- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Suppression of Human Dendritic Cells by Regulatory T Cells
(*contributed equally to this work) Published: Vol 11, Iss 21, Nov 5, 2021 DOI: 10.21769/BioProtoc.4217 Views: 4620
Reviewed by: Scott McCombSaskia F. ErttmannAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation and Ex Vivo Testing of CD8+ T-Cell Division and Activation Using Mouse Splenocytes
Melissa Dolan [...] John M.L. Ebos
Aug 20, 2025 3859 Views

Detection of Autophagy in Human Peripheral Blood Mononuclear Cells Using Guava® Autophagy and Flow Cytometry
Melanie Scherer [...] Jörg Bergemann
Sep 20, 2025 1413 Views

Protocol for the Isolation and Analysis of Extracellular Vesicles From Peripheral Blood: Red Cell, Endothelial, and Platelet-Derived Extracellular Vesicles
Bhawani Yasassri Alvitigala [...] Lallindra Viranjan Gooneratne
Nov 5, 2025 1442 Views
Abstract
Regulatory T cells (Tregs) suppress immune responses via a variety of mechanisms and can be used as a cellular therapy to induce tolerance. The function of Tregs is commonly assessed in vitro using assays that measure suppression of effector T cell proliferation and/or cytokine production. However, Tregs can also suppress the function of antigen presenting cells, creating a need for methodology to routinely measure this aspect of their function. This protocol describes a method to measure human Treg-mediated suppression of CD80 and CD86 expression on mature, monocyte-derived dendritic cells. Representative data show suppression mediated by polyclonal Tregs as well as antigen-specific Tregs generated using chimeric antigen receptor (CAR) technology. This method can be used in parallel to T cell suppression assays to measure the functional activity of human Tregs.
Keywords: Regulatory T cellsBackground
Regulatory T cells (Tregs) are immunosuppressive cells that play a fundamental role in maintaining peripheral tolerance. Tregs inhibit the action of many immune cells, including effector T cells and antigen presenting cells (APC), via cell contact-dependent and contact-independent mechanisms. The suppressive function of Tregs is typically assessed in vitro by measuring their ability to inhibit the proliferation of polyclonally stimulated T cells. However, methods to measure how Tregs suppress APCs are limited.
One strategy Tregs use to inhibit the function of APCs is the removal of co-stimulatory molecules from the APC, thereby reducing their ability to stimulate effector T cells. Tregs achieve this by expressing CTLA-4, which binds CD80 and CD86 with a high affinity and allows the Treg to physically remove these molecules from the APC cell surface (Walker and Sansom, 2011). We have also previously reported the ability of human Tregs to suppress the expression of co-stimulatory molecules on both immature and mature monocyte-derived DCs (moDCs) (Wang et al., 2011).
This protocol describes a method to test Treg-mediated suppression of CD80 and CD86 and is modified from a previously published mouse-based protocol (Onishi et al., 2008). Our protocol focuses on the ability of human Tregs to suppress the expression of CD80 and CD86 by moDCs. In this assay, polyclonal Tregs transduced with a truncated nerve growth factor receptor (ΔNGFR) reporter can reduce CD80 and CD86 expression in moDCs. Furthermore, when using HLA-A2+ target moDCs, antigen-specific Tregs expressing an HLA-A2-specific chimeric antigen receptor (CAR) are more potent than polyclonal Tregs (Dawson et al., 2020; Fung et al., 2021).
Materials and Reagents
Materials
5 ml polystyrene round-bottom tubes (Corning, catalog number: 352052)
TC-coated 6-well plates (Corning, catalog number: 353502)
TC-coated 12-well plates (Corning, catalog number: 353503)
96-well U- and V-bottom plates (Corning, catalog numbers: 353077, 3894)
1.5 ml microcentrifuge tubes (Fisher Scientific, catalog number: 229442)
15 ml and 50 ml conical tubes (Corning, catalog numbers: 352096, 352070)
Sterile 1 ml or 3 ml syringe (BD, catalog numbers: 309628, 309657)
Media and Buffers
LymphoprepTM (STEMCELL Technologies, catalog number: 07801)
Dulbecco’s Phosphate Buffered Saline (DPBS; Gibco, catalog number: 14190), 1×
X-VIVO 15 (Lonza, catalog number: BEBP02-054Q)
Human serum (Wisent Bio Products, catalog number: 022210)
Penicillin/streptomycin (Gibco, catalog number: 15140-122)
GlutaMAX (Gibco, catalog number: 35050-061)
Sodium pyruvate (Gibco, catalog number: 11360-070)
Fetal bovine serum (Gibco, catalog number: 12483020)
Ethylenediaminetetraacetic acid solution (EDTA) (Sigma-Aldrich, catalog number: 03690)
EasySep Buffer (see Recipes)
Dendritic Cell Medium (see Recipes)
Reagents
Stericup-GP Sterile Vacuum Filtration System (Millipore, catalog number: SCGPU05RE), 500 ml
EasySep Human CD14 Positive Selection Kit II (STEMCELL Technologies, catalog number: 17858)
Acridine Orange/Propidium Iodide (AO/PI; Nexcelom, catalog number: NEX-CS201065ML)
Cytokines
Recombinant human granulocyte-macrophage colony-stimulating factor (GM-CSF) (STEMCELL Technologies, catalog number: 78015), 20 μg/ml
Recombinant human interleukin (IL)-4 (STEMCELL Technologies, catalog number: 78045), 20 μg/ml
Recombinant human tumour necrosis factor (TNF)-α (eBioscience, catalog number: 14-8329-63), 20 μg/ml
Prostaglandin E2 (PGE2) (Tocris, catalog number: 2296),100 mM
Recombinant human IL-1β (STEMCELL Technologies, catalog number: 78041), 20 μg/ml
Recombinant human IL-6 (STEMCELL Technologies, catalog number: 78148), 20 μg/ml
Recombinant human interferon (IFN)-γ (eBioscience, catalog number: 14-8319-80), 20 μg/ml
Recombinant human IL-2 (Proleukin) (Novartis, DIN# 02130181)
Antibodies
Fc Receptor Binding Inhibitor Polyclonal Antibody (eBioscience, catalog number: 14-9161-73)
Fixable Viability Dye (FVD) (eBioscience, catalog number: 65-0865-18)
Anti-human CD3 (UCHT1) APC (BD Biosciences, catalog number: 564465)
Anti-human CD3 (UCHT1) BB515 (BD Biosciences, catalog number: 564465)
Anti-human CD4 (OKT4) APC (eBioscience, catalog number: 17-0048-42)
Anti-human CD4 (RPA-T4) BV711 (BioLegend, catalog number: 300558)
Anti-human CD8a (RPA-T8) BV711 (BioLegend, catalog number: 301044)
Anti-human CD11c (B-ly6) PE (BD Biosciences, catalog number: 555392)
Anti-human CD14 (M5E2) BV421 (BioLegend, catalog number: 301830)
Anti-human CD14 (M5E2) BV786 (BD Biosciences, catalog number: 563698)
Anti-human CD40 (5C3) PE-Cy7 (BD Biosciences, catalog number: 561215)
Anti-human CD56 (CMSSB) PE (eBioscience, catalog number: 12-0567-42)
Anti-human CD69 (FN50) BV785 (BioLegend, catalog number: 310932)
Anti-human CD70 (113-16) PerCP-Cy5.5 (BioLegend, catalog number: 355107)
Anti-human CD80 (L307.4) FITC (BD Biosciences, catalog number: 557226)
Anti-human CD83 (HB15e) BV421 (BioLegend, catalog number: 305324)
Anti-human CD86 (2331 (FUN-1)) APC (BD Biosciences, catalog number: 555660)
Anti-human CD86 (HA5.2B7) PerCP-Cy5.5 (Beckman Coulter, catalog number: B30647)
Equipment
Type II Biosafety cabinet (NuAire, model: LabGard ES NU-540)
Centrifuge, microcentrifuge (Eppendorf, models: 5810R and 5452)
STEMCELL EasySepTM magnet (STEMCELL Technologies, catalog number: 18000)
Cell counter (Nexcelom, model: Cellometer Auto 2000)
37°C incubator with 5% (v/v) CO2 (Sanyo, model: MCO-18AIC)
Flow cytometer (BD LSRFortessa X-20; alternative instruments can be used)
Software
FlowJo software (BD Biosciences, v10.7)
Procedure
Overview
Day 0: Prepare peripheral blood mononuclear cells (PBMCs).
Day 0: Isolate CD14+ monocytes from PBMCs by positive selection.
Day 0: Differentiate monocytes into dendritic cells by culturing in the presence of GM-CSF and IL-4.
Day 3: Replenish GM-CSF and IL-4.
Day 5: Mature dendritic cells by adding TNF-α, PGE2, IL-1β, and IL-6 to the culture.
Day 6: Mature dendritic cells by adding IFN-γ to the culture.
Day 7: Confirm maturation of the DCs by flow cytometry and set up the suppression assay.
Day11: Collect cells, stain, perform flow cytometry, and analyse.
Day 0: Prepare PBMCs
Prepare PBMCs: Either freshly isolated from blood or thawed PBMCs. PBMCs can be isolated from human peripheral blood by density gradient centrifugation using LymphoprepTM, according to the manufacturer’s protocol.
Note: Using batch-frozen PBMCs from one donor to differentiate moDCs will reduce donor-to-donor variability. Expect 5-10% fresh/frozen PBMCs or 50-100% CD14+ cells to become moDCs. See Notes for further details.
Optional: Set aside ~5 × 103-10 × 103 PBMCs for purity check (see Table 1, Figure 1, and Notes for further information).
Table 1. Day 0 Purity Check Panel. Stain PBMCs in parallel, if desired. See Figure 1 for representative data.
Marker Dilution Clone Fluorophore Fc Receptor Binding Inhibitor (preincubate for 10 min and do not wash) 1:5 Polyclonal N/A FVD 1:1,000 N/A eF780 CD3 1:100 UCHT1 BB515 CD4 1:100 OKT4 APC CD8a 1:100 RPA-T8 BV711 CD14 1:200 M5E2 BV421 CD56 1:25 CMSSB PE 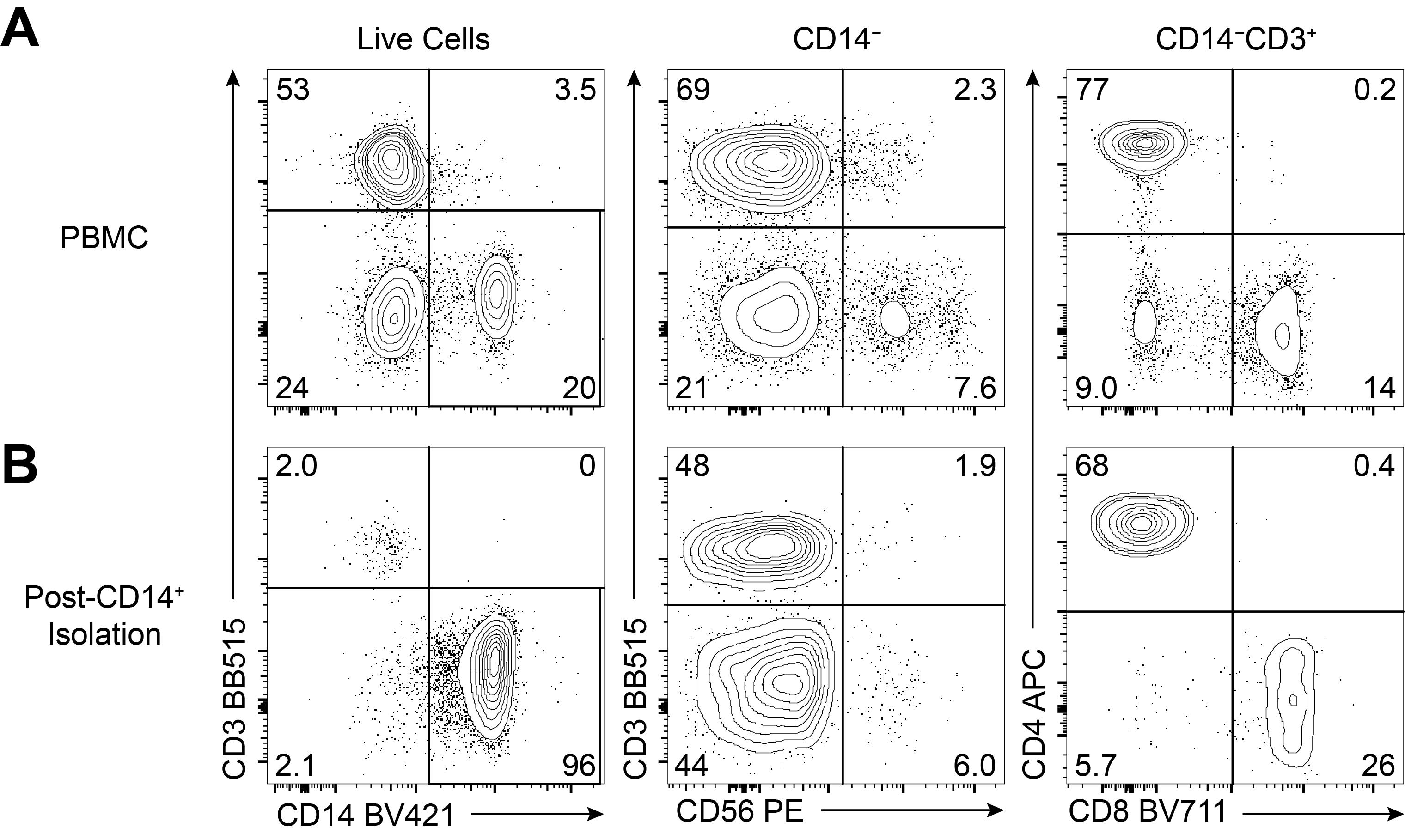
Figure 1. Day 0 CD14+ Purity Check. (A) Approximately 5 × 103-10 × 103 total PBMCs and (B) CD14-enriched cells were stained and analysed by flow cytometry to evaluate the purity of the CD14-enriched cells and the extent of CD3+ cell contamination. Contaminating T cells can affect downstream results if OKT3 (anti-CD3) is included during assays (see Notes). Cells were stained as described in Table 1 and in accordance with “Guidelines for the use of flow cytometry and cell sorting in immunological studies” (Cossarizza et al., 2019); they were acquired with a BD LSRFortessa X-20 and data analysed using FlowJo. The proportion of total live cells that were CD14+CD3‒ (CD14+ cell purity) is shown on the left. From the live CD14‒ cells, CD56 expression was analysed (middle) to identify NK/NKT cells (CD56med/hi) and a subset of monocytes (CD56lo). The contaminating live CD14‒CD3+ T cells were further analysed for their expression of CD4 and CD8 (right). See Notes for more information.
Day 0: Isolate CD14+ monocytes using the STEMCELL EasySep Human CD14 Positive Selection Kit II, as per the manufacturer’s protocol and described below, unless stated otherwise:
Resuspend PBMCs at 100 × 106/ml in EasySep Buffer (0.1-2 ml). The total starting number of PBMCs should be between 10 × 106 and 200 × 106 cells. For x number of CD14+ monocytes, start with 10× (fresh) or ~20× (frozen) PBMCs.
Transfer PBMCs to a 5 ml polystyrene round-bottom tube.
Add 100 μl of Selection Cocktail per millilitre of cells and incubate 10 min at room temperature (RT).
Vortex RapidSpheres for 30 s, add 100 μl of RapidSpheres per ml cells, mix tube by gentle rotation, and incubate for 3 min at RT.
Top up tube with EasySep Buffer to 2.5 ml, mix by pipetting, place in an EasySepTM magnet, and incubate for 3 min at RT.
Gently pour tube while in magnet to discard supernatant. The round-bottom tube contains CD14+ cells (keep).
Wash remaining RapidSpheres by removing the round-bottom tube from the magnet and topping up tube with 2.5 ml of EasySep Buffer. Mix by pipetting to wash layer off tube wall, replace the tube back in the magnet, and incubate for 3 min at RT. Pour to discard supernatant and pipette last drop from tube while inverted.
Repeat Step B7 so that the tube of cells is incubated in the magnet a total of three times.
Optionally repeat Step B7 two more times (total of five times on magnet) for potentially higher enrichment.
Note: This step is not part of the original STEMCELL Technologies protocol.
Transfer the cells from the round-bottom tube into a 15 ml conical tube by washing the walls of the round-bottom tube with ~3 ml of DPBS. Centrifuge at 450 × g for 5 min and discard supernatant.
Resuspend the enriched CD14+ cells in Dendritic Cell Medium (see Recipes).
Count cells (see protocols from Nexcelom Cellometer Auto 2000 for details; alternatively, a hemocytometer and trypan blue can be used).
Optional: set aside ~5 × 103-10 × 103 monocytes for purity check (see Table 1, Figure 1, and Notes for further information).
Day 0: Culture and differentiate enriched CD14+ cells into dendritic cells
Adjust cells to 2 × 106 cells per millilitre with Dendritic Cell Medium.
Plate 2 ml cells per well in a 6-well plate (4 × 106 cells per well).
Note: Plate multiple wells of cells as needed.
Add GM-CSF (final concentration: 50 ng/ml) and IL-4 (final concentration: 100 ng/ml).
Incubate cells at 37°C (5% v/v CO2) for 3 days.
Note: Cytokines can be stored at 4°C for ~1 week.
Day 3: Change Dendritic Cell Medium and replenish cytokines
Change Dendritic Cell Medium: without disturbing the cells at the bottom of the well, collect 1.5 ml of medium into sterile 1.5 ml microcentrifuge tube, centrifuge at 450 × g for 5 min, discard the supernatant, resuspend the pelleted cells in 1.5 ml of fresh Dendritic Cell Medium, and re-plate into original well.
Fully replenish the cytokines by adding fresh GM-CSF (final concentration: 50 ng/ml) and IL-4 (final concentration: 100 ng/ml), assuming that no cytokines from day 0 remain in the culture.
Incubate at 37°C (5% v/v CO2) for 2 days.
Note: Cytokines can be stored at 4°C for ~1 week.
Alternative medium change method: Centrifuge plate at 450 × g for 5 min, gently replace 1.5 ml of Dendritic Cell Medium, and fully replenish the GM-CSF and IL-4 as detailed above.
Day 5: Mature dendritic cells
Collect cells: pipette up and down to detach cells and collect either into a 15 ml or 50 ml conical tube. Detach remaining cells using a 1 ml or 3 ml syringe rubber in gentle, one-way movements. Rinse well twice with DPBS to collect as many cells as possible.
Wash cells by filling the conical tube with DPBS, centrifuge cells at 450 × g for 5 min, and discard the supernatant.
Resuspend cells in fresh Dendritic Cell Medium, count cells, and adjust volume so that cells are at a final concentration of 1 × 106 cells/ml in Dendritic Cell Medium.
Add GM-CSF (final concentration: 50 ng/ml) and IL-4 (final concentration: 100 ng/ml).
Plate cells in a new well by adding 2 ml of cells per well in a new 6-well plate (2 × 106 cells per well) or 1 ml of cells per well in a new 12-well plate (1 × 106 cells per well).
Mature moDCs by adding TNF-α (final concentration: 50 ng/ml), PGE2 (final concentration: 1 μg/ml, ~2.837 µM), IL-1β (final concentration: 10 ng/ml), and IL-6 (final concentration: 100 ng/ml).
Note: If immature moDCs are desired, skip this step. Keeping a minimum of 1 × 106 immature moDCs (GM-CSF and IL-4 with no additional cytokines) is useful for evaluating the moDC maturation at the end (day 7, see Procedure G).
Incubate at 37°C (5% v/v CO2) for 1 day.
Day 6: Continue to mature dendritic cells
Add IFN-γ (final concentration: 50 ng/ml).
Note: Skip this step for immature moDCs.
Incubate at 37°C (5% v/v CO2) for 1 day.
Day 7: Confirm maturation of the DCs before setting up the suppression assay
Collect cells by pipetting to resuspend and transfer to a 15 ml conical tube. Detach remaining cells using a 1 ml or 3 ml syringe rubber in gentle, one-way movements. Wash wells twice with DPBS and add this to the conical tube. Centrifuge cells at 450 × g for 5 min, resuspend in fresh Dendritic Cell Medium, count cells, and adjust cell concentration to 5 × 105 cells/ml in Dendritic Cell Medium.
Confirm moDC maturation by collecting ~5 × 106-10 × 106 mature moDCs and immature moDCs for staining for flow cytometry (see Table 2, Figure 2).
Table 2. Day 7 moDC Maturation Check Panel. Immature moDC must be stained in parallel. See Figure 2 for representative data.
Marker Dilution Clone Fluorophore Fc Receptor Binding Inhibitor (preincubate for 10 min and do not wash) 1:5 Polyclonal N/A FVD 1:1,000 N/A eF780 CD3 1:50 UCHT1 APC CD11c 1:50 B-ly6 PE CD14 1:100 M5E2 BV785 CD80 1:50 L307.4 FITC CD83 1:50 HB15e BV421 CD86 1:50 HA5.2B7 PerCp-Cy5.5 CD40 1:100 5c3 PECy-7 HLA-DR 1:50 L243 BV510 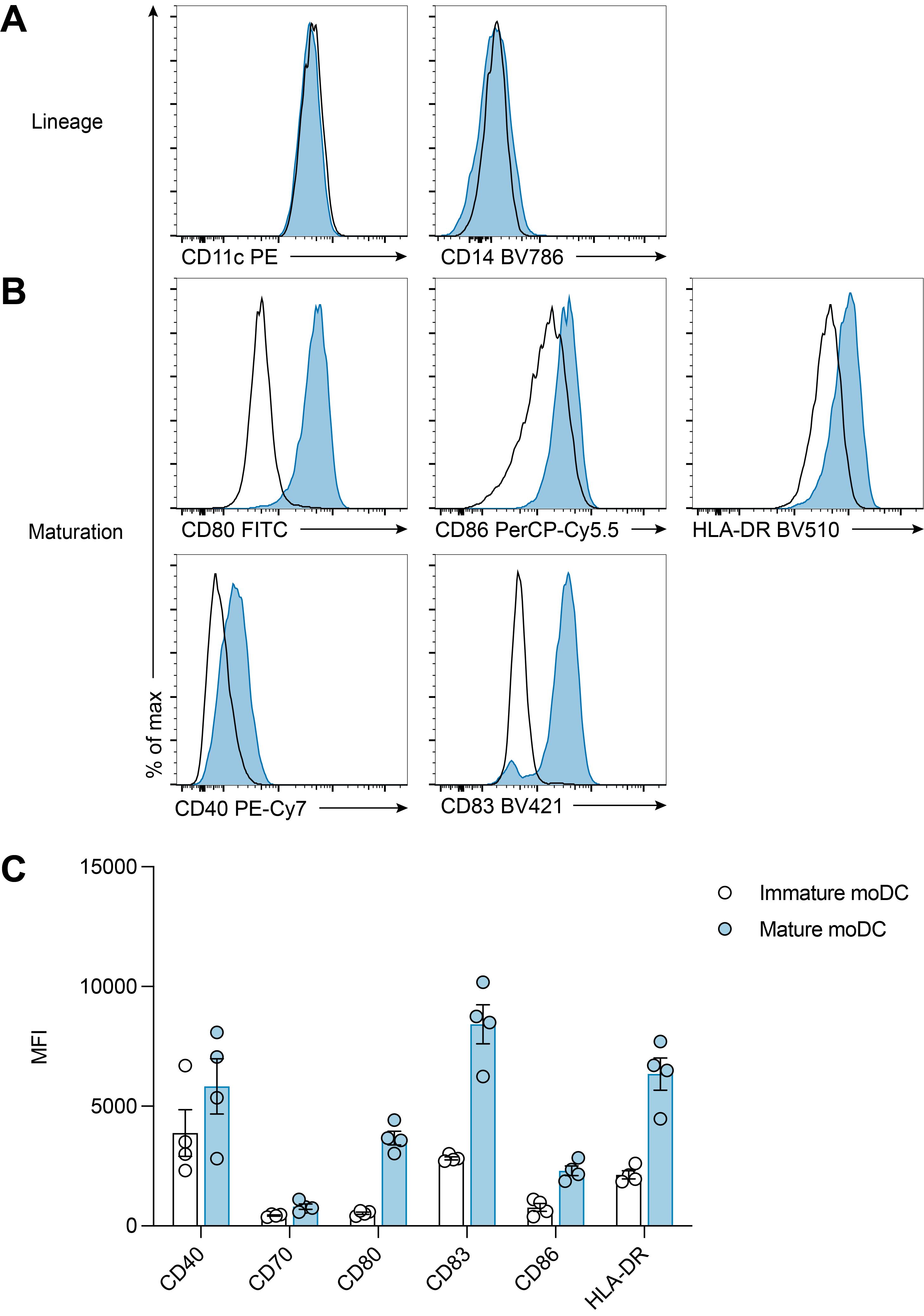
Figure 2. Day 7 phenotype of immature and mature moDCs. (A-C) Immature (black line) and mature (blue) moDCs were analysed by flow cytometry for expression of CD11c, CD14, CD80, CD86, CD40, CD83, and HLA-DR. Cells were stained as described in Table 2 and in accordance with “Guidelines for the use of flow cytometry and cell sorting in immunological studies” (Cossarizza et al., 2019); they were acquired with a BD LSRFortessa X-20 and data analysed using FlowJo. moDCs (gated as live, single cells) expressed similar levels of lineage markers (A) but upregulated expression of CD80, CD86, HLA-DR, CD40, and CD83 following maturation (B-C). MFI, geometric mean fluorescence intensity.
Day 7: Set up DC suppression assay
Plate 100 µl moDC in a 96-well U-bottom plate (50,000 cells per well).
Collect rested Tregs, count cells, and resuspend at 2.5 × 106 cells per ml in Dendritic Cell Medium with 100 IU/ml IL-2.
Note: Tregs should be expanded and rested using protocols established by the user’s lab. Details of how Tregs were expanded and rested for this protocol are provided in Dawson et al. (2020). Briefly, Tregs were isolated from peripheral blood and polyclonally expanded with artificial APCs in the presence of 1000 IU/ml IL-2 for 7 days. Tregs were then rested by culturing in fresh medium with 100 IU/ml IL-2 overnight. CAR Tregs were generated by lentivirally transducing polyclonal Tregs one day post-stimulation.
Set up moDC and Treg co-culture in a 1-to-5 ratio (50,000 DCs to 250,000 Tregs per well): add 100 μl Treg cell suspension into the wells containing moDCs from Step H1.
Control wells: set up one well of moDC alone and another well of Treg alone as controls in separate wells with a final volume of 200 µl per well.
Co-culture cells in Dendritic Cell Medium with 50 IU/ml IL-2 at 37°C (5% v/v CO2) for 4 days.
Day 11: Flow cytometry
Collect cells: centrifuge the cell culture plate at 970 × g for 3 min, remove from centrifuge, and discard half the volume (100 μl) supernatant by pipetting. Resuspend the cells in the remaining supernatant and transfer into a new 96-well V-bottom plate. Wash the wells in the cell culture plate with 100 μl DPBS and add this to the respective wells in the V-bottom plate.
Centrifuge the plate at 970 × g for 3 min, discard the supernatant, and resuspend the cells in 200 µl DPBS to wash.
Centrifuge the plate at 970 × g for 3 min and discard the supernatant.
Stain cells for analysis by flow cytometry using Table 3. See Figure 3 for example data.
Table 3. Day 11 moDC Suppression Assay Panel. moDC-alone and Treg-alone controls must be stained in parallel. See Figure 3 for representative data.
Marker Dilution Clone Fluorophore Fc Receptor Binding Inhibitor
(preincubate for 10 min and do not wash)1:5 Polyclonal N/A FVD 1:1,000 N/A eF780 CD4 1:100 RPA-T4 BV711 CD69 1:50 FN50 BV785 CD11c 1:50 B-ly6 PE CD70 1:50 113-16 PerCP-Cy5.5 CD80 1:50 L307.4 FITC CD83 1:50 HB15e BV421 CD86 1:50 FUN-1 APC CD40 1:100 5c3 PE-Cy7 HLA-DR 1:50 L243 BV510 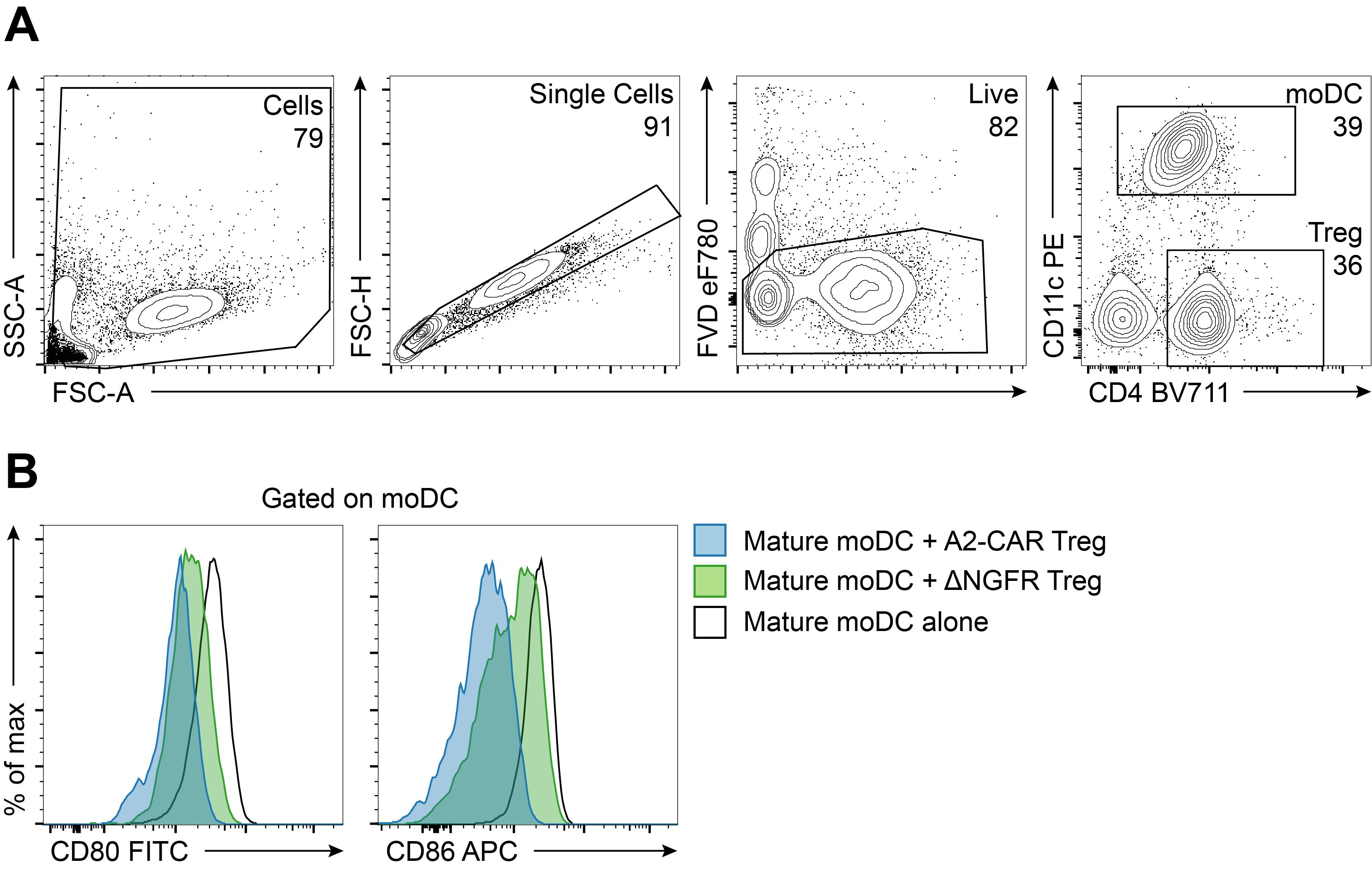
Figure 3. Day 11 analysis of CD80 and CD86 on mature moDCs following co-culture with Tregs. (A-B) Mature moDCs and Tregs were co-cultured for 4 days, and then cells were stained as described in Table 3 and in accordance with “Guidelines for the use of flow cytometry and cell sorting in immunological studies” (Cossarizza et al., 2019); they were acquired with a BD LSRFortessa X-20 and data analysed using FlowJo. (A) Gating strategy to identify mature moDCs. Live singlet cells were gated, and moDCs were identified as CD11c+CD4‒ cells. (B) Expression of CD80 (left) and CD86 (right) following co-culture with Tregs. This assay was performed using HLA-A2+ moDCs co-cultured with either polyclonal ΔNGFR-transduced Tregs or HLA-A2-specific CAR-transduced Tregs. Compared to moDCs cultured alone (black line), polyclonal Tregs exert a moderate level of suppression (green), as determined by the decrease in CD80 and CD86 expression. This suppression is greater when co-cultures are performed with A2-CAR Tregs (blue). See Dawson et al. (2020) for more examples.
Notes
PBMC: Fresh and frozen PBMCs are similar and suitable for moDC differentiation. Frozen PBMCs yield slightly fewer CD14+ monocytes than fresh PBMCs. Using frozen PBMCs from a single donor can reduce donor-to-donor moDC variability. Fresh PBMCs can be prepared the day of (proceed to CD14+ isolation immediately) or the day before running the experiment [store overnight at 4°C in 25 ml 10% (v) fetal bovine serum-supplemented medium of choice (e.g., RPMI 1640) mixed with 25 ml DPBS in a 50 ml tube placed horizontally in a fridge].
CD14+ selection performance: Post-selection (3× magnet-isolation, as per manufacturer’s protocol), 90-95% are CD14+ cells and 0.5-5% are CD3+ T cells (see Figure 1). Detection of contaminating T cells can be obscured if FSC/SSC voltages and thresholds are set too low. T cell contamination persists to day 7 and can affect results from moDC-T cell co-cultures (e.g., if OKT3 is added). Thus, an anti-CD3 antibody can be added to the day 7 moDC maturation panel to evaluate purity since the cells have expanded (instead of day 0).
moDC lineage markers: moDCs should be CD11c+ and CD14–. moDC maturation markers: mature moDCs should be CD80hi, CD86hi, HLA-DRhi (Ag presentation and T cell co-stimulation), CD40+ (promote CD40L+ T cell maturation and cytokine secretion), and CD83+ (function less clear).
Co-culture: Confirmation of moDC maturation is important before setting up suppression co-culture.
Recipes
EasySep Buffer
DPBS supplemented with 2% (v/v) fetal bovine serum, 1 mM EDTA. Contents were sterile-filtered with a Stericup-GP vacuum filtration flask.
Dendritic Cell Medium
X-VIVO 15 supplemented with 5% (v/v) human serum, 1% (v/v) penicillin-streptomycin, 1% (v/v) GlutaMAX, 1% (v/v) sodium pyruvate. Contents were sterile-filtered with a Stericup-GP vacuum filtration flask.
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research (CIHR) FDN-154304 and TxCell. AJL and NAJD were supported by a CIHR doctoral award and MKL received a salary award from the BC Children's Hospital Research Institute. This method is derived from the original publication by Dawson et al. (2020) (DOI: 10.1126/scitranslmed.aaz3866).
Competing interests
The authors of this manuscript have received research funding from Sangamo Therapeutics (formerly TxCell SA) to partially support this work. MKL has also received research funding from Takeda, Bristol Myers Squibb, Pfizer, and CRISPR Therapeutics for work not related to this study.
Ethics
For all studies, healthy volunteers gave written informed consent according to protocols approved by the University of British Columbia Clinical Research Ethics Board and Canadian Blood Services.
References
- Cossarizza, A, Chang, HD, Radbruch, A, Acs, A, Adam, D, Adam-Klages, S, Agace, WW, Aghaeepour, N, Akdis, M and Allez, M. (2019). Guidelines for the use of flow cytometry and cell sorting in immunological studies (second edition). Eur J Immunol 49(10): 1457-1973.
- Dawson, N. A. J., Rosado-Sanchez, I., Novakovsky, G. E., Fung, V. C. W., Huang, Q., McIver, E., Sun, G., Gillies, J. and Speck, M. (2020). Functional effects of chimeric antigen receptor co-receptor signaling domains in human regulatory T cells. Sci Transl Med 12(557): eaaz3866.
- Fung, V. C. W., Rosado-Sanchez, I. and Levings, M. K. (2021). Transduction of Human T Cell Subsets with Lentivirus. Methods Mol Biol 2285227-254.
- Onishi, Y., Fehervari, Z., Yamaguchi, T. and Sakaguchi, S. (2008). Foxp3+ natural regulatory T cells preferentially form aggregates on dendritic cells in vitro and actively inhibit their maturation. Proc Natl Acad Sci U S A 105(29): 10113-10118.
- Walker, L. S. and Sansom, D. M. (2011). The emerging role of CTLA4 as a cell-extrinsic regulator of T cell responses. Nat Rev Immunol 11(12): 852-863.
- Wang, A. Y., Crome, S. Q., Jenkins, K. M., Medin, J. A., Bramson, J. L. and Levings, M. K. (2011). Adenoviral-transduced dendritic cells are susceptible to suppression by T regulatory cells and promote interleukin 17 production. Cancer Immunol Immunother 60(3): 381-388.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Huang, Q., Lam, A. J., Boardman, D. A., Dawson, N. A. J. and Levings, M. K. (2021). Suppression of Human Dendritic Cells by Regulatory T Cells. Bio-protocol 11(21): e4217. DOI: 10.21769/BioProtoc.4217.
- Dawson, N. A. J., Rosado-Sanchez, I., Novakovsky, G. E., Fung, V. C. W., Huang, Q., McIver, E., Sun, G., Gillies, J. and Speck, M. (2020). Functional effects of chimeric antigen receptor co-receptor signaling domains in human regulatory T cells. Sci Transl Med 12(557): eaaz3866.
Category
Immunology > Immune cell differentiation > T cell
Cell Biology > Cell-based analysis > Flow cytometry
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link