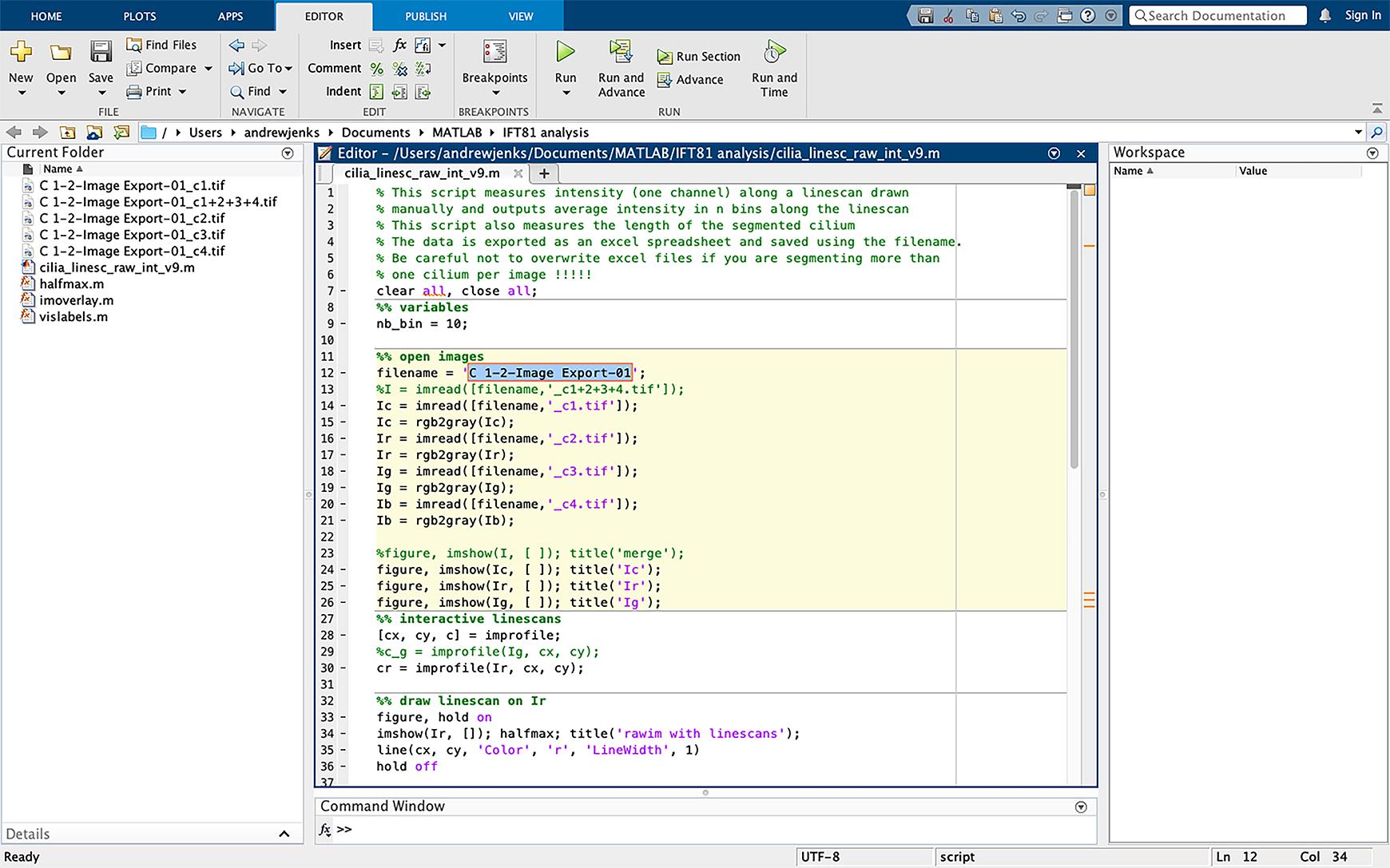
Figure 3. Changing script file name. Change the filename in script (line 12, highlighted in blue) to the same name as the files to be analyzed. The TIFF extension should not be included in the filename.Click on the dash sign (-) next to line 28 in the script; this will add a breakpoint in the script, which allows the script to be paused at this point (Figure 4).
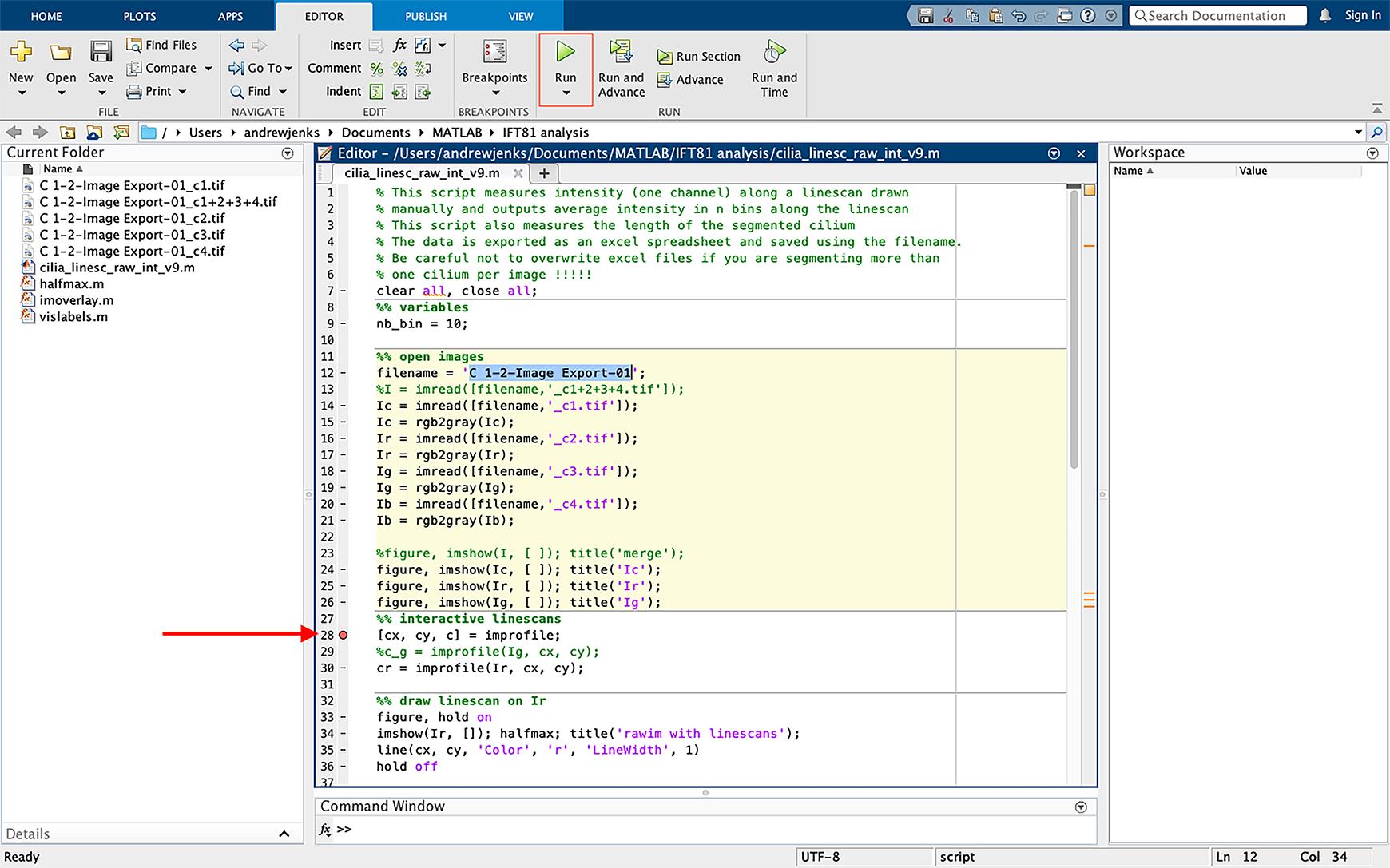
Figure 4. Selecting file to be analyzed and inserting a breakpoint in the script. Insert a breakpoint in the script by clicking on the dash sign next to line 28; this will convert the dash to a red circle (highlighted with an arrow). Run the script by clicking “run” (green icon boxed-in in red).Defining cilia and analyzing IFT81 pixel intensity
Run the script by pressing the green icon on top of “run,” highlighted in Figure 4. An image of acetylated tubulin will appear in a new window. Click on “tools>zoom in” and draw a square around the cilia to be analyzed.
Press “continue” on script (“continue” is found in the same place as the run icon).
Using the cursor, draw a line along the center of the cilia starting from the base to the tip of the cilia. If the cilia bends, stop, click, and continue. When the line reaches the cilia tip, double click with the left mouse button (Figure 5).
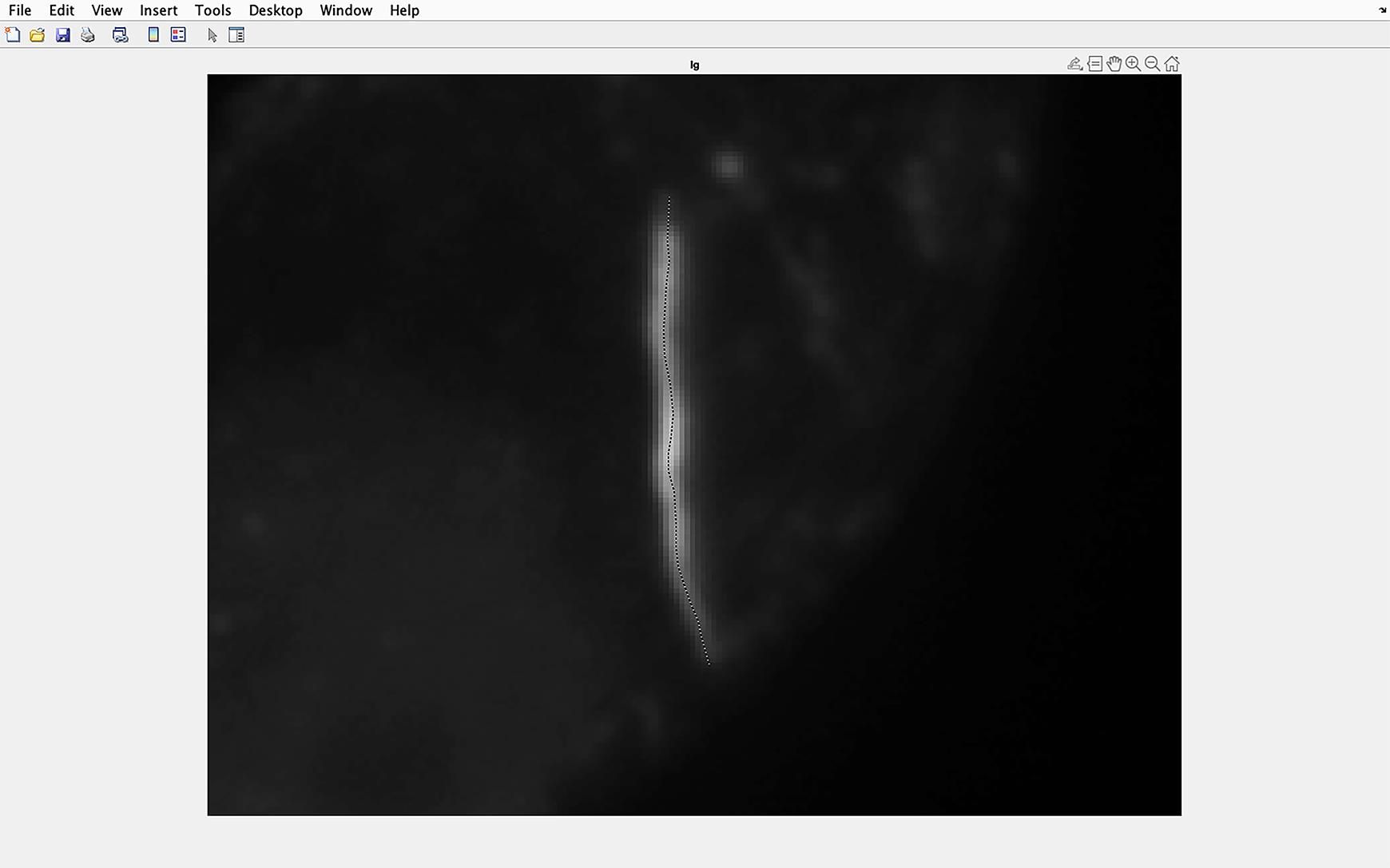
Figure 5. Identifying the cilium to be analyzed. Draw a line along the center of the magnified cilium, ensuring the line goes from the base to the tip of the cilium. Double click when the line is at the cilium tip. This example uses A204 cells.The line drawn along the cilia will be automatically overlaid onto the IFT81 staining image, and pixel intensity will be measured along this line (measurements extend several pixels at the tip and in the lateral direction throughout the cilia, to ensure pixel intensity of the cilia is measured) (Figure 6).
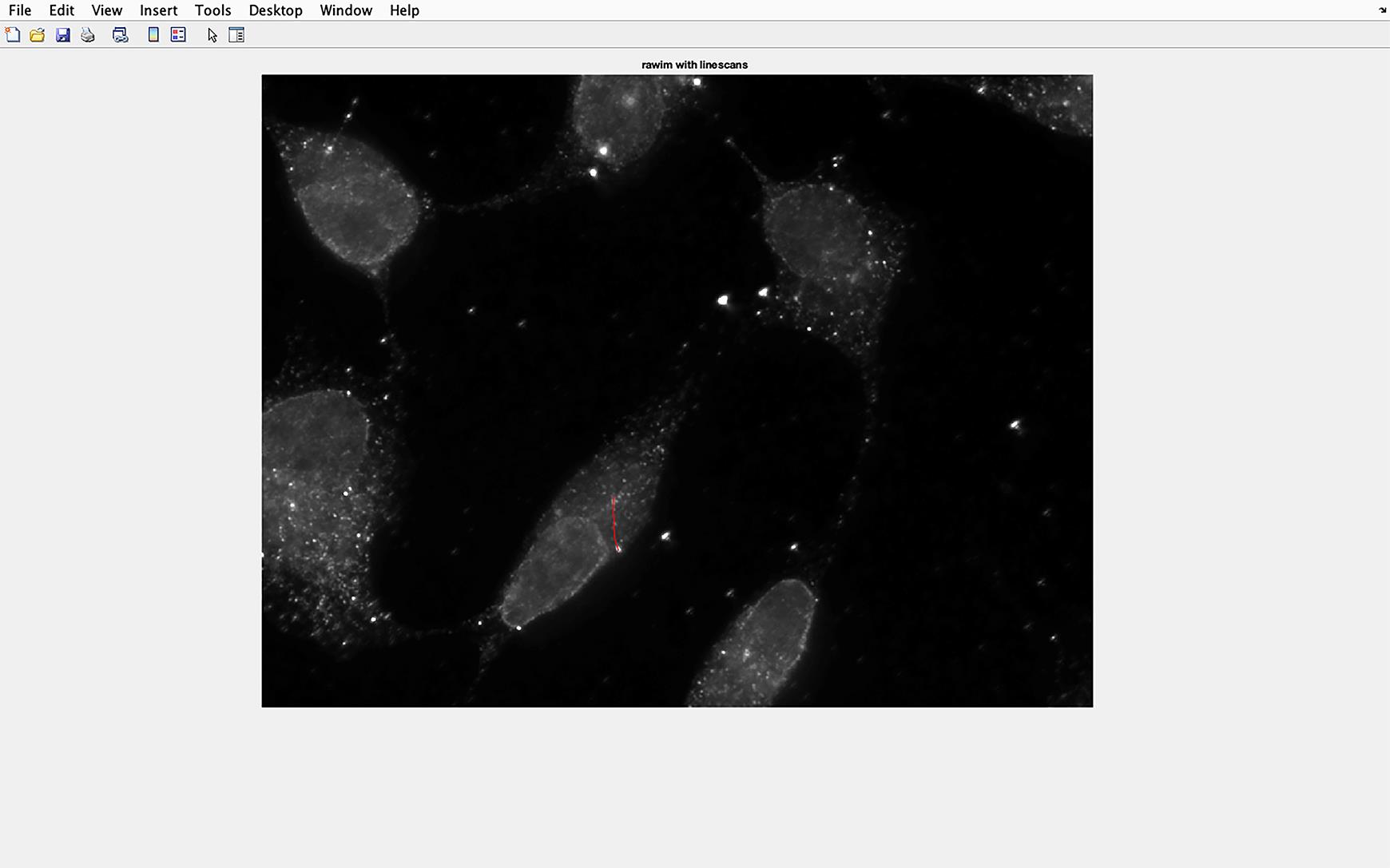
Figure 6. Cilium line overlaid on to IFT81 image. The line drawn along the cilium will be automatically transferred to the IFT81 image, and the pixel intensity of this section of the image will be measured.A histogram of the IFT81 staining is displayed (Figure 7).
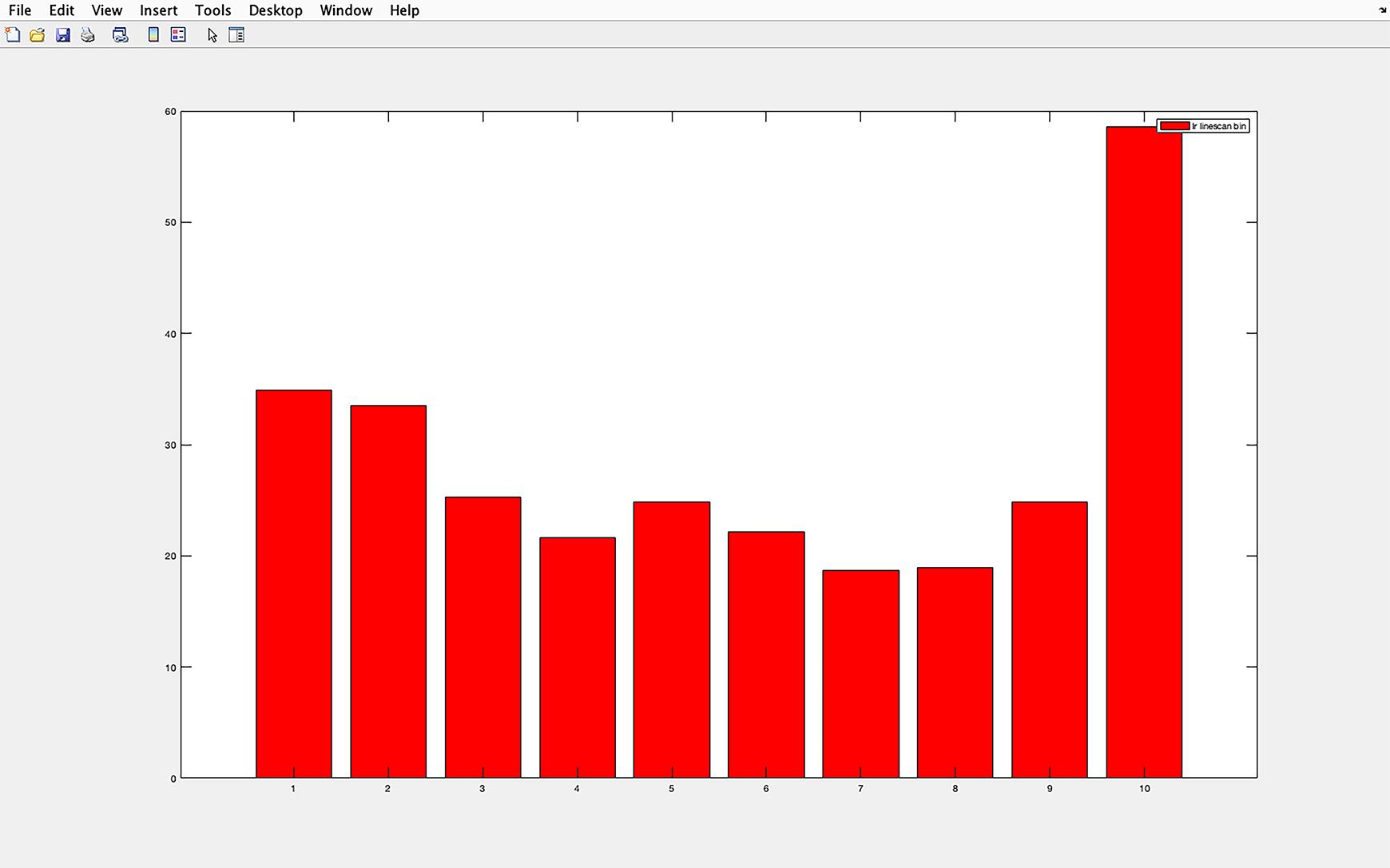
Figure 7. IFT81 pixel intensity histogram. Example of the histogram produced with the script. The average pixel intensity for each section of a cilium (1-10, proximal to distal) is shown. In the y-axis is the average pixel intensity, and the x-axis shows the cilium divided into 10 equal sections. Section 1 represents the cilium base and section 10 the tip. The base of the cilium was determined using the centrosomal marker γ-tubulin.An Excel spreadsheet of the IFT81 staining of the cilia is shown in the current folder side bar. This spreadsheet presents the average pixel intensity of the cilia divided into 10 sections (1 proximal-10 distal), and it also includes the cilia length. If multiple cilia are to be analyzed per image, it is important to change the name of the file so that the file is not overwritten when subsequent cilia are analyzed (Figure 8).
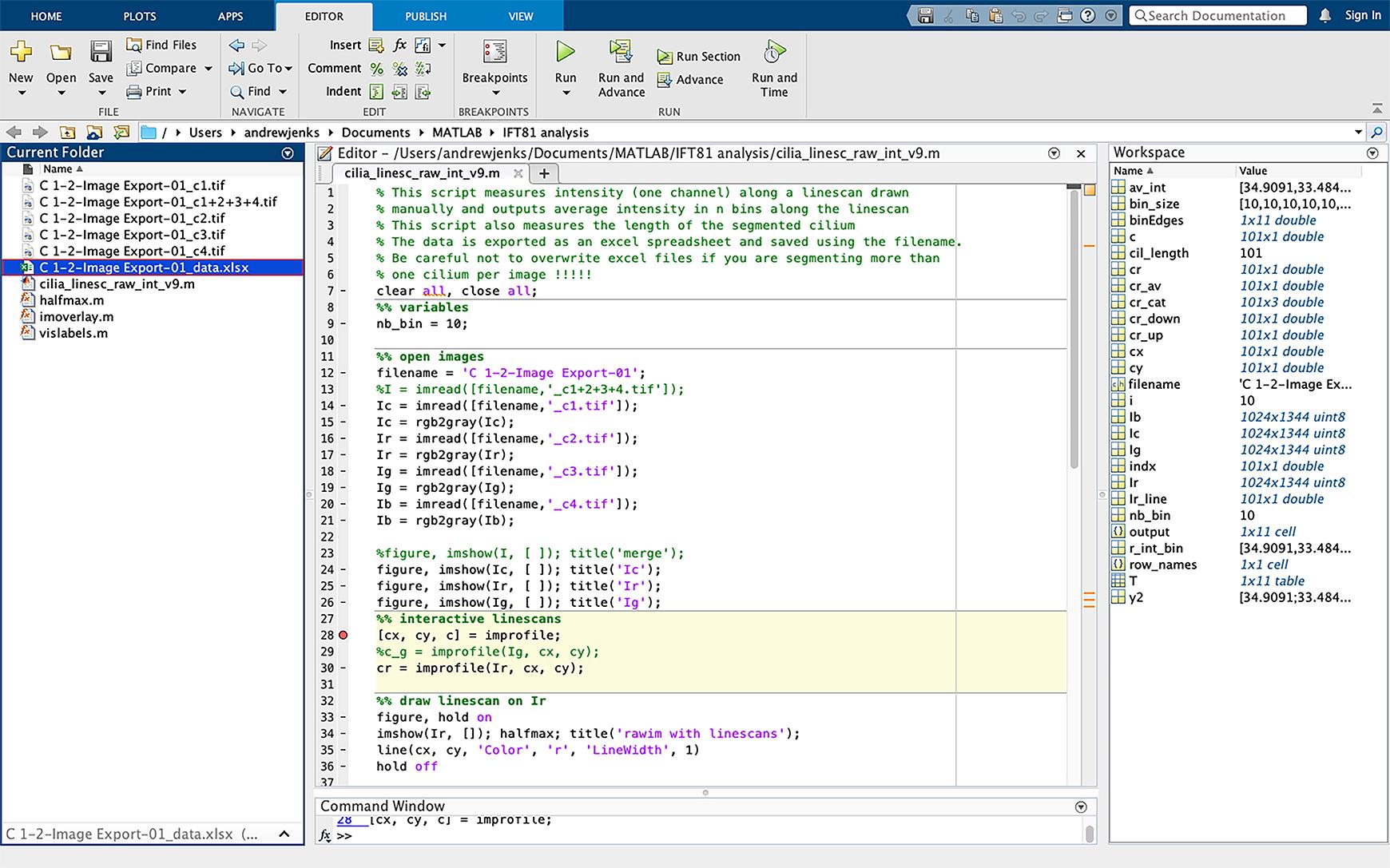
Figure 8. Excel spreadsheet with data. The Excel spreadsheet of IFT81 pixel intensity will appear in the current folder side bar (highlighted in blue). If multiple cilia are to be analyzed per image, change the name of the file to avoid overwriting this file.
Data analysis
In a typical experiment, pixel intensity is analyzed using 150 cilia for each experimental condition. The data is then presented as pixel intensity along proximal to distal sections of the cilium. Statistical significance is calculated using a Student’s t-test. In this example, our analysis showed that dasatinib resistant A204 cells had lower IFT81 protein levels along the proximal and distal ends of the cilia (Jenks et al., 2018). This method can be used for analyzing the distribution of any signal given by a protein or biological molecule that can be imaged using immunofluorescence. This highlights the versatility of this analysis tool, which enables the accurate quantification of the distribution of cilia-localized proteins.
This detailed protocol will allow the user to:
Analyze the ciliary distribution of any protein that can be observed using immunofluorescence.
Compare the distribution of ciliary proteins in different groups of cell lines (e.g., drug resistant cell lines compared to drug sensitive cell lines).
Quantitatively analyze the levels of protein localization to the cilium.
Measure cilium length.
Present data for publication.
Four additional MATLAB scripts were designed for analyzing cilia protein localization. Each script uses a similar workflow to the example described (Figures 1-8). CILIA 2 can automatically identify and quantify cilia length and intensity. CILIA 2 and CILIA 3 require the combined images to have 4 fluorescent channels. In order for CILIA 2 (cilia_linesc_aut_v6) to automatically quantify cilia from an image, the order of the exported TIFF fluorescent channels must be as follows: channel 1-centriolar marker, channel 2-signal to be analyzed, channel 3-Arl13B or cilia marker of choice, and channel 4-DAPI or alternative nuclear marker. An overlay of all channels must be included in order for this script to automatically identify and orientate cilia. CILIA 3 also requires channel 2 to be the signal to be analyzed and channel 3 to be the cilia marker; however, the remaining channels can be changed as the cilia is manually defined. CILIA 2 (cilia_linesc_aut_v6) (protocol detailed in Figures 9-15) and CILIA 3 (cilia_linesc_int_v9) (protocol detailed in Figures 9, 16-22) present staining intensity as average pixel intensity of each of the 10 cilia sections divided by the average pixel intensity of the entire cilia. The remaining two additional scripts, CILIA 4 (cilia_linesc_raw_int_3c_v9) and CILIA 5 (cilia_linesc_int_3c_v9), only need three fluorescent channels and require exported TIFF fluorescent channels to be organized as follows: channel 1-Arl13B or cilia marker of choice and channel 2-signal to be analyzed. For the example shown, DAPI was used in channel 3 to define the nucleus (this staining can be changed if required). CILIA 4 (cilia_linesc_raw_int_3c_v9) and CILIA 5 (cilia_linesc_int_3c_v9) only differ in how the staining intensity is presented. CILIA 4 (cilia_linesc_raw_int_3c_v9) (protocol detailed in Figures 23-30) produces results as average pixel intensity, and CILIA 5 (cilia_linesc_int_3c_v9) (protocol detailed in Figures 31-37) produces results as average pixel intensity of each of the 10 cilia sections divided by the average pixel intensity of the entire cilia. These additional scripts are further described below, using a composite image of cilia stained with acetylated tubulin and Arl13B (Figure 9).
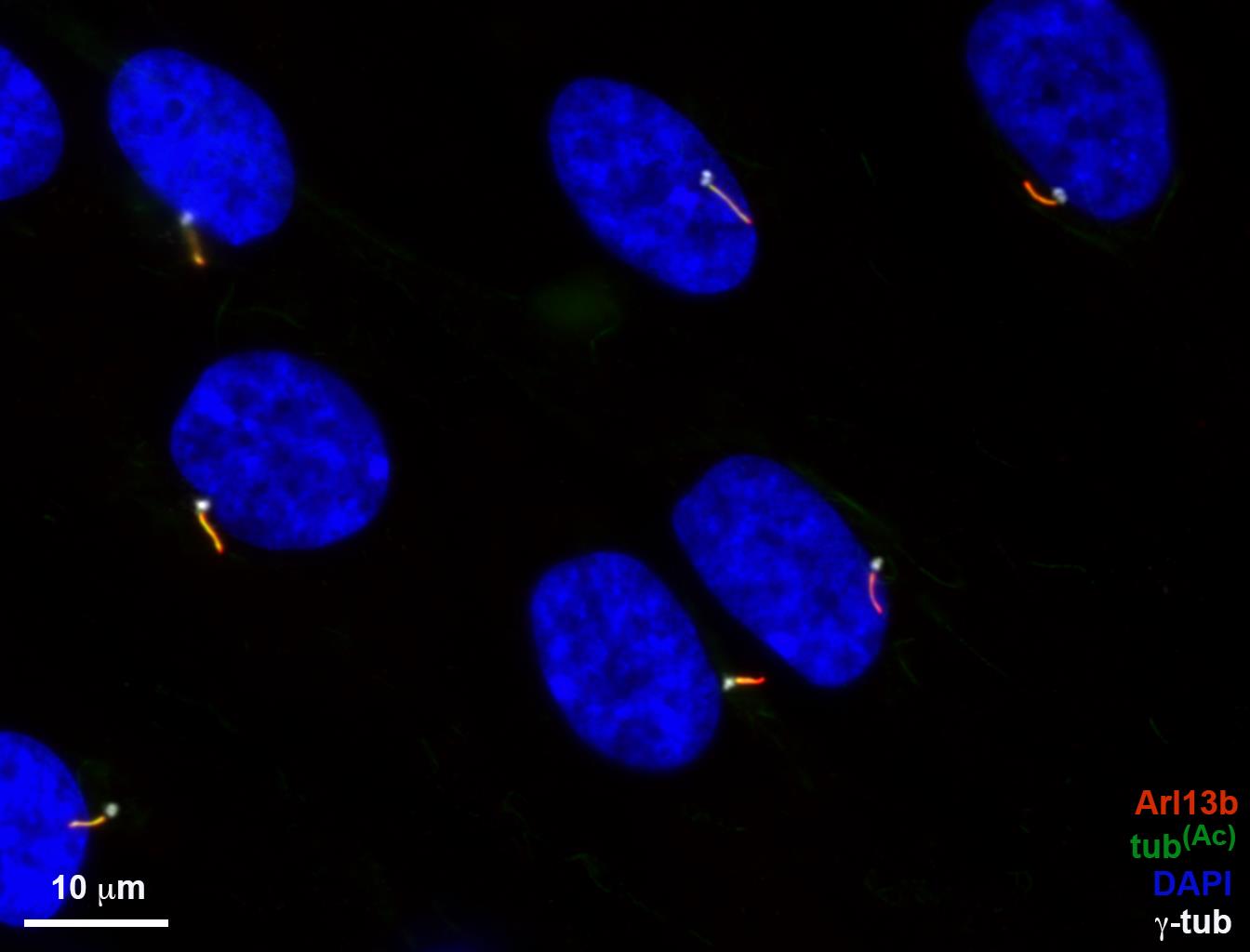
Figure 9. RPE-1 cilia image used in the analyses with CILIA 2 and CILIA 3 scripts. RPE-1 cells were serum starved for 48 h to induce ciliogenesis, then fixed and stained with antibodies for Arl13B (red), acetylated tubulin (green), and γ-tubulin (white). Nuclear stain is DAPI (blue). Scale bar = 10 μm.
CILIA 2 – Automated four channel automated script (cilia_linesc_aut_v6):
Four (4) fluorescent channels. For the example shown, C1 is the centriole marker (γ-tubulin), C2 is the protein of interest (acetylated tubulin), C3 is the cilia marker (Arl13B), and C4 is the nuclear stain (DAPI). Cilia are automatically defined by the script and divided into 10 equal sections. Data is presented as channel 2 pixel intensity of each cilia section divided by the average channel 2 pixel intensity along the entire cilia. This ratio will be presented as 10 cilia sections (bins), running from the base to the cilia tip. In the example shown in Figures 10-15, it is used to quantify acetylated tubulin. To detect all cilia, it might be necessary to adjust the channel 2 pixel intensity threshold on line 10 of the script (level).
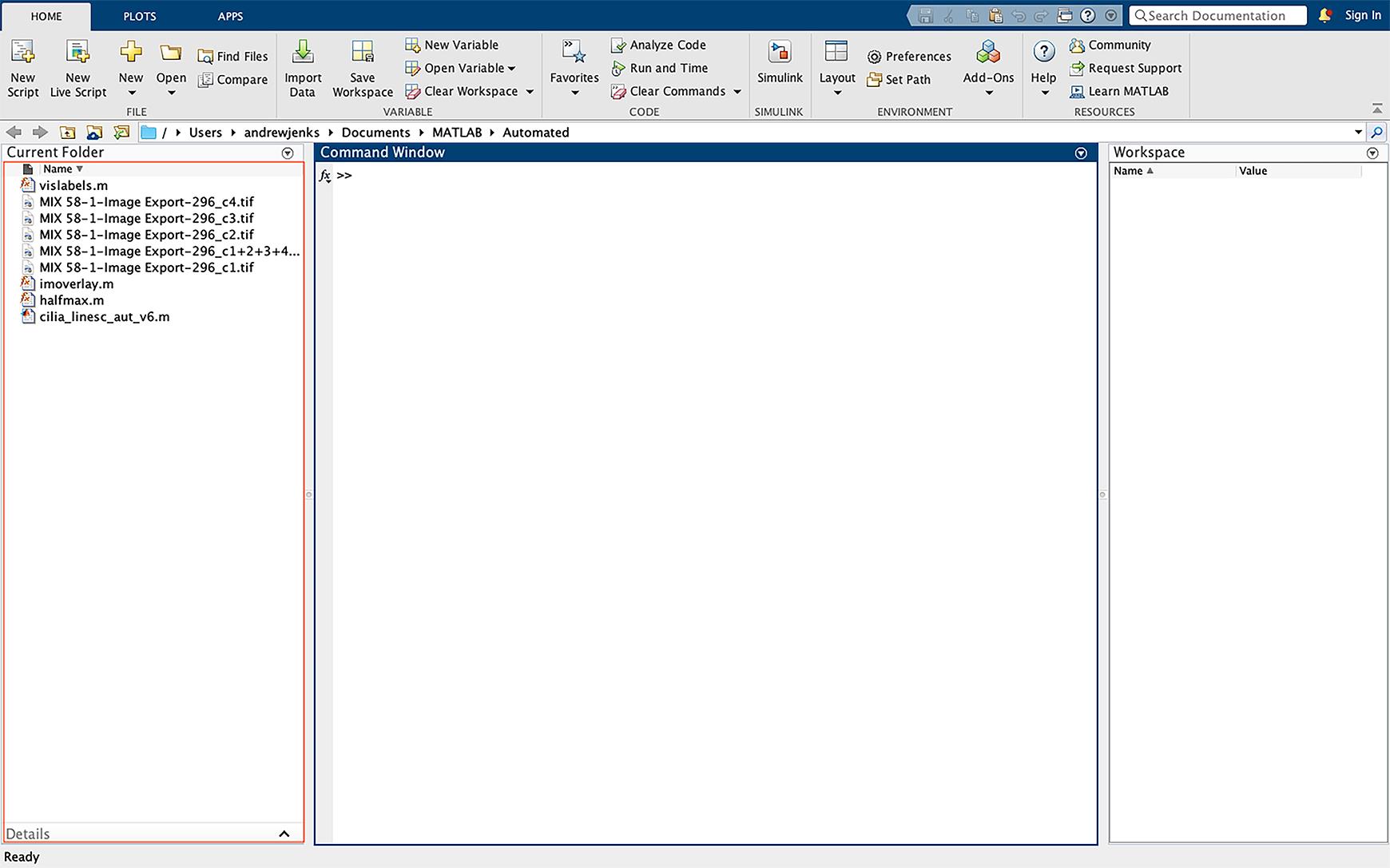
Figure 10. Importing data into MATLAB. Drag the script files and images to be analyzed into the MATLAB far-left window of the operating interface. Images must be imported as the individual fluorescent channels as TIFF files.
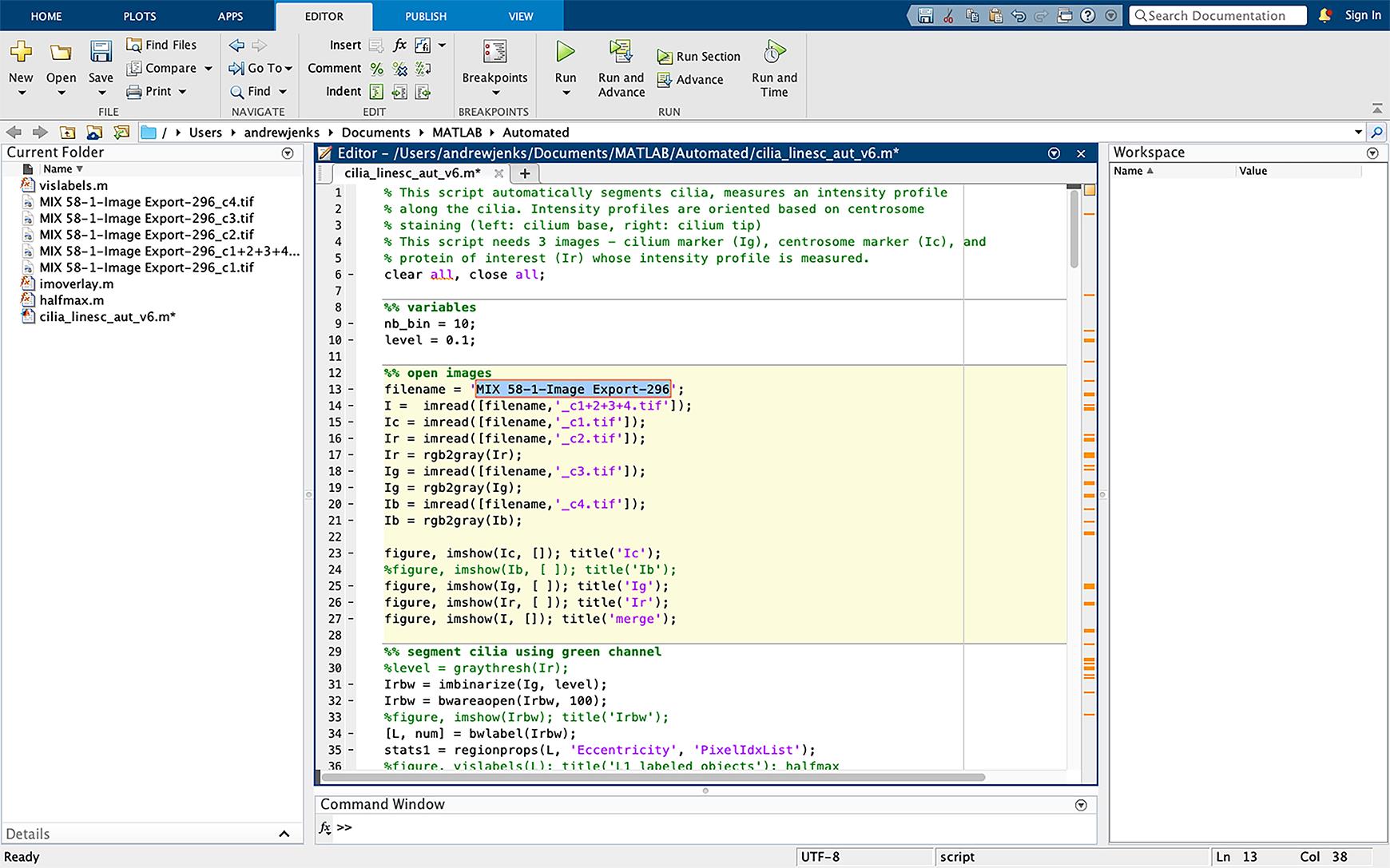
Figure 11. Changing the script file name. Change the filename in script (line 13, highlighted in blue) to the same name as the files to be analyzed. The TIFF extension should not be included in the filename.
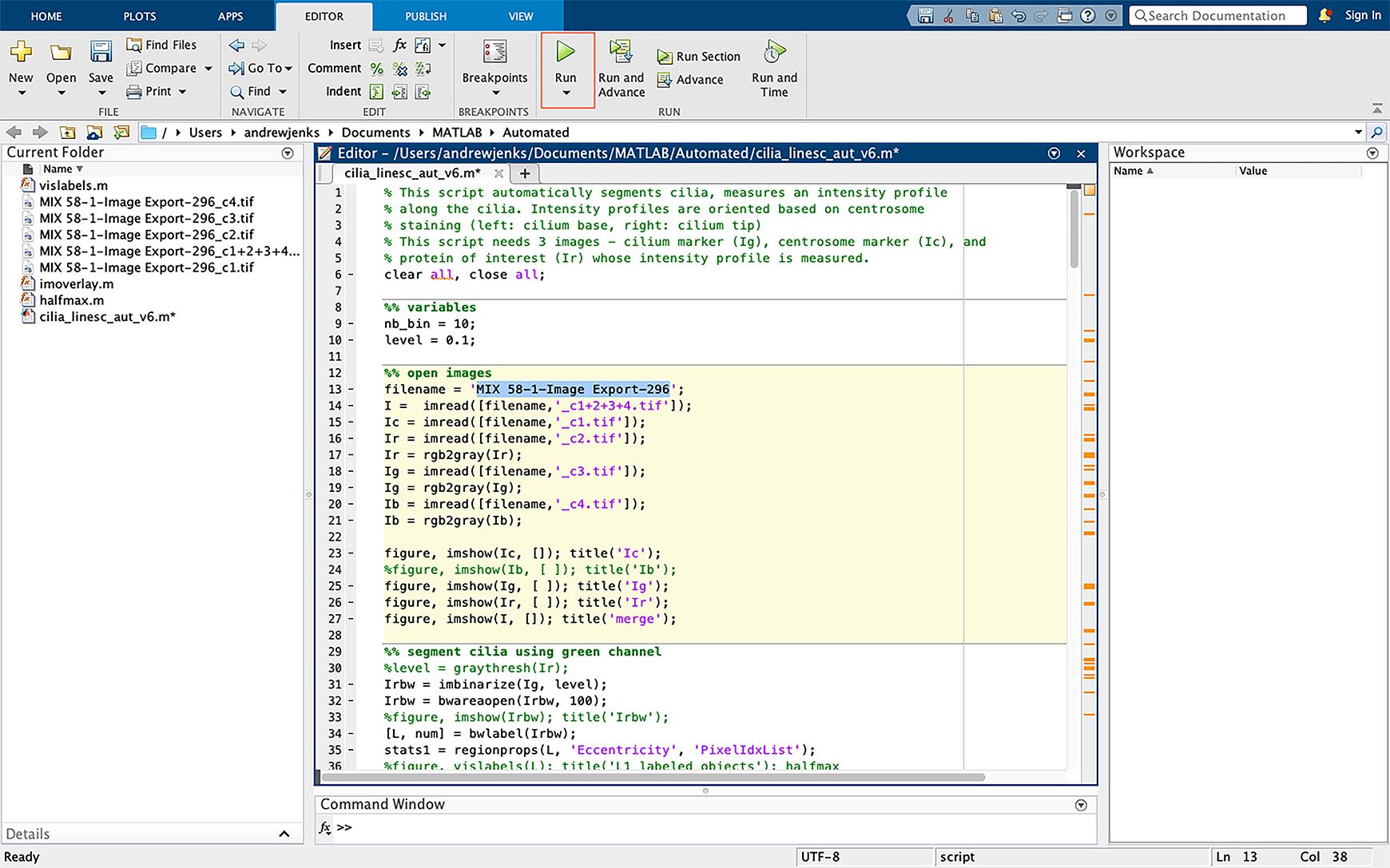
Figure 12. Running the script. Start the script by pressing “run” (green icon highlighted on the top).
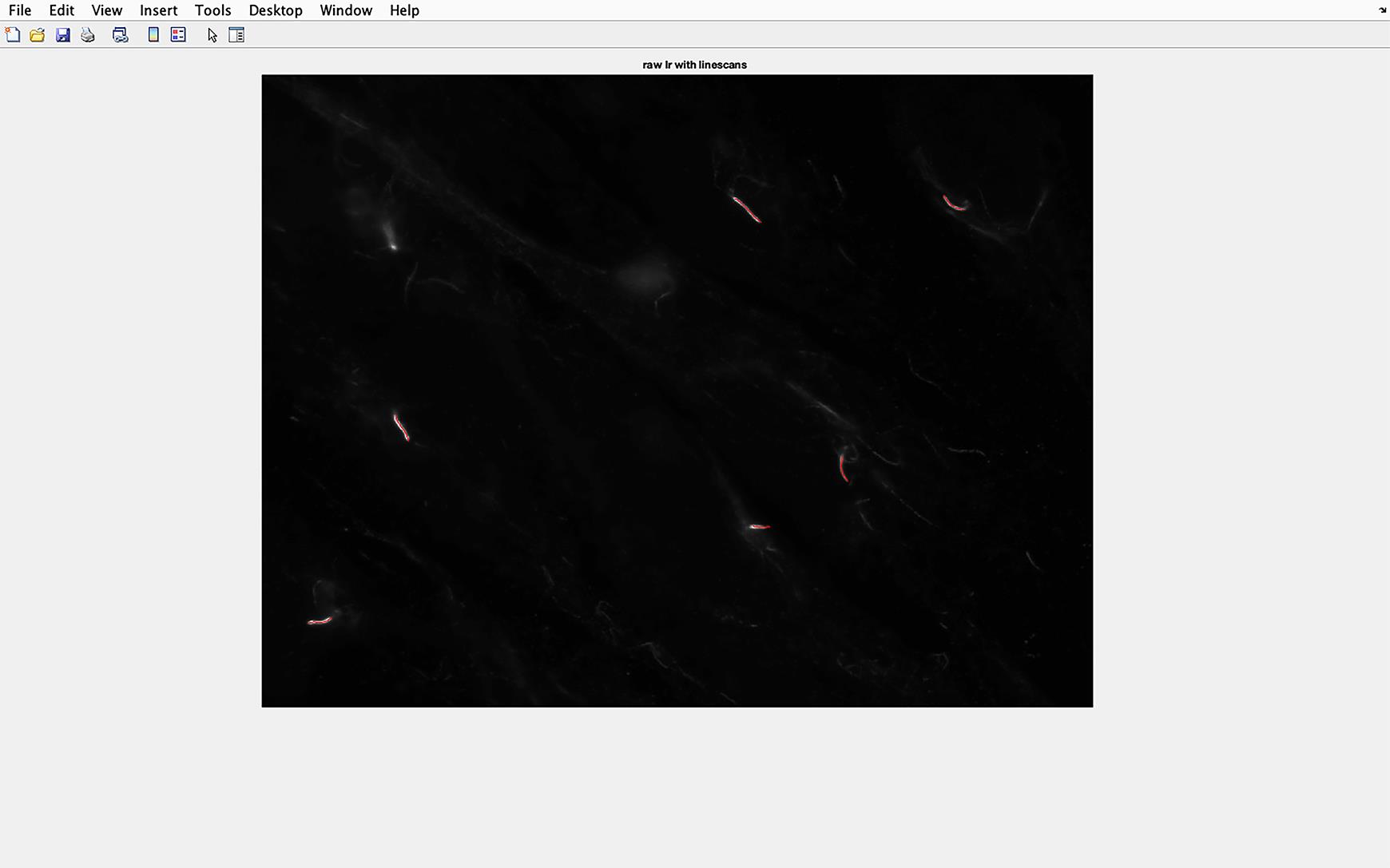
Figure 13. Cilia line overlaid on to acetylated tubulin image. Cilia will be automatically defined using Arl13B staining and γ-tub to orientate cilia direction. These cilia will be transferred to the acetylated tubulin image, and the pixel density of these sections of the image will be analyzed.
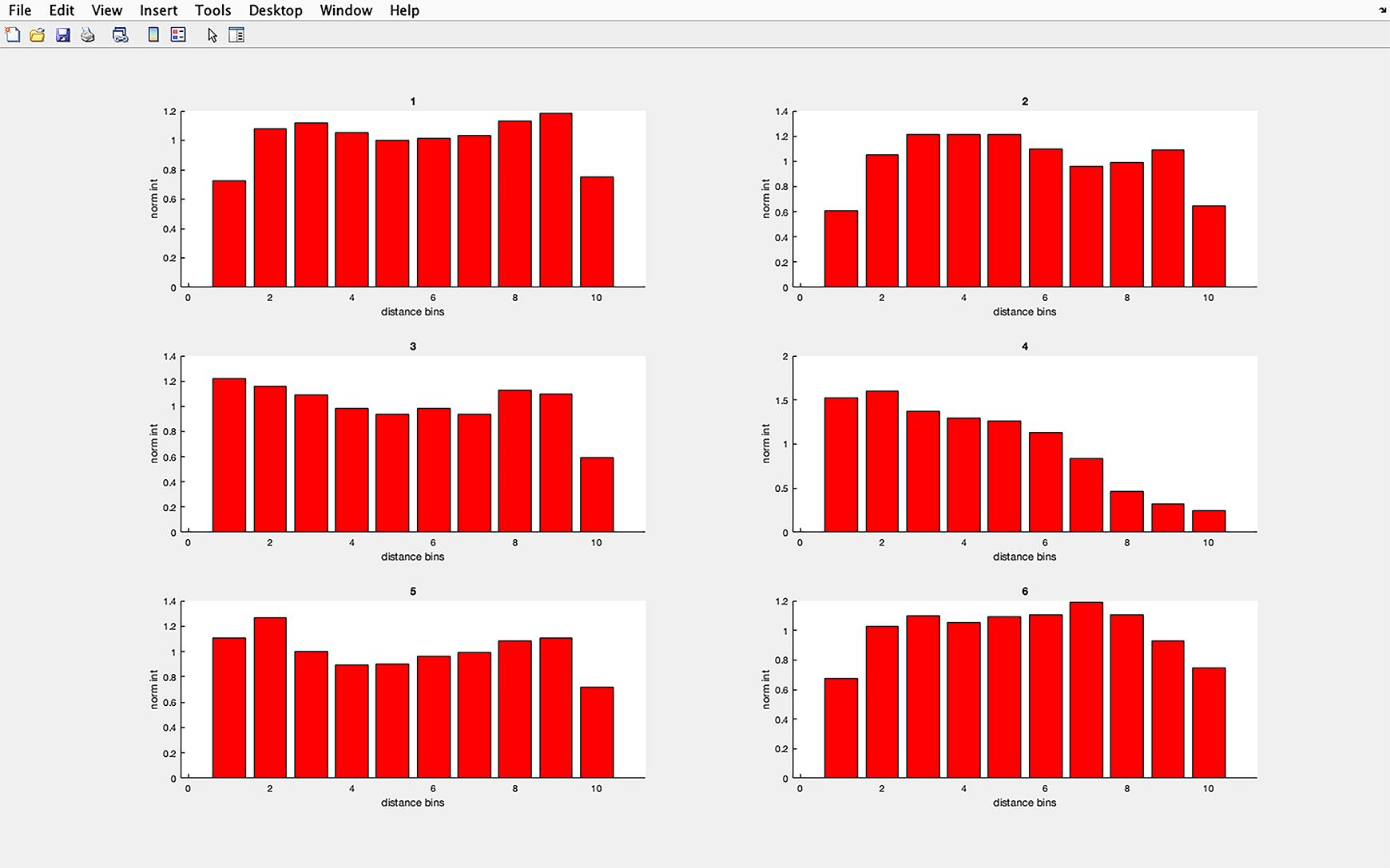
Figure 14. Acetylated tubulin pixel intensity histogram. Example of the histograms produced from an image containing six cilia. The y-axis denotes pixel intensity as a ratio of average pixel intensity divided by the average pixel intensity of the entire cilia. Section 1 represents the cilia base and section 10 the cilia tip. The base of the cilia was determined using the centrosomal marker γ-tubulin.
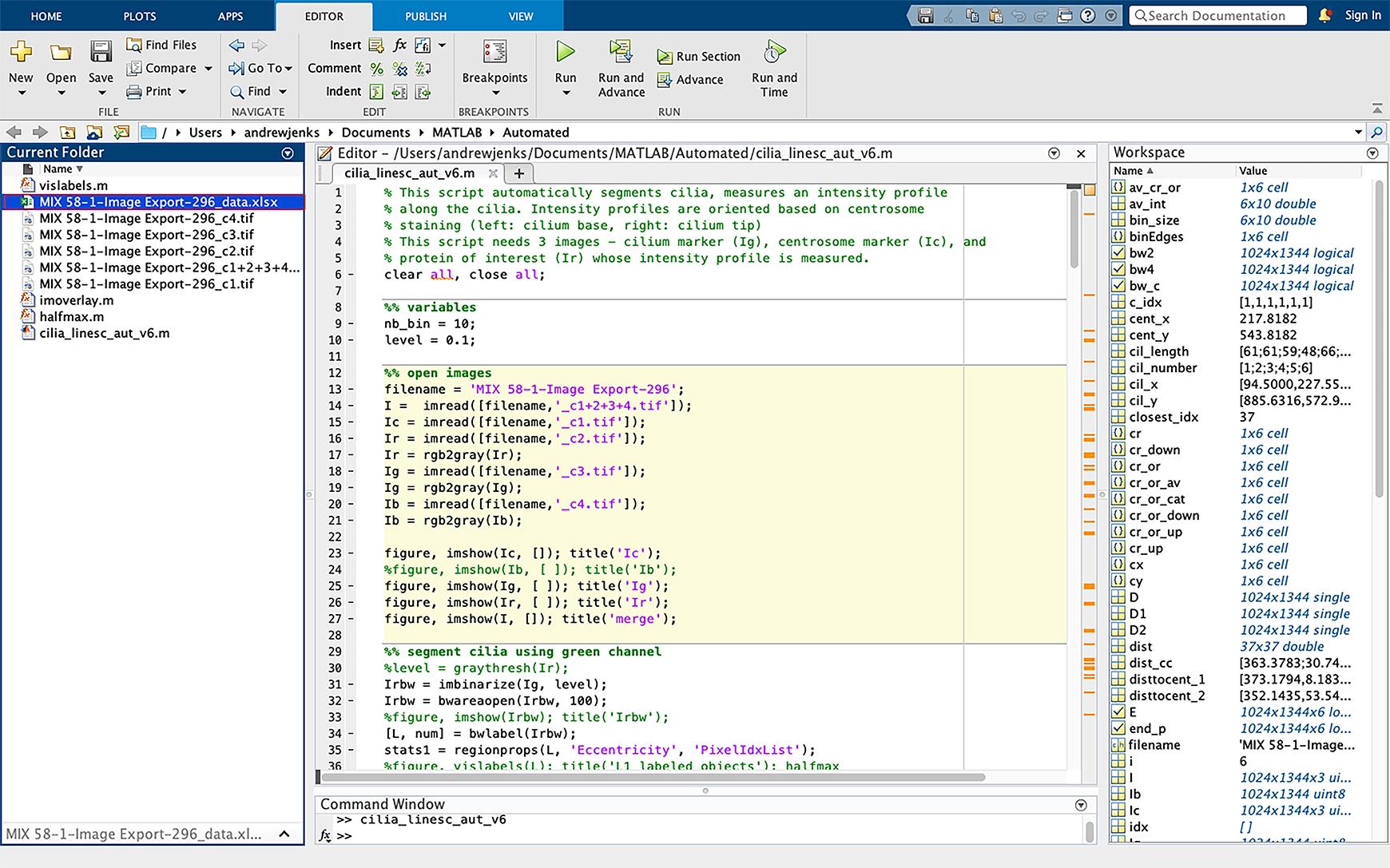
Figure 15. Example of pixel ratio excel spreadsheet. The Excel spreadsheet of acetylated tubulin pixel ratio will appear in the current folder side bar (highlighted in blue).
CILIA 3 – Four channel pixel ratio (cilia_linesc_int_v9):
Four (4) fluorescent channels. For the example shown, C1 is the centriole marker (γ-tub), C2 is the protein of interest (acetylated tubulin), C3 is the cilia marker (Arl13B), and C4 is the nuclear stain (DAPI). Once cilia are manually defined, the script will then divide cilia into 10 equal sections. Channel 2 pixel intensity of each section will be divided by the average channel 2 pixel intensity of the entire cilia. This ratio will be presented as 10 cilia sections running from the base to the cilia tip. In the example shown in Figures 16-22, it is used to quantify acetylated tubulin.
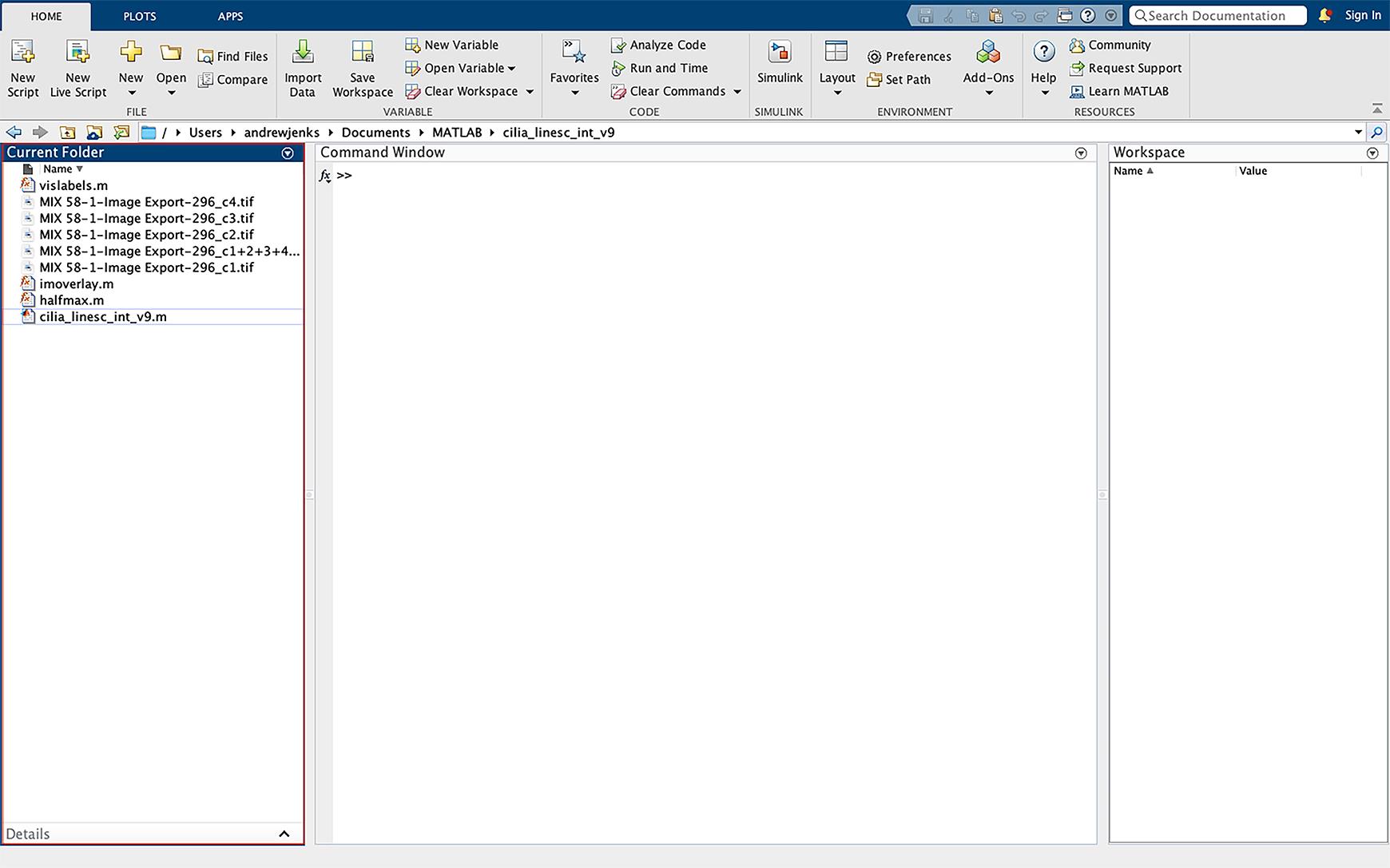
Figure 16. Importing data into MATLAB. Drag the script files and images to be analyzed into the MATLAB directory left sidebar of the operating interface. Images must be imported as the individual fluorescent channels as TIFF files.
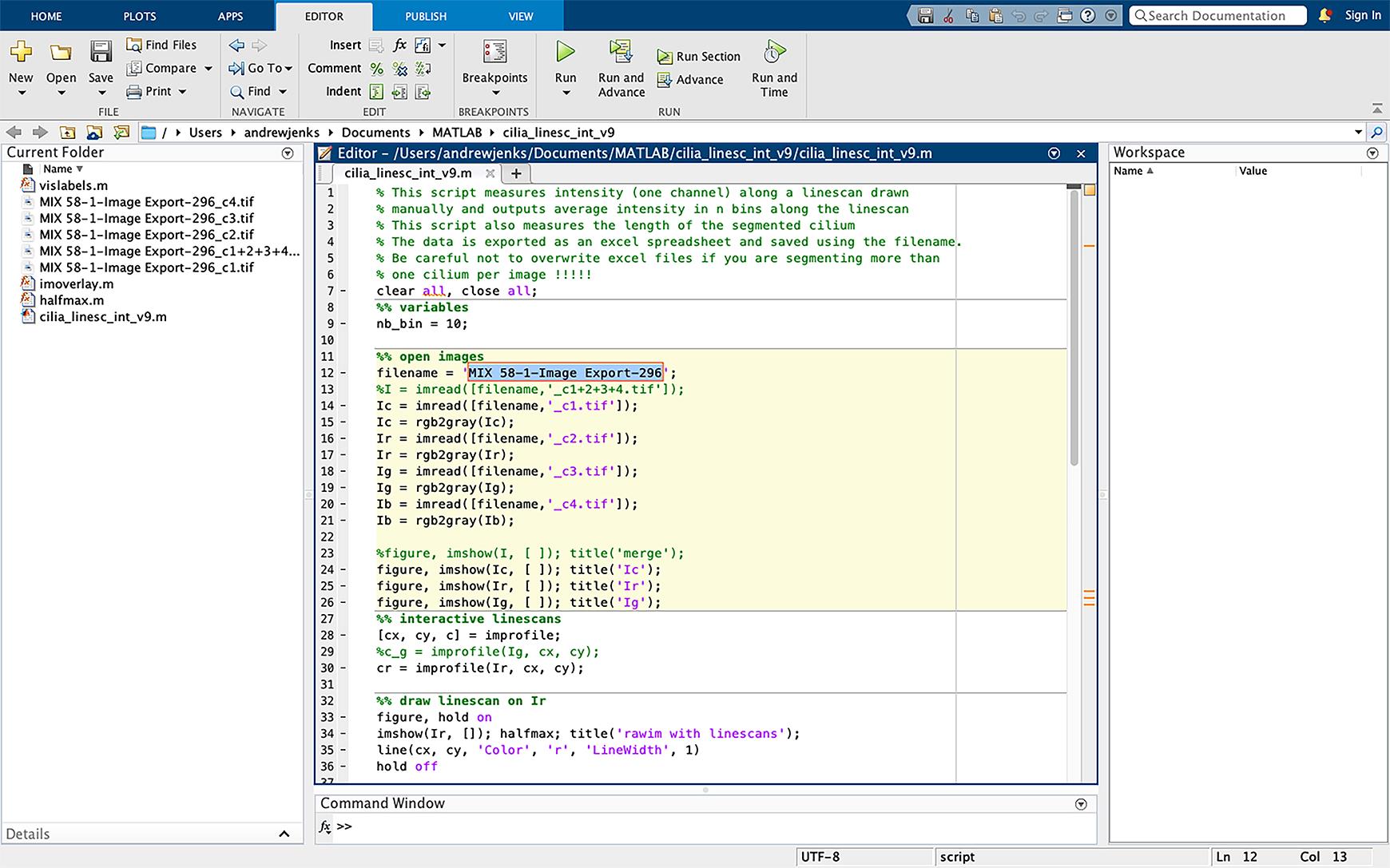
Figure 17. Changing the script file name. Change the filename in the script (line 12, highlighted in blue) to the same name as the files to be analyzed. The TIFF extension should not be included in the file name.
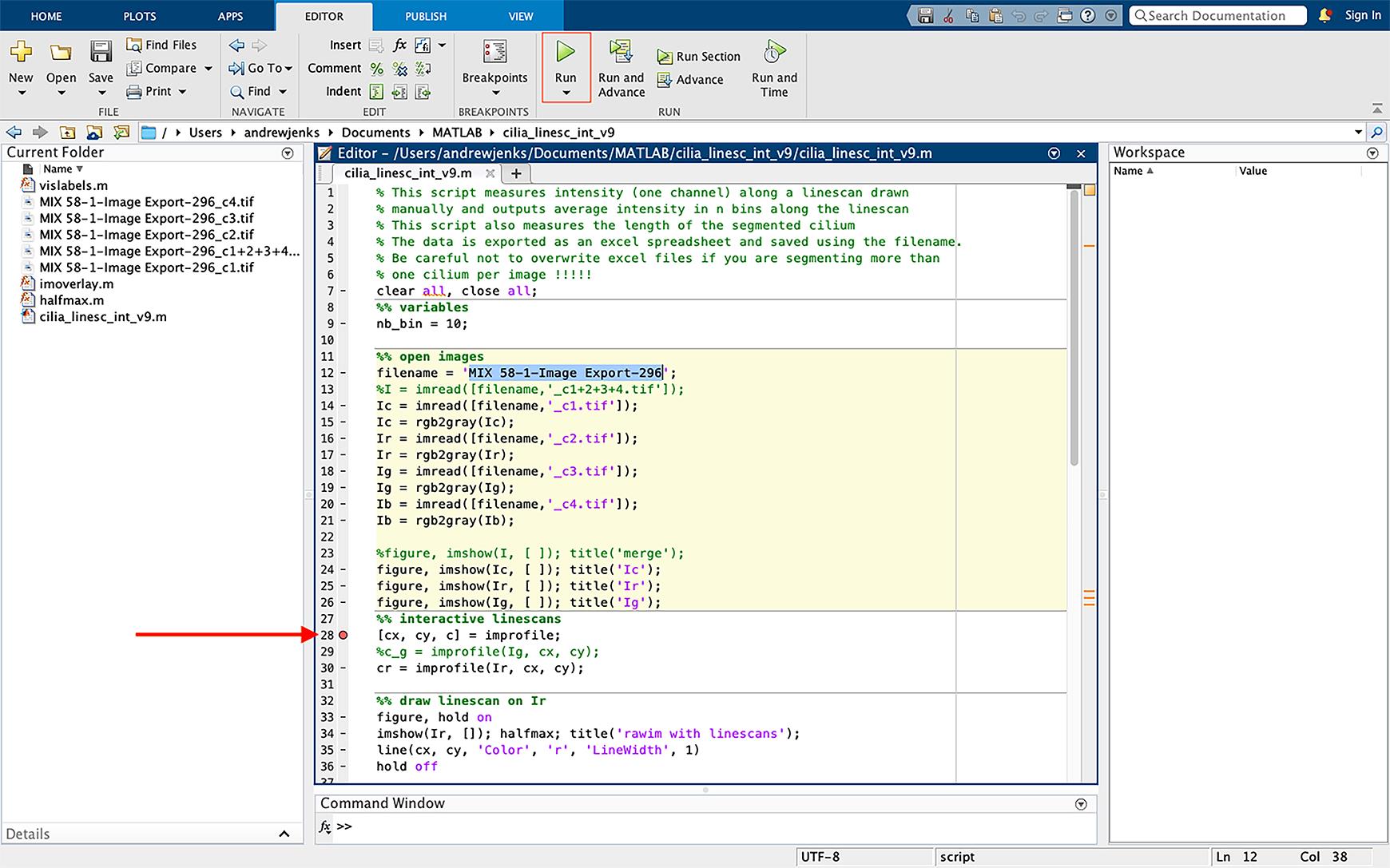
Figure 18. Selecting the file to be analyzed and inserting a breakpoint in the script. Insert a breakpoint in the script by clicking on the dash sign next to line 28. This will convert the dash to a red circle (highlighted with an arrow). Start the script by pressing “run” (green icon highlighted on the top).
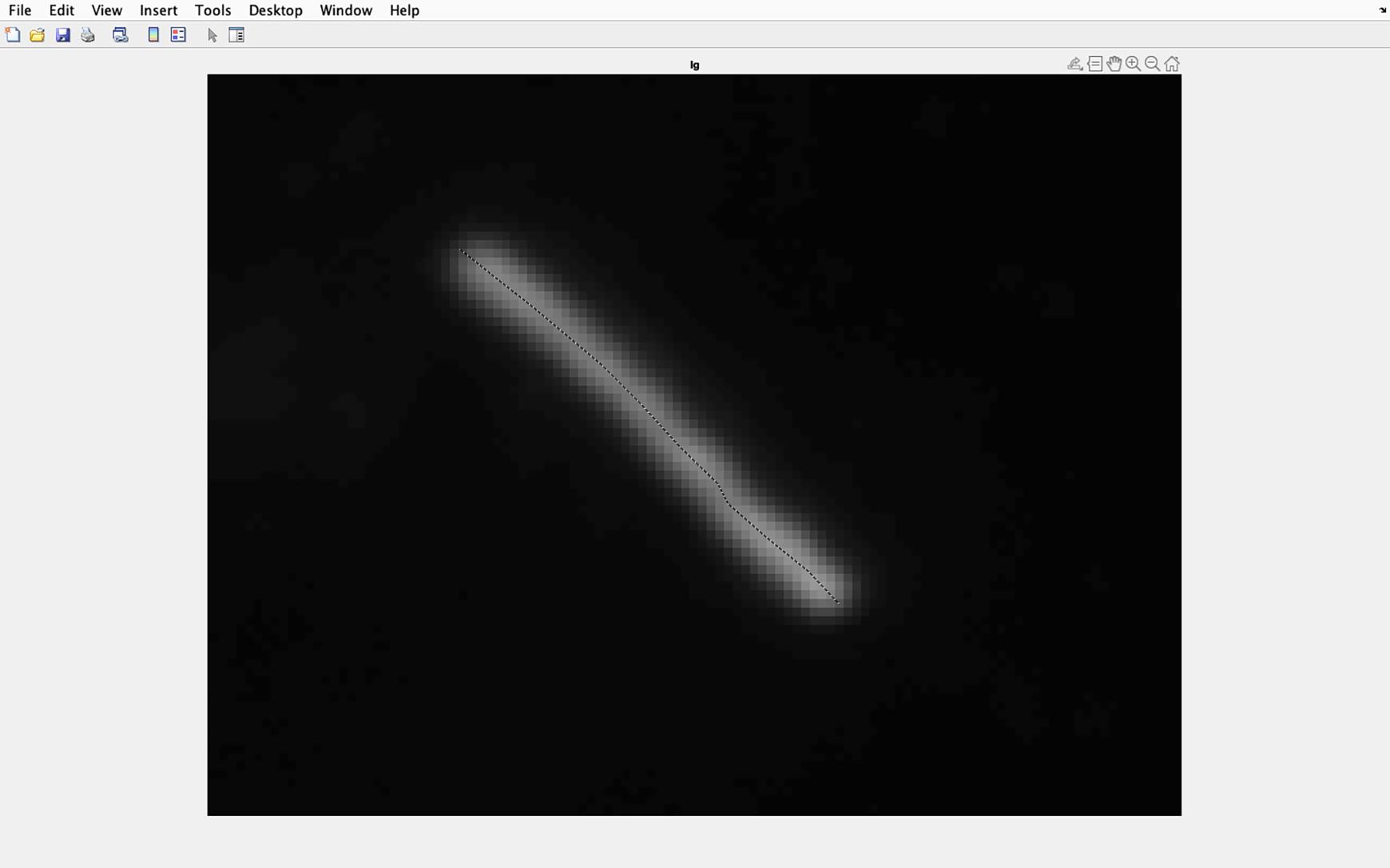
Figure 19. Identifying cilia to be analyzed. Draw a line along the center of the magnified cilia, ensuring the line goes from the base to the tip of the cilia. Double click when the line is at the cilia tip.
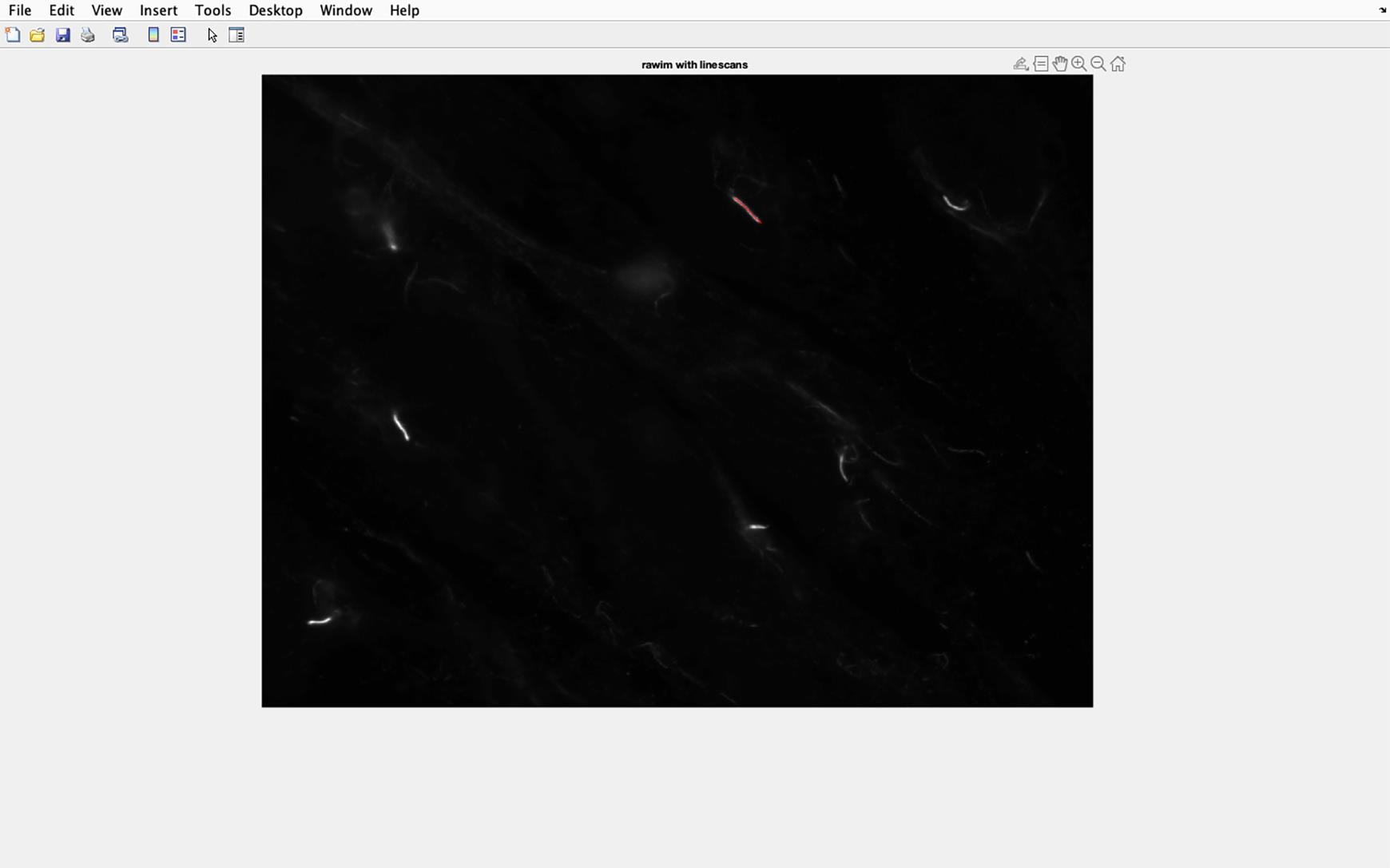
Figure 20. Cilia line overlaid on to acetylated tubulin image. The line drawn along the cilia will be transferred to the acetylated tubulin image. Then, the pixel intensity of this section of the image will be analyzed.
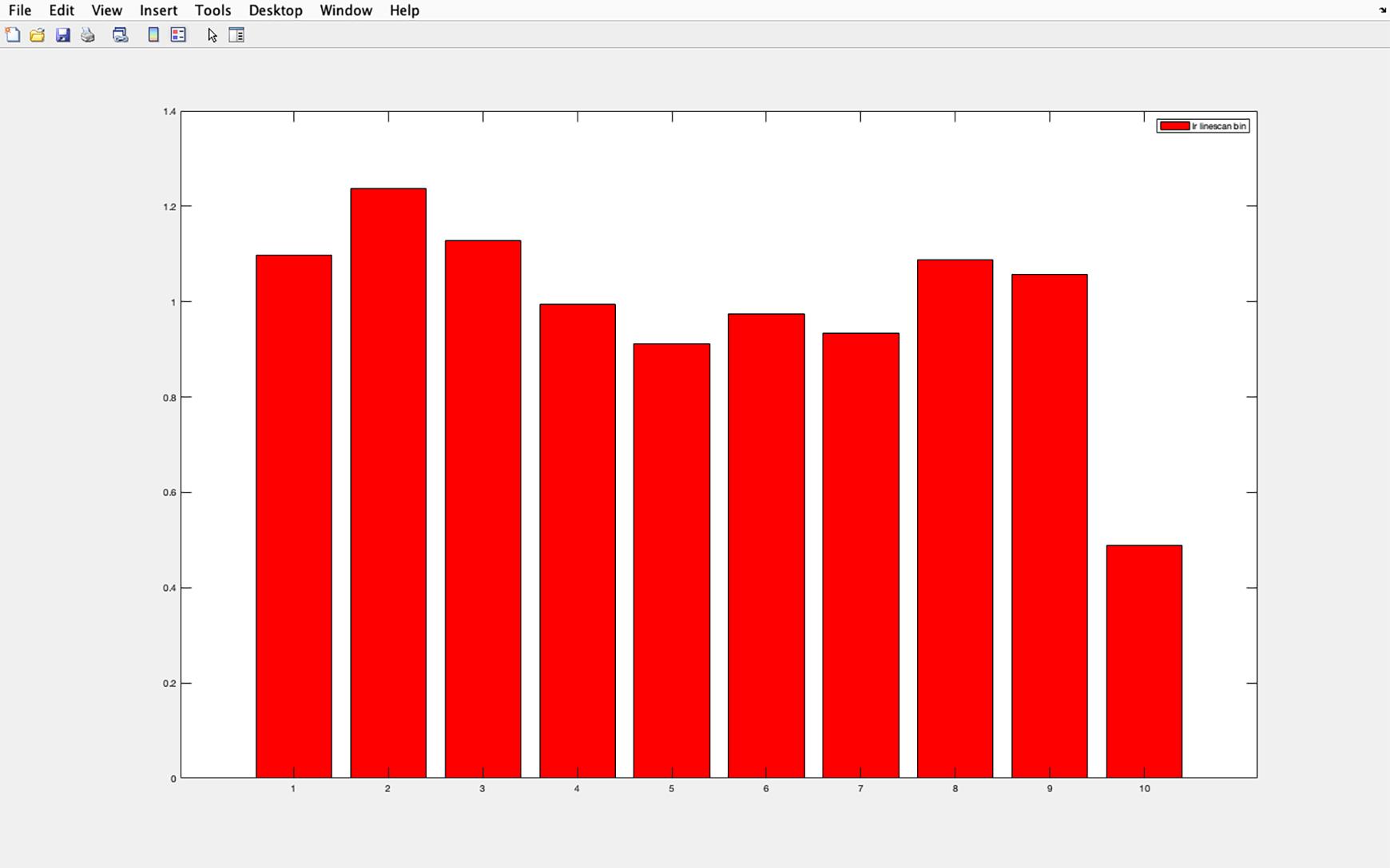
Figure 21. Acetylated tubulin pixel intensity histogram. Example of the histogram produced for average pixel intensity in each section of a cilia (1 proximal to 10 distal). The y-axis shows pixel intensity as a ratio of average pixel intensity of each section divided by the average pixel intensity of the entire cilia. Section 1 represents the cilia base and section 10 the cilia tip. The base of the cilia was determined using the centrosomal marker γ-tubulin.
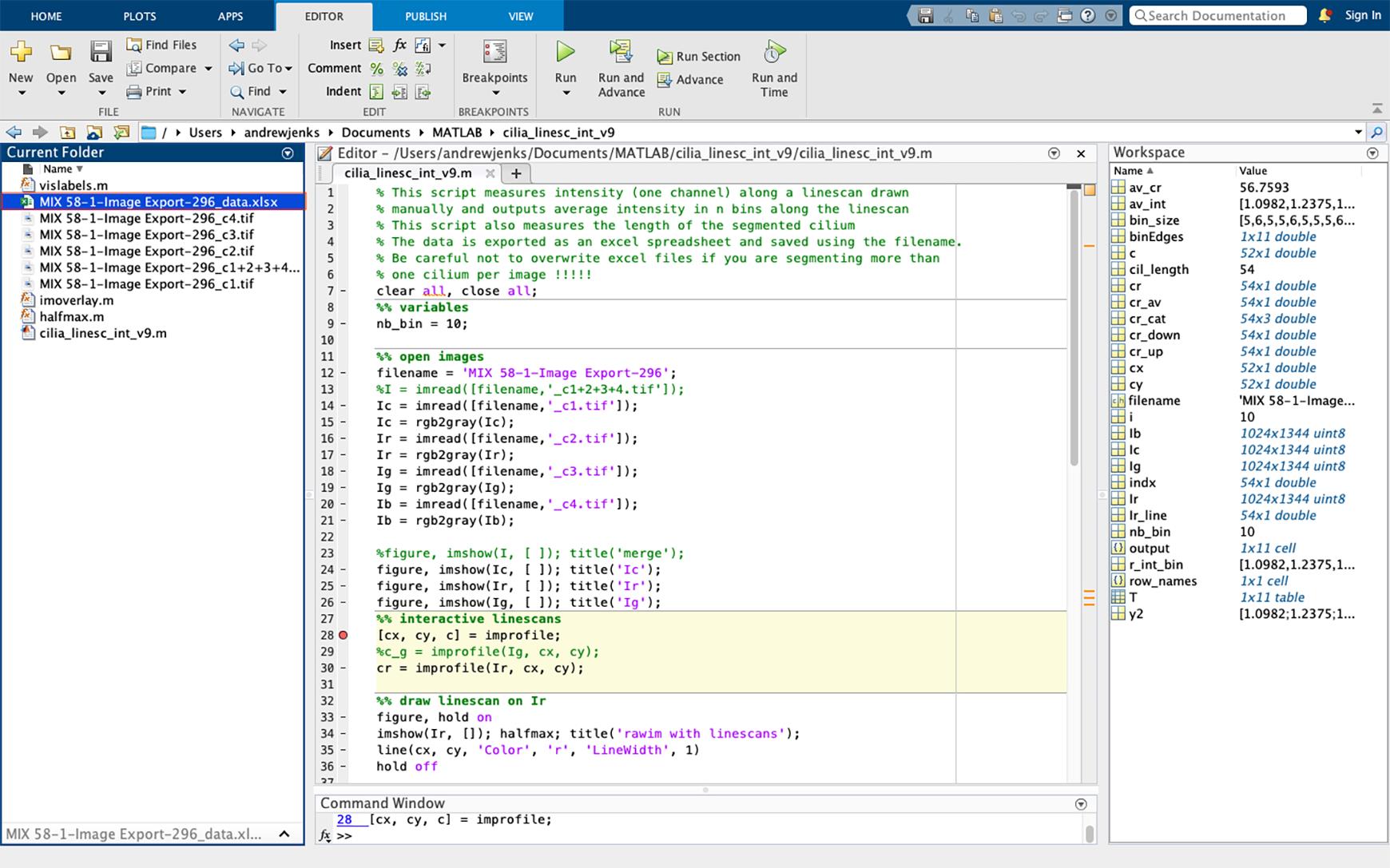
Figure 22. Example of average pixel intensity excel spreadsheet. The Excel spreadsheet of acetylated tubulin pixel intensity will appear in the current folder side bar (highlighted in blue). If multiple cilia are to be analyzed per image, change the name of the file to avoid overwriting this file.
Finally, for the example of CILIA 4 and CILIA 5 scripts, we used the image shown in Figure 9 without the γ-tubulin staining (Figure 23).
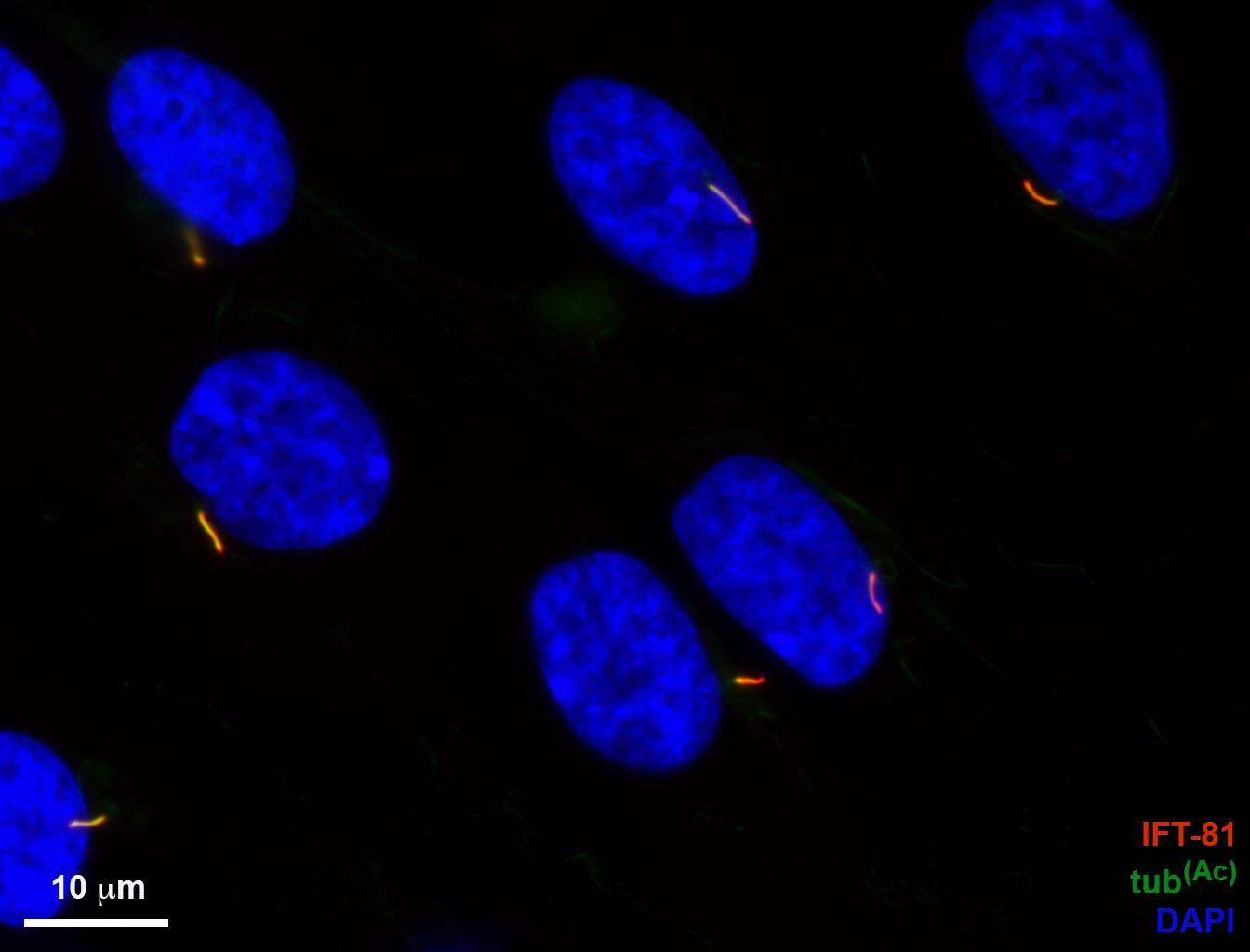
Figure 23. RPE-1 cilia image used in the analyses with CILIA 4 and CILIA 5 scripts.> RPE-1 cells were serum starved for 48 h to induce ciliogenesis, then fixed and stained with antibodies for Arl13B (red) and acetylated tubulin (green). Nuclear stain is DAPI (blue). Scale bar = 10 μm.
CILIA 4 – Three channel raw intensity script (cilia_linesc_raw_int_3c_v9):
This script uses three (3) fluorescent channels. For the example shown, C1 is the cilia marker (Arl13B), C2 is the protein of interest (acetylated tubulin), and C3 is the nuclear stain (DAPI). Once cilia are manually defined, the script will divide cilia into 10 equal sections, and the raw pixel intensity of channel 2 will be measured along the defined cilia and presented as 10 cilia sections running from the base to the cilia tip. In the example shown in Figures 24-30, this is used to quantify acetylated tubulin.
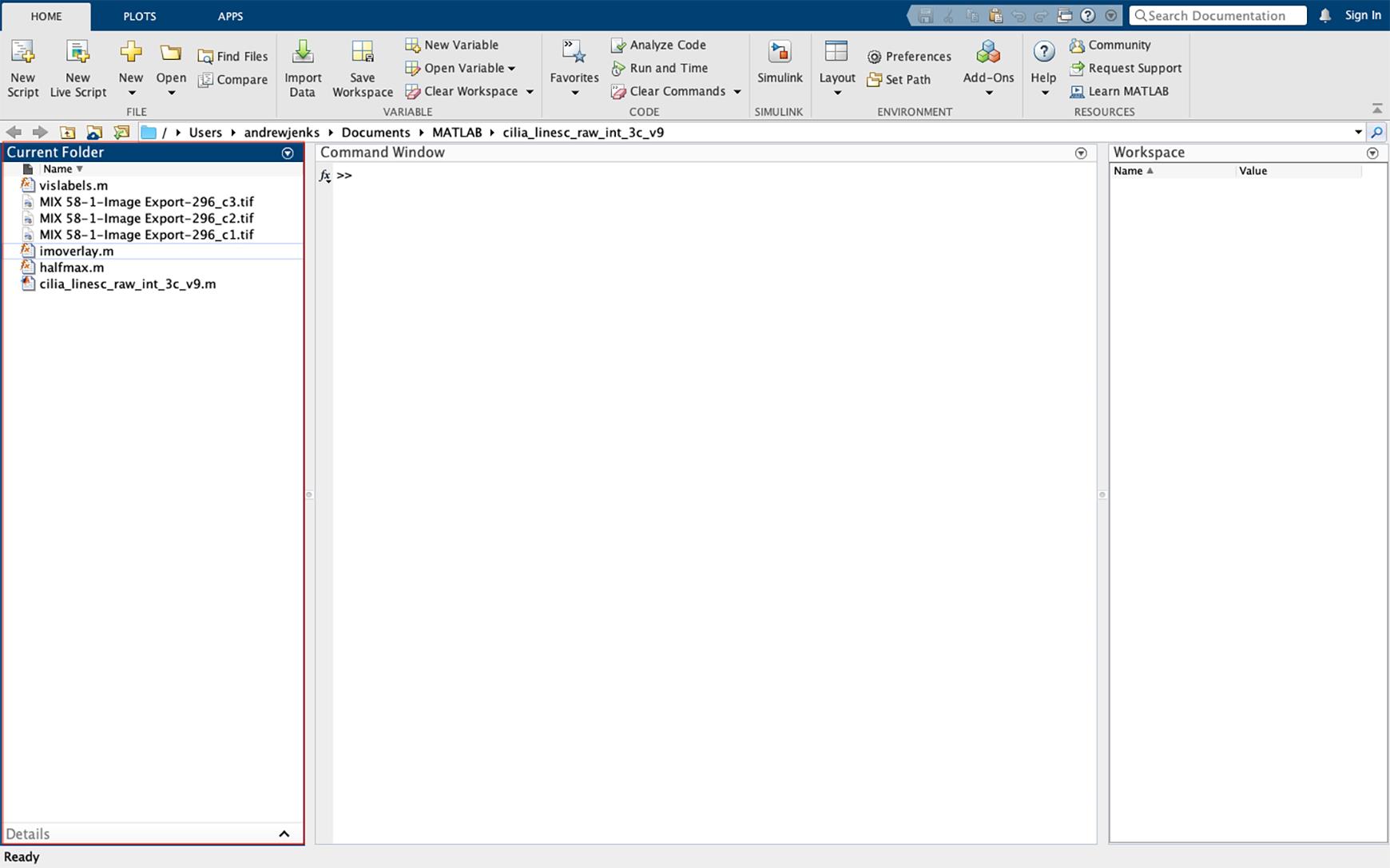
Figure 24. Importing data into MATLAB. Drag the script files and images to be analyzed into the MATLAB far-left window of the operating interface. Images must be imported as TIFF files of the individual fluorescent channels.
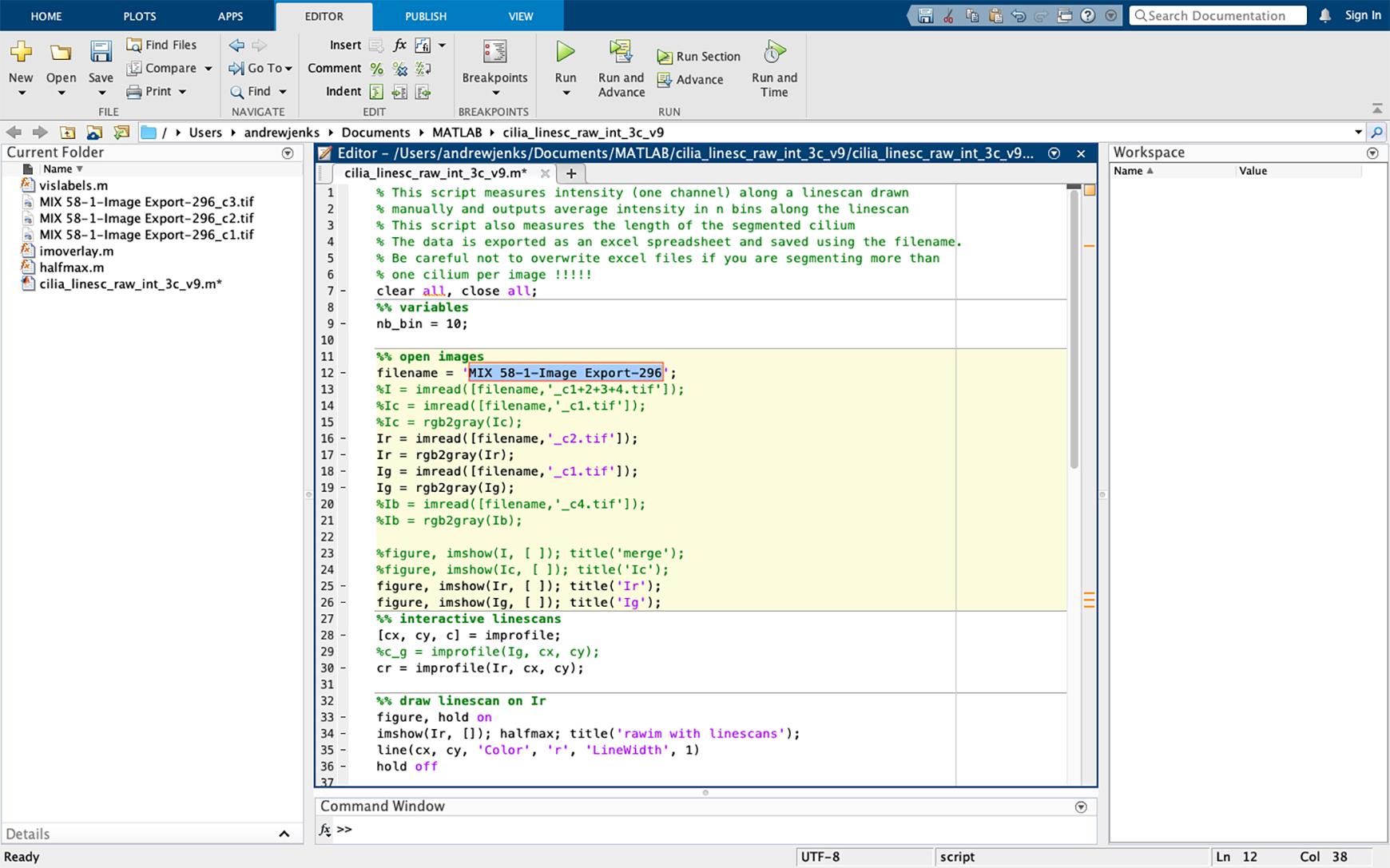
Figure 25. Changing the script file name. Change the filename in the script (line 12, highlighted in blue) to the same as the files to be analyzed. The TIFF extension should not be included in the file name.
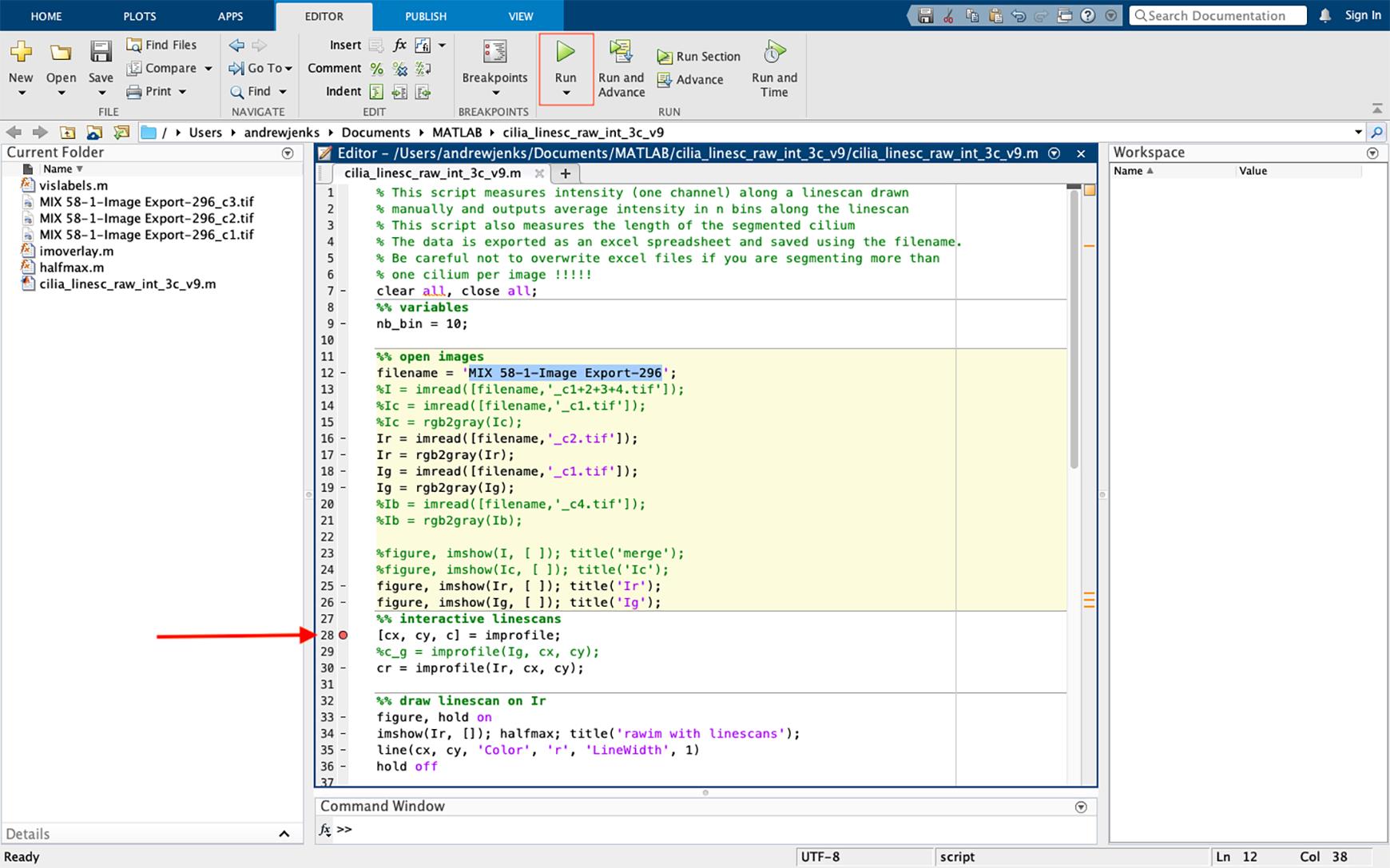
Figure 26. Selecting the file to be analyzed and inserting a breakpoint in the script. Insert a breakpoint in the script by clicking on the dash sign next to line 28, which will convert the dash to a red circle (highlighted using an arrow). Start the script by pressing “run” (green icon “highlighted” on the top).
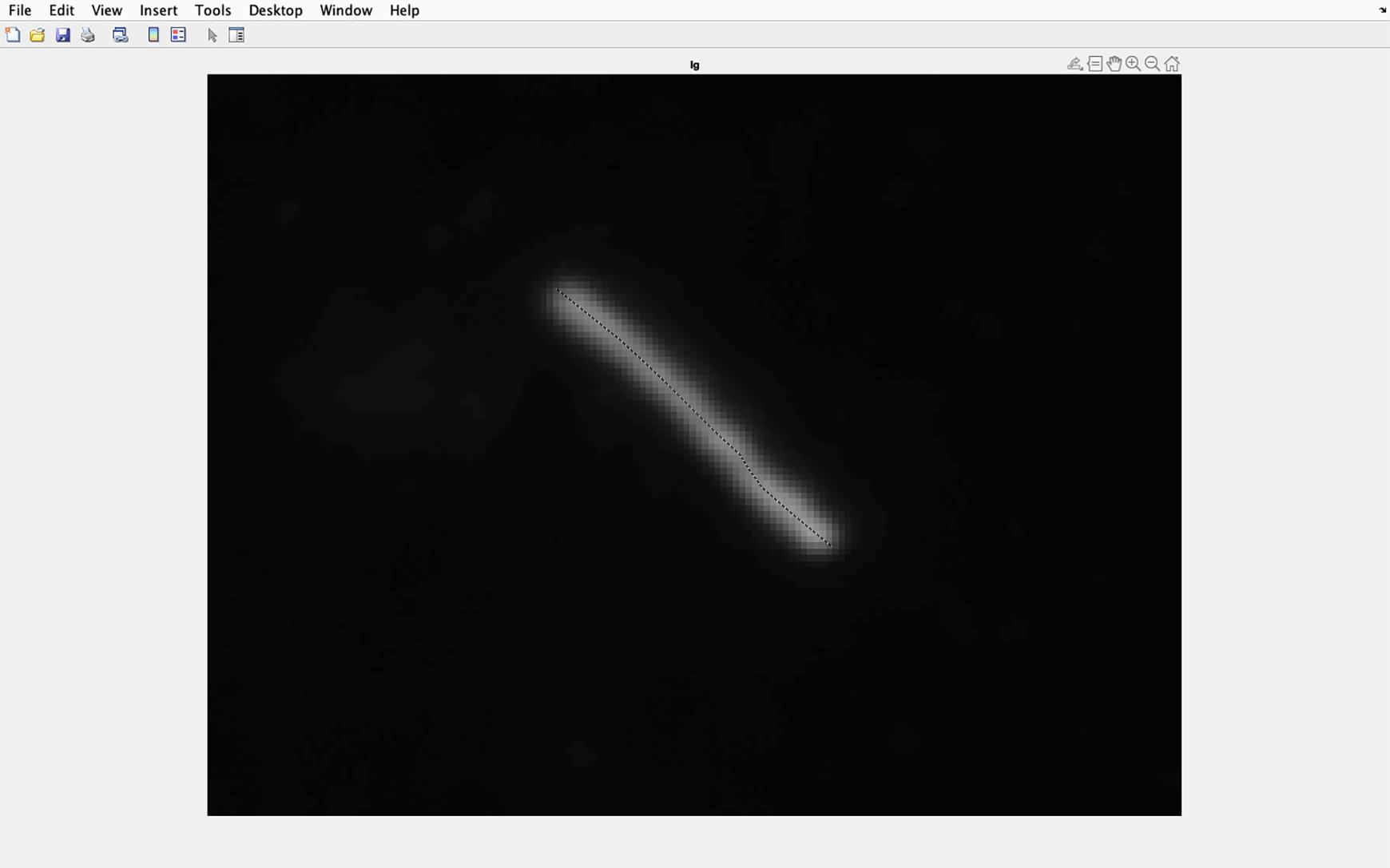
Figure 27. Identifying cilia to be analyzed. Draw a line along the center of the magnified cilia going from the base to the tip of the cilia. Double click when the line is at the cilia tip.
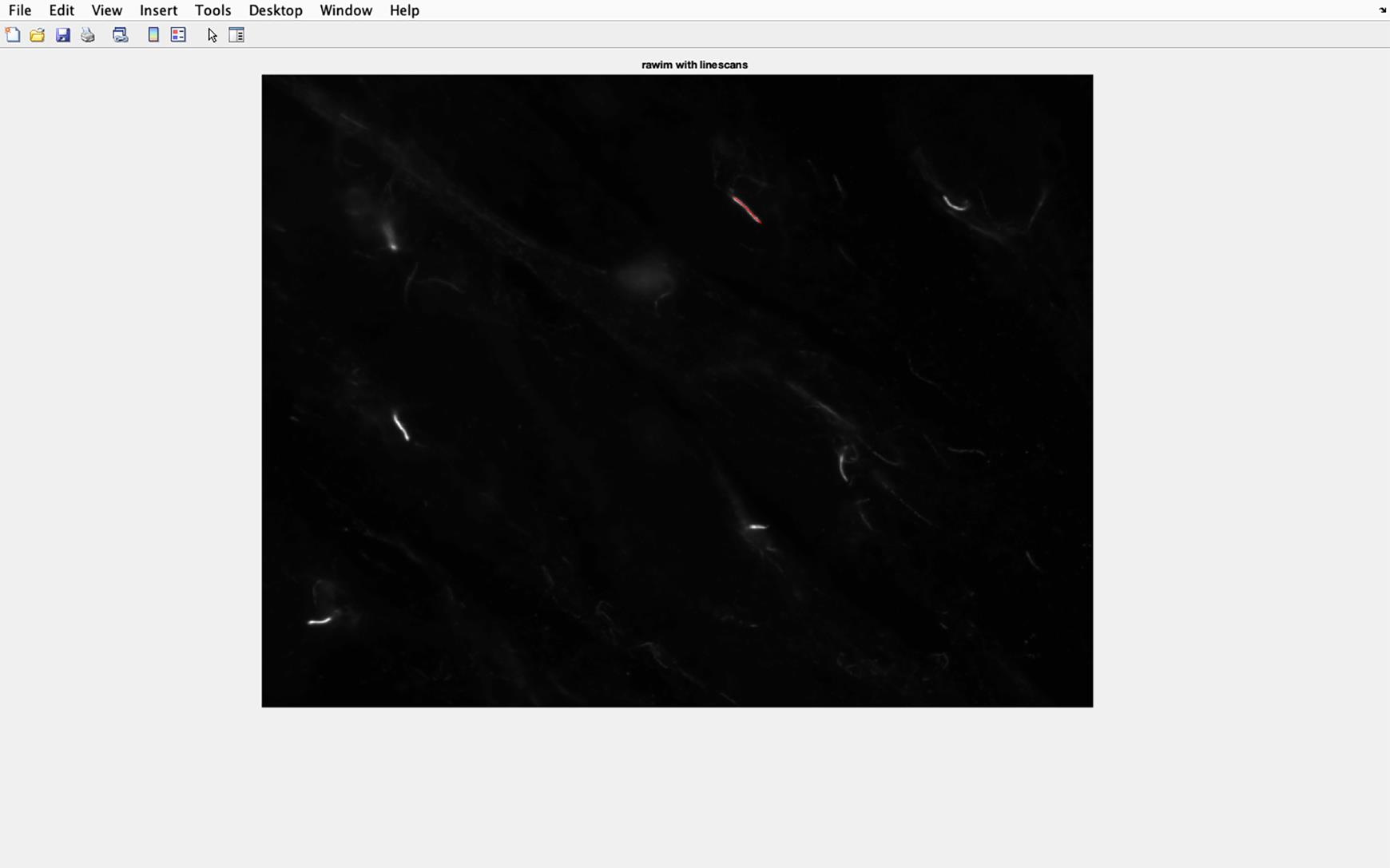
Figure 28. Cilia line overlaid on to acetylated tubulin image. The line drawn along the cilia will be transferred to the acetylated tubulin image, and the pixel intensity of this section in the image will be analyzed.
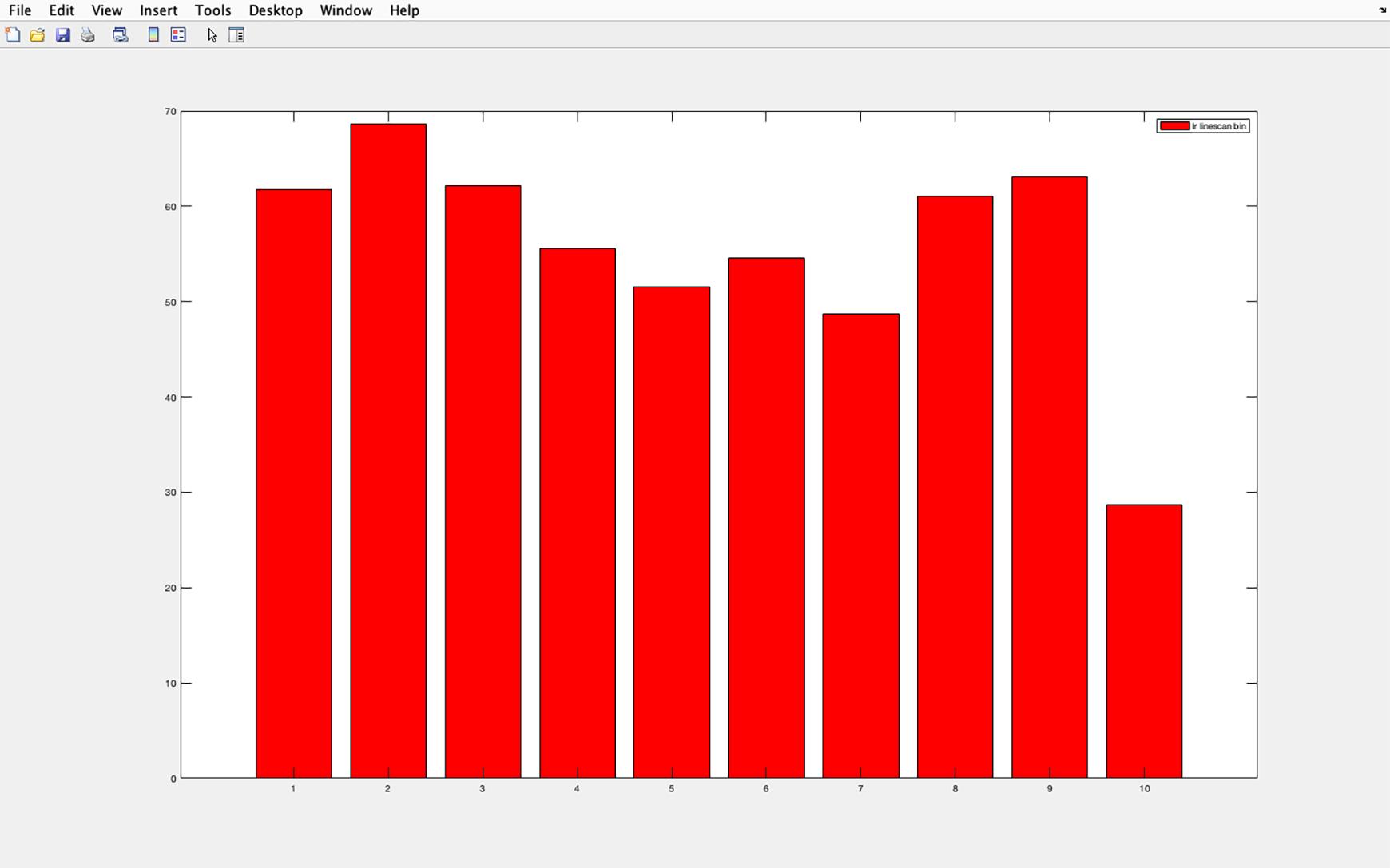
Figure 29. Acetylated tubulin pixel intensity histogram. Example of the histogram produced for average pixel intensity in each section of a cilia (from 1 proximal to 10 distal). The y-axis shows pixel intensity, and the x-axis shows the cilia divided into 10 equal sections. Section 1 represents the cilia base and section 10 the cilia tip.
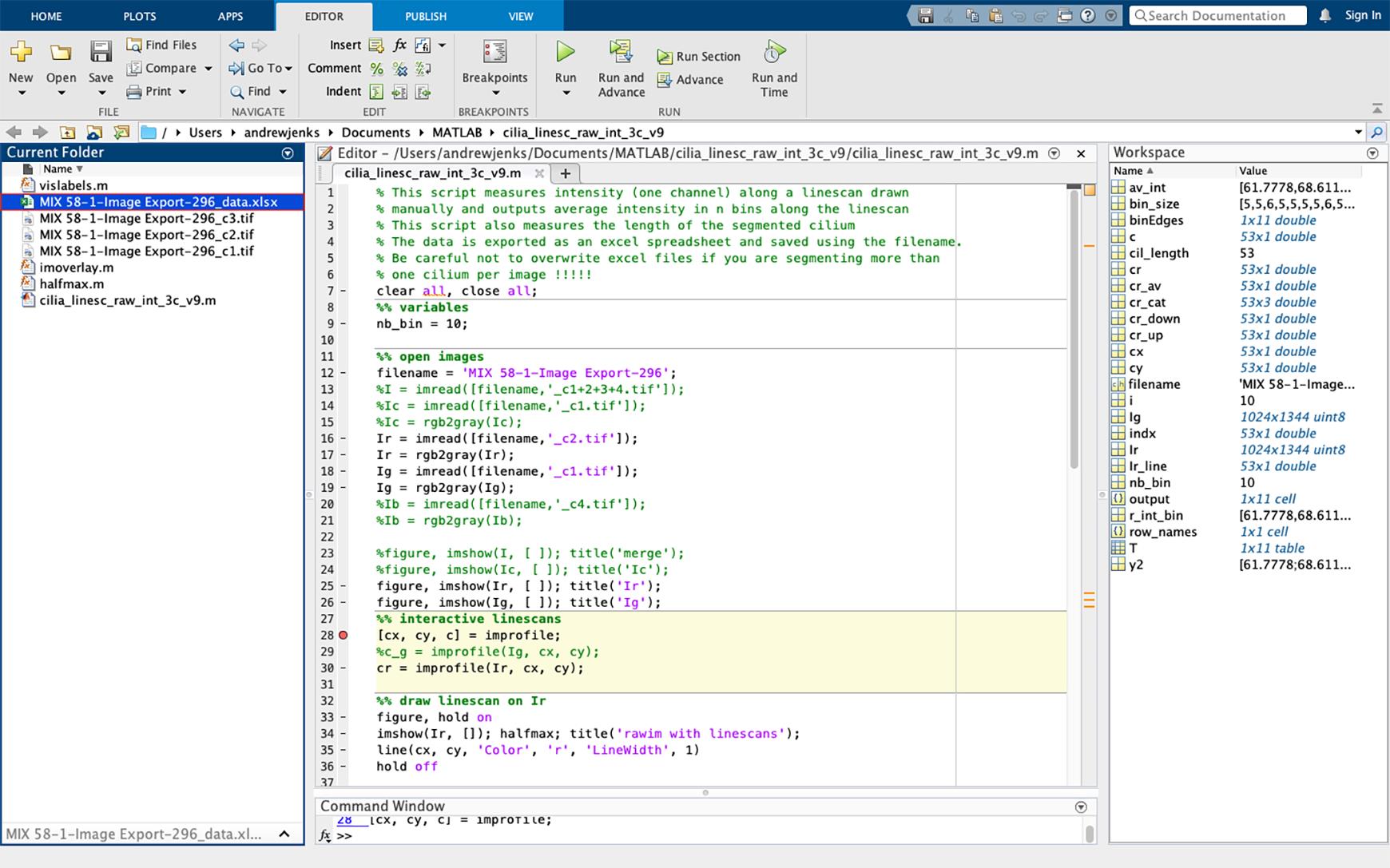
Figure 30. Example of average pixel intensity excel spreadsheet. The Excel spreadsheet of acetylated tubulin pixel intensity will appear in the current folder side bar (highlighted in blue). If multiple cilia are to be analyzed per image, change the name of the file to avoid overwriting.
CILIA 5 – Three channel pixel ratio (cilia_linesc_int_3c_v9):
This script uses three (3) fluorescent channels. For the example shown, C1 is the cilia marker (Arl13B), C2 is the protein of interest (acetylated tubulin), and C3 is the nuclear stain (DAPI). Once cilia are manually defined, the script will divide cilia into 10 equal sections, and channel 2 pixel intensity of each section will be divided by the average channel 2 pixel intensity of the entire cilia. This ratio will be presented as 10 cilia sections running from the base to the cilia tip. In the example shown in Figures 31-37, this is used to quantify acetylated tubulin.
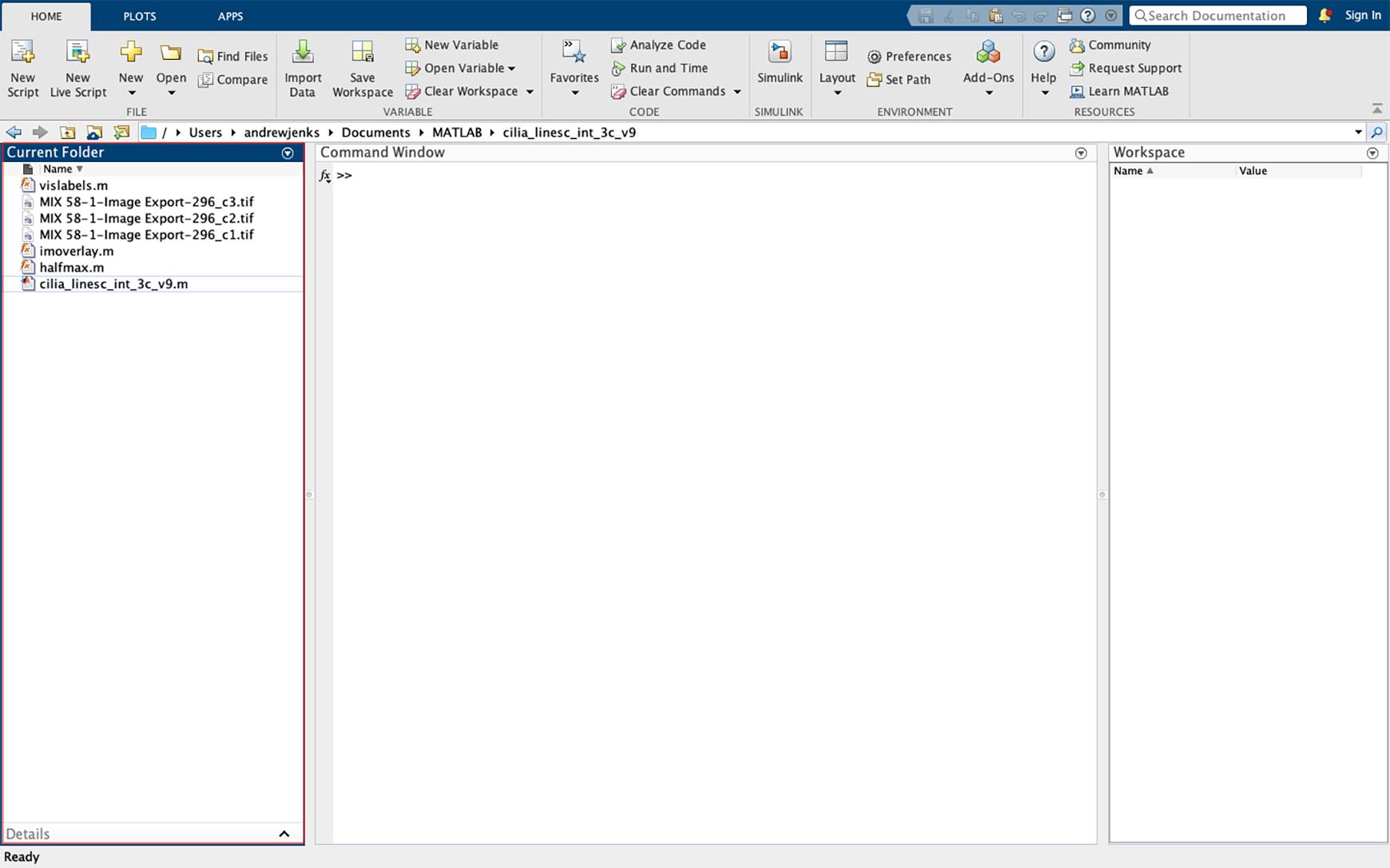
Figure 31. Importing data into MATLAB. Drag the script files and images to be analyzed into the MATLAB left sidebar window of the operating interface. Images must be imported as TIFF files from the individual fluorescent channels.
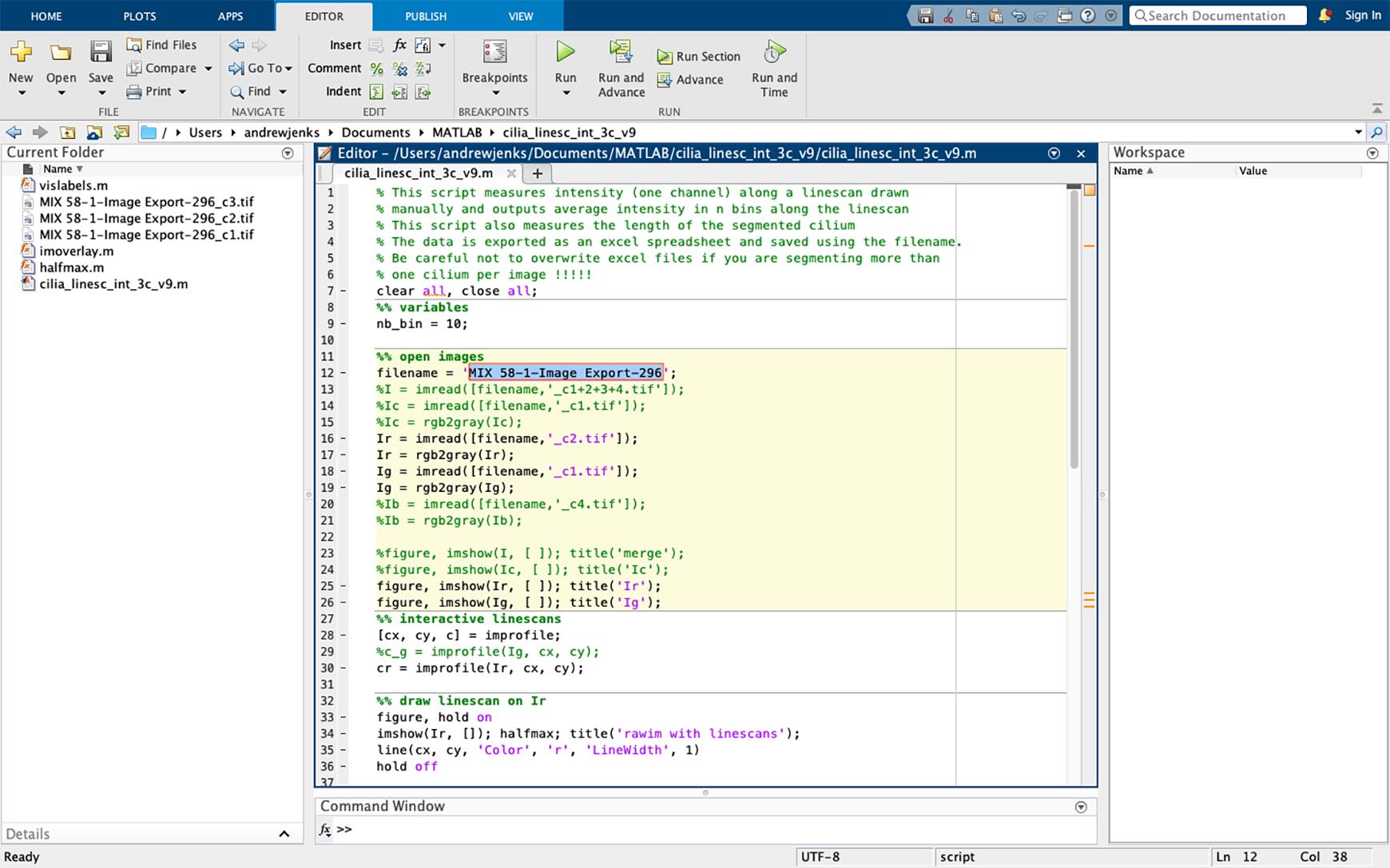
Figure 32. Changing script file name. Change the filename in the script (line 12, highlighted in blue) to the same name as the file to be analyzed. The TIFF extension should not be included in the file name.
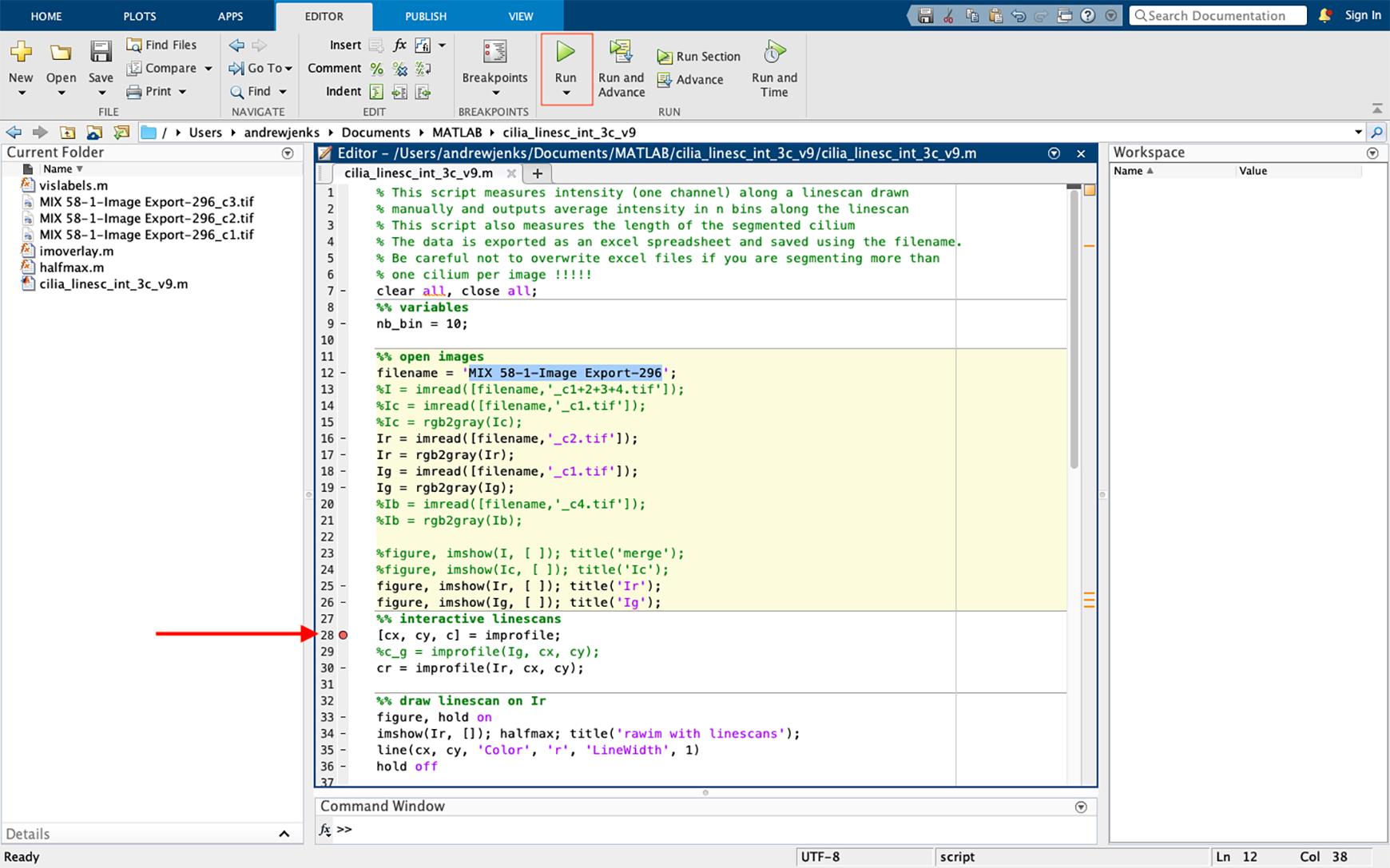
Figure 33. Selecting the file to be analyzed and inserting a breakpoint in the script. Insert a breakpoint in the script by clicking on the dash sign next to line 28, converting the dash to a red circle (highlighted using an arrow). Start the script by pressing “run” (green icon “highlighted” on the top).
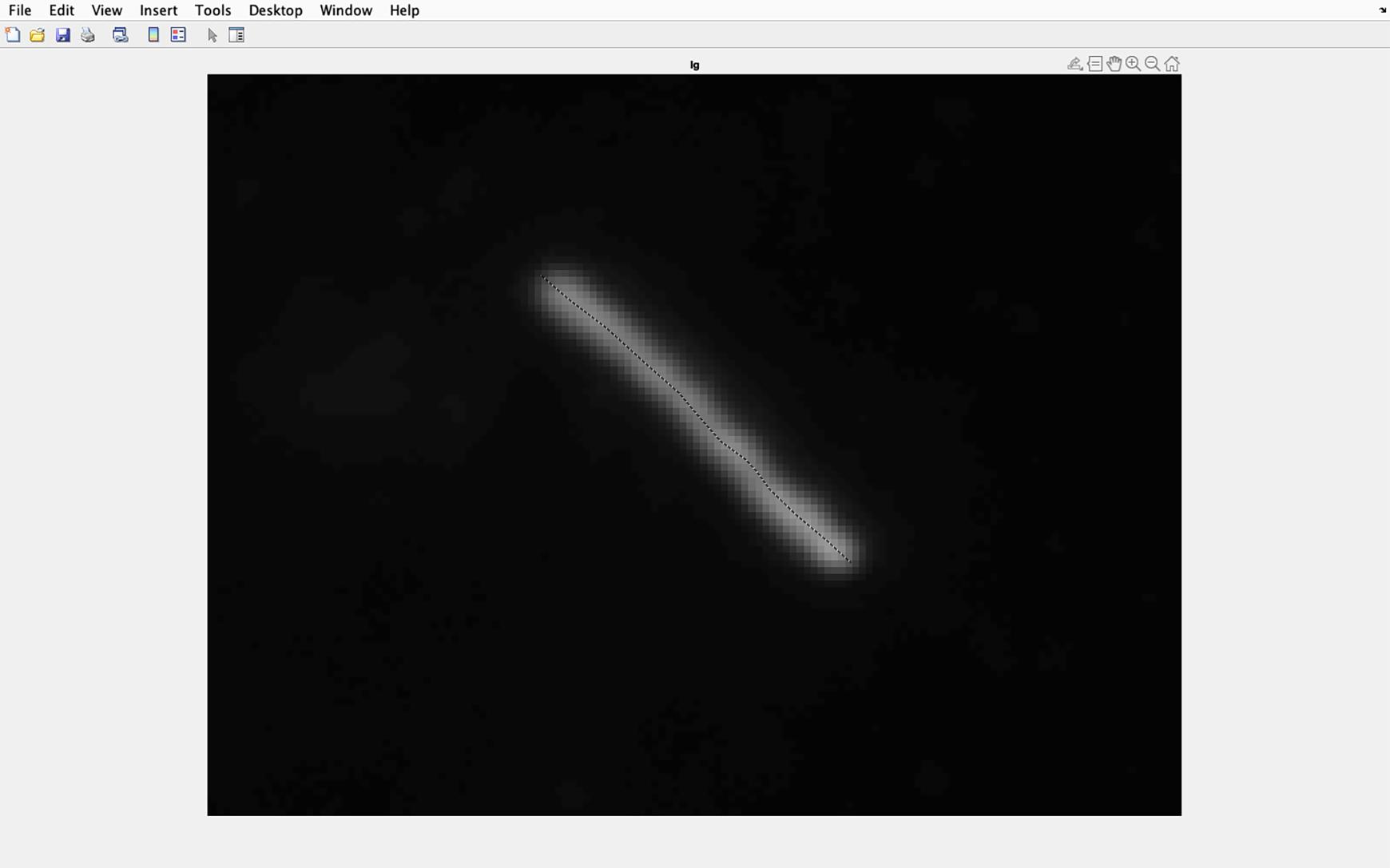
Figure 34. Identifying cilia to be analyzed. Draw a line along the center of the magnified cilia, ensuring the line goes from the base to the tip of the cilia. Double click when the line is at the cilia tip.
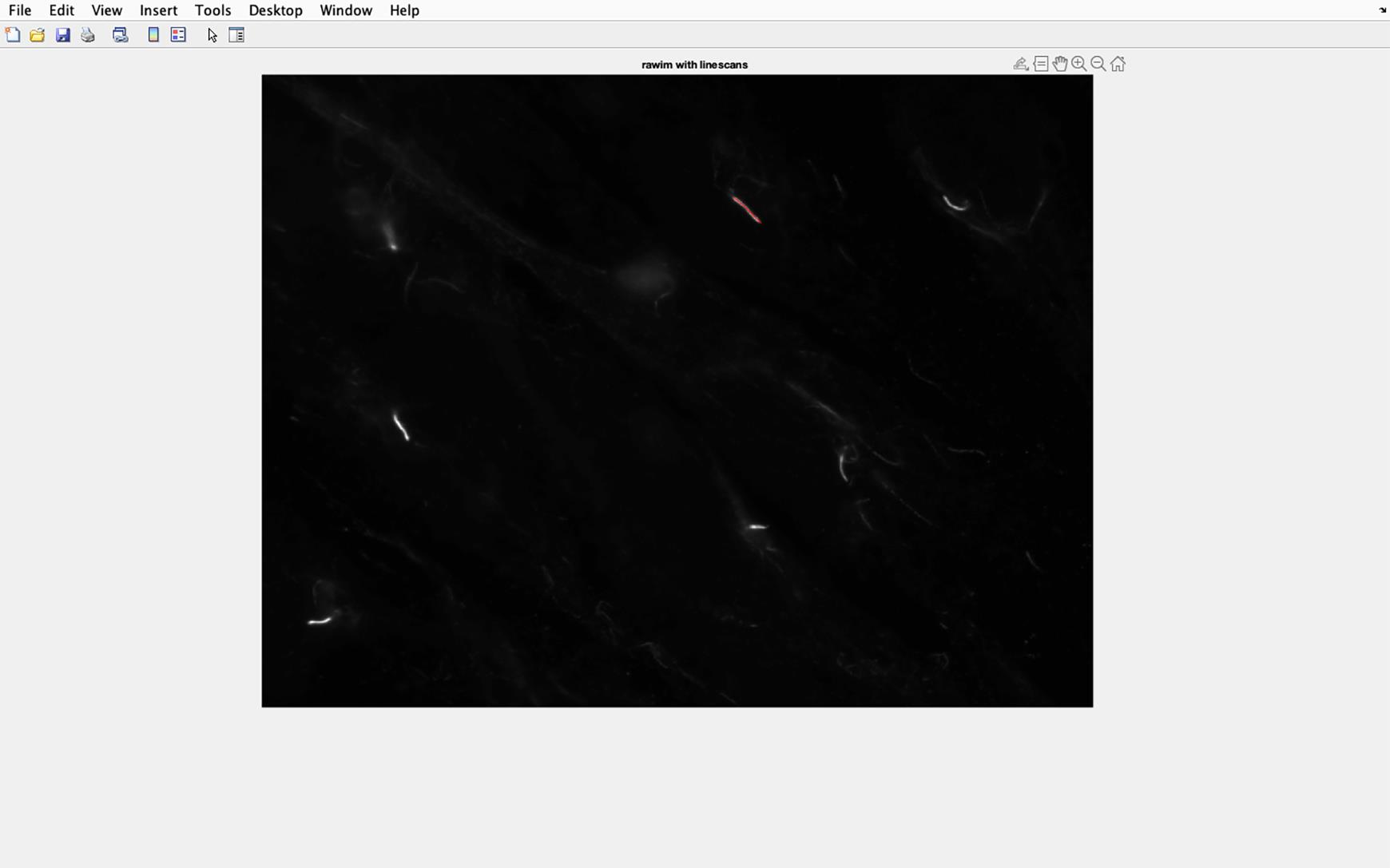
Figure 35. Cilia line overlaid on to acetylated tubulin image. The line drawn along the cilia will be transferred to the acetylated tubulin image, and the pixel intensity in this section of the image will be analyzed.
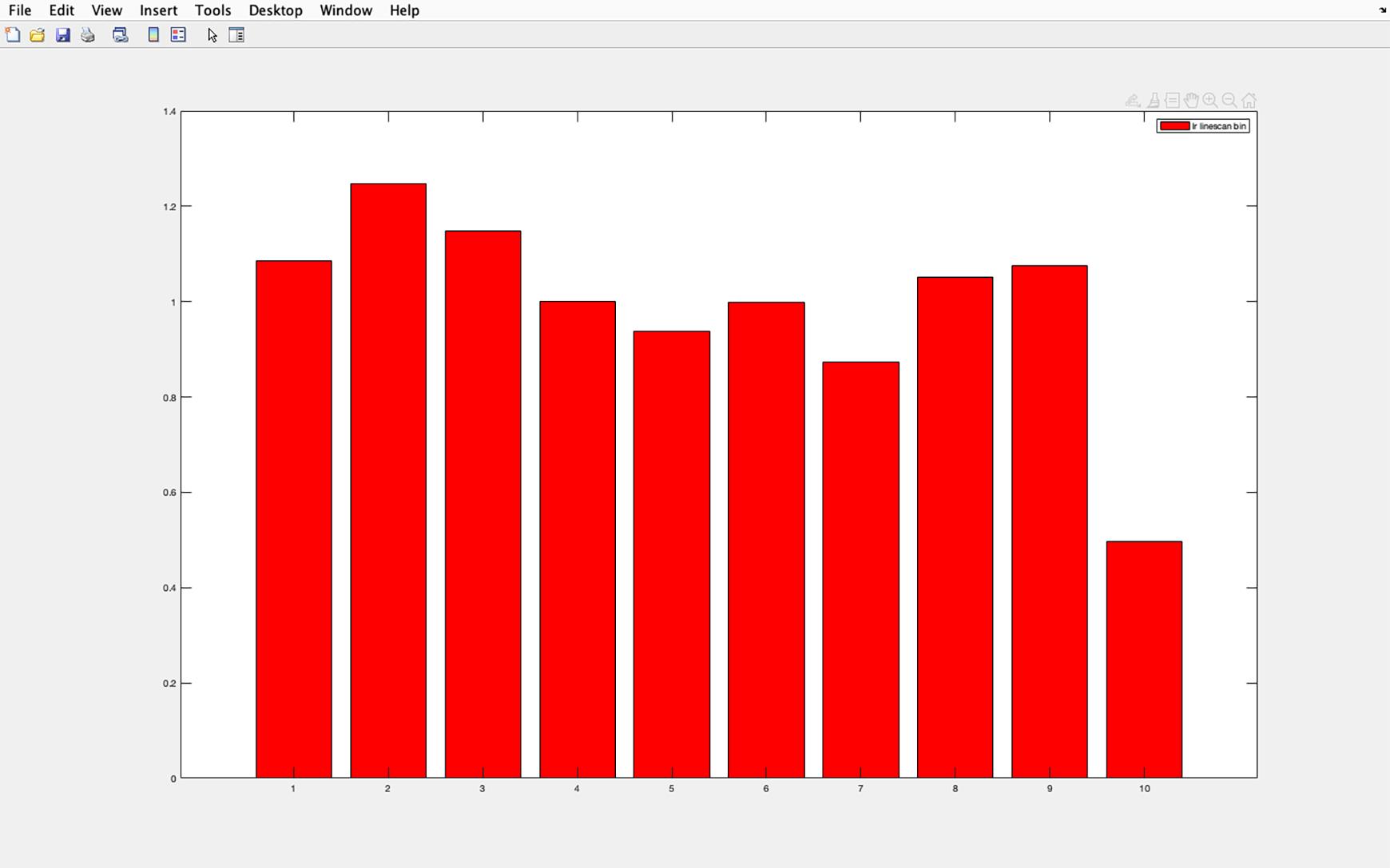
Figure 36. Acetylated tubulin pixel intensity histogram. Example of a histogram calculated with CILIA 5 script. The y-axis shows pixel intensity as a ratio of average pixel intensity of each section divided by the average pixel intensity of the entire cilia. Section 1 represents the cilia base and section 10 the cilia tip.

Figure 37. Example of pixel intensity excel spreadsheet. The Excel spreadsheet of acetylated tubulin pixel intensity will appear in the current folder side bar (highlighted in blue). If multiple cilia are to be analyzed per image, change the name of the file to avoid overwriting.
Recipes
- PTEM Buffer
20 mM PIPES, pH 6.8
0.2% Triton X-100
10 mM EGTA
1 mM MgCl2 - 1× Phosphate Buffered Saline (1× PBS)
Dissolve 10 g/L NaCl, 0.25 g/L KCl, 0.25 g/L KH2PO4 and 1.437 g/L Na2HPO4.
Adjust pH to 7.2 using HCl. - 1× PBST
1× PBS
0.1% v/v Triton-X-100 - 1× PBSBT
1× PBS
0.1% (v/v) Triton-X-100
3% (w/v) bovine serum albumin
0.05% (w/v) NaN3
Acknowledgments
This research was partly funded by the Institute of Cancer Research and by a Sarcoma UK grant (to B.E.T.). Special thanks to Adam Tyson (UCL) for critically reading this manuscript.
Competing interests
There are no conflicts of interest or competing interests.
References
- Anvarian, Z., Mykytyn, K., Mukhopadhyay, S., Pedersen, L. B. and Christensen, S. T. (2019). Cellular signalling by primary cilia in development, organ function and disease. Nat Rev Nephrol 15(4): 199-219.
- Christensen, S. T., Clement, C. A., Satir, P. and Pedersen, L. B. (2012). Primary cilia and coordination of receptor tyrosine kinase (RTK) signalling. J Pathol 226(2): 172-184.
- Goetz, S. C. and Anderson, K. V. (2010). The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet 11(5): 331-344.
- Hansen, J. N., Rassmann, S., Stuven, B., Jurisch-Yaksi, N. and Wachten, D. (2021). CiliaQ: a simple, open-source software for automated quantification of ciliary morphology and fluorescence in 2D, 3D, and 4D images. Eur Phys J E Soft Matter 44(2): 18.
- Jenks, A. D., Vyse, S., Wong, J. P., Kostaras, E., Keller, D., Burgoyne, T., Shoemark, A., Tsalikis, A., de la Roche, M., Michaelis, M., Cinatl, J., Jr., Huang, P. H. and Tanos, B. E. (2018). Primary Cilia Mediate Diverse Kinase Inhibitor Resistance Mechanisms in Cancer. Cell Rep 23(10): 3042-3055.
- Lauring, M. C., Zhu, T., Luo, W., Wu, W., Yu, F. and Toomre, D. (2019). New software for automated cilia detection in cells (ACDC). Cilia 8: 1.
- Schou, K. B., Pedersen, L. B. and Christensen, S. T. (2015). Ins and outs of GPCR signaling in primary cilia. EMBO Rep 16(9): 1099-1113.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Jenks, A. D., Fivaz, M. and Tanos, B. E. (2021). Quantitative Determination of Primary Cilia Protein Distribution Using Immunofluorescence Staining and MATLAB Analysis. Bio-protocol 11(23): e4248. DOI: 10.21769/BioProtoc.4248.
Category
Cell Biology > Cell imaging > Fluorescence
Biological Sciences > Biological techniques
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









