- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Enrichment of Cytoplasmic RNA Granules from Arabidopsis Seedlings
Published: Vol 11, Iss 21, Nov 5, 2021 DOI: 10.21769/BioProtoc.4212 Views: 3301
Reviewed by: Wenrong HeYuan WangYe Xu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Rapid Miniprep of Intact Chloroplasts from Arabidopsis thaliana Leaves
Brenda A. Carranza-Correa [...] Manuel Gutiérrez-Aguilar
May 20, 2025 2637 Views

Effective Gene Silencing in Plants by Synthetic Trans-Acting siRNAs Derived From Minimal Precursors
Adriana E. Cisneros [...] Alberto Carbonell
Oct 20, 2025 1806 Views

Turbo-RIP: A Protocol for TurboID-based RNA Immunopurification to Map RNA Landscapes in Plant Biomolecular Condensates
Zhi Zhang [...] Panagiotis Nikolaou Moschou
Feb 5, 2026 213 Views
Abstract
RNA granules (RGs) are membraneless intracellular compartments that play important roles in the post-transcriptional control of gene expression. Stress granules (SGs) are a type of RGs that form under environmental challenges and/or internal cellular stresses. Stress treatments lead to strong mRNAs translational inhibition and storage in SGs until the normal growth conditions are restored. Intriguingly, we recently showed that plant stress granules are associated with siRNA bodies, where the RDR6-mediated and transposon-derived siRNA biogenesis occurs (Kim et al., 2021). This protocol provides a technical workflow for the enrichment of cytoplasmic RGs from Arabidopsis seedlings. We used the DNA methylation-deficient ddm1 mutant in our study, but the method can be applied to any other plant samples with strong RG formation. The resulting RG fractions can be further tested for either RNAs or proteins using RNA-seq and mass spectrometry-based proteomics.
Keywords: RNA granuleBackground
RNA granules (RGs) are non-membraneous cellular architectures that are relevant to a variety of biological processes. Of these, stress granules (SGs) contain non-translating mRNAs and various RNA-binding proteins, and serve as the assorting sites of mRNAs for storage, translational reinitiation, or degradation (Anderson and Kedersha, 2009). Recently, we demonstrated that plant SGs include numerous transposon RNAs in DNA methylation-deficient mutants (Kim et al., 2021). Being natural endogenous mutagens in genomes, transposons are counteracted by the host’s epigenetic silencing mechanisms, which are primarily mediated by siRNAs. Several studies have suggested that transposon-derived siRNAs are produced in the siRNA bodies, which are often in association with SGs (McCue et al., 2012 and 2013). The transcriptome of SGs in yeast and human have been characterized in detail (Jain et al., 2016; Khong et al., 2017), revealing that SG-located RNAs are depleted of ribosomes and relatively longer in length. Consistently, our latest work also showed for the first time in a plant system that SGs contain weakly translating RNAs, the majority of which are derived from transposons (Kim et al., 2021). Given the importance and prevalence of RGs in a wide variety of biological processes, the identification of their RNA and protein components is a critical first step towards understanding RG-mediated gene expression control. Therefore, we describe here a versatile method for the enrichment of the RGs from Arabidopsis seedlings.
Materials and Reagents
Whatman filter paper (Merk, catalog number: WHA1001150)
Pipette tip 1,000 μl (Axygen, catalog number: T-1000-C-L-R-S)
Pipette tip 10 ml (Eppendorf, catalog number: 0030000781)
1.5 ml microcentrifuge tube (Axygen, catalog number: MCT-150-C-ZX)
50 ml centrifuge tube (Corning, catalog number: 430828)
Petri dish (any brand, 47 mm diameter)
Funnel (any brand, 100 mm diameter)
Arabidopsis ddm1-2 mutant in the Columbia-0 background
Ethanol (Merk, catalog number: 51976)
Triton X-100 (Merk, catalog number: T8787)
Murashige and Skoog basal medium with Vitamins (PhytoTech, catalog number: M519)
Distilled water, generated using the RSJ Water Purification system (Tanon, catalog number: RODI-220B1)
Liquid nitrogen
Miracloth (Sigma-Aldrich, catalog number: 475855)
Tris base (Fisher Scientific, catalog number: BP152-500)
Potassium hydroxide (KOH) (Sigma-Aldrich, catalog number: 221473)
Hydrochloric acid (HCl) (Fisher Scientific, catalog number: A466-250)
Potassium acetate (KOAc) (Sigma-Aldrich, catalog number: P1190)
Magnesium acetate (MgOAc) (Sigma-Aldrich, catalog number: 63052)
Dithiothreitol (DTT) (Fisher Scientific, catalog number: R0861)
Nonylphenyl-polyethylene Glycol (NP-40) (Fisher Scientific, catalog number: 49-201)
cOmpleteTM, EDTA-free protease inhibitor cocktail (Sigma-Aldrich, catalog number: 11873580001)
RNasin® Plus RNase inhibitor (Promega, catalog number: N2615)
Sterilization solution (see Recipes)
Half-strength MS-medium plate (see Recipes)
RG lysis buffer (see Recipes)
Equipment
Pipette 1,000 μl, 10 ml (Eppendorf, catalog numbers: 3123000063, 4720000011)
Vortexer (any brand)
Clean bench (any brand)
Refrigerator (4°C) (any brand)
Plant growth chamber (any model with temperature and light control)
Mortar and pestle (any brand)
Sorvall LYNX 4000 Superspeed Centrifuge (ThermoFisher, catalog number: 75006580)
Microcentrifuge 5424R (Eppendorf, model: 5424R)
Centrifuge 5810R (Eppendorf, model: 5810R, catalog number: 022625101)
Procedure
Overview
This protocol allows for the simple and fast enrichment of RG fractions (albeit crude) from plant samples. Briefly, the RG fractions are separated and dissolved by continuous centrifugation with RG lysis buffer. The enriched RG fractions can be subsequently subjected to RNA-seq and protein analysis (Figure 1).
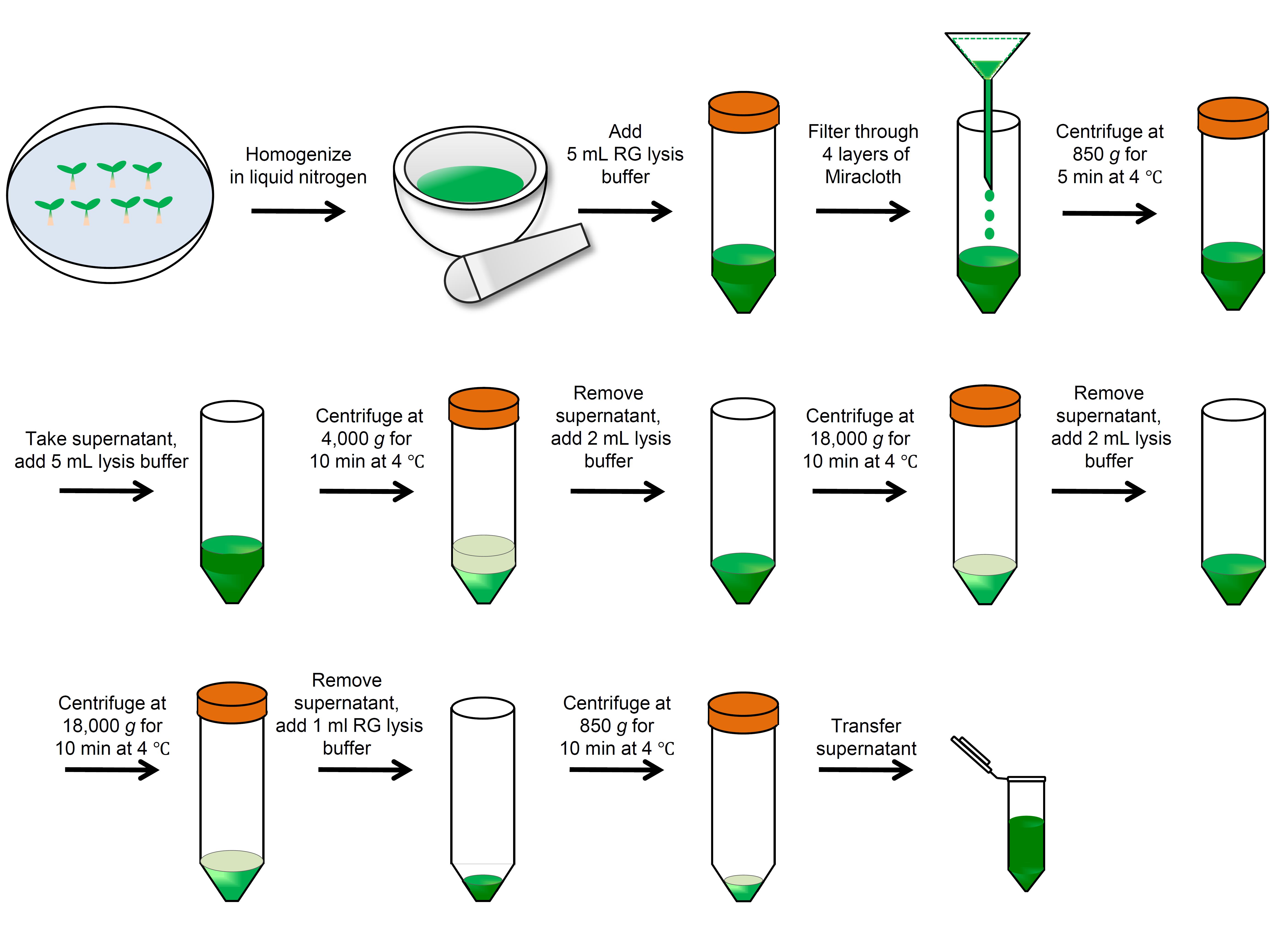
Figure 1. Scheme of RNA granule enrichmentPlant preparation
Place 100 μl of ddm1-2 seeds in a 1.5 ml microcentrifuge tube.
Add 1 ml of sterilization solution to seeds and vortex for 4 min (see Recipe 1).
Discard the solution.
Wash the seed with 1 ml of 100% ethanol.
Vortex for 1 min and discard the ethanol.
Repeat Steps B4 and B5.
Prepare a sheet of filter paper and pipet the seeds onto the paper.
Let the ethanol evaporate for 3 min.
Pick up the paper and sprinkle the seeds onto half-strength Murashige and Skoog medium (see Recipe 2).
Wrap the plate with parafilm and place it into a 4°C chamber for 2 days.
Transfer the plate into a growth chamber set at 22°C and under 16 h light/8 h dark cycles for 10 days.
Notes:
Any plant samples in addition to Arabidopsis seedlings can be used for this protocol.
Perform seed sterilization on a clean bench (wipe down with 70% ethanol before use).
RNA granule enrichment
Grind 2 g of seedlings into a fine powder in liquid nitrogen using a precooled mortar and pestle.
Collect the samples (approximately 5 ml) into a 50 ml tube and resuspend in 5 ml of RG lysis buffer (see Recipe 3).
Filter the resulting slurry through four layers of Miracloth in a funnel to a 50 ml conical tube and centrifuge at 850 × g for 5 min at 4°C to pellet cell debris.
Transfer the supernatant to a new 50 ml tube and add 5 ml of RG lysis buffer.
Centrifuge at 4,000 × g for 10 min at 4°C and discard the supernatant.
Resuspend the pellet in 2 ml of RG lysis buffer. Centrifuge at 18,000 × g for 10 min at 4°C.
Resuspend the pellet in 2 ml of RG lysis buffer, vortex, and centrifuge at 18,000 × g at 4°C for 10 min.
Discard the supernatant and resuspend the pellets gently in 1 ml of RG lysis buffer. Centrifuge at 850 × g for 10 min at 4°C.
Transfer the supernatant (enriched with RGs) into a 1.5 ml microcentrifuge tube without disturbing any residue and keep it in a freezer until use.
Note: We recommend using the fluorescence-tagged RG-marker plant lines to quickly check for successful RG enrichment.
RNA analysis
The final RG fraction resulting from the protocol described above can be subjected to regular RNA extraction and subsequently tested for either targeted RNA analyses using RT-qPCR or transcriptome-wide profiling with RNA-seq.
Note: Refer to Kim et al. (2021) for suggestions on any RG-specific marker genes.
Data analysis
RNA-seq data generated from the RG enrichment fractions can be analyzed as detailed in the original paper (Kim et al., 2021; https://doi.org/10.1038/s41477-021-00867-4).
Recipes
Sterilization solution
70% ethanol
0.05% Triton X-100
Half-strength MS-medium plate
2.2 g/L Murashige and Skoog basal medium with Vitamins
Adjust pH to 5.7 with KOH
7 g/L plant agar
Sterilize by autoclaving
RG lysis buffer
50 mM Tris-HCl, pH 7.4
100 mM KOAc
2 mM MgOAc
0.5% NP-40
0.5 mM DTT
One tablet (in 50 ml) of protease inhibitor cocktail (cOmpleteTM, EDTA-free Protease Inhibitor Cocktail)
1 U/μl RNasin Plus RNase Inhibitor
1 M Tris-HCl, pH 7.4 2.5 ml 1 M KOAc 5 ml 1 M MgOAc 0.1 ml 10% NP-40 2.5 ml 1 M DTT 25 μl Protease inhibitor cocktail One tablet 40,000 U/ml RNasin Plus RNase Inhibitor 1.25 ml Distilled water Top up to 50 ml
Notes:
Prepare all solutions and buffers with distilled water.
Add DTT and RNase Inhibitor right before use.
Acknowledgments
The work was supported by grants from the National Natural Science Foundation of China (31970518), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB27030209), and the General Program of Natural Science Foundation of Shanghai (21ZR1470700). E. Y. Kim is the recipient of a President’s International Fellowship Initiative (PIFI) young staff fellowship (2021FYB0001) from CAS. This protocol was adapted from our previously reported work (Kim et al., 2021).
Competing interests
The authors declare no conflicts of interest.
References
- Anderson, P. and Kedersha, N. (2009). RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol 10(6): 430-436.
- Jain, S., Wheeler, J. R., Walters, R. W., Agrawal, A., Barsic, A. and Parker, R. (2016). ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164(3): 487-498.
- Khong, A., Matheny, T., Jain, S., Mitchell, S. F., Wheeler, J. R. and Parker, R. (2017). The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol Cell 68(4): 808-820e805.
- Kim, E. Y., Wang, L., Lei, Z., Li, H., Fan, W. and Cho, J. (2021). Ribosome stalling and SGS3 phase separation prime the epigenetic silencing of transposons. Nat Plants 7(3): 303-309.
- McCue, A. D., Nuthikattu, S., Reeder, S. H. and Slotkin, R. K. (2012). Gene expression and stress response mediated by the epigenetic regulation of a transposable element small RNA. PLoS Genet 8(2): e1002474.
- McCue, A. D., Nuthikattu, S. and Slotkin, R. K. (2013). Genome-wide identification of genes regulated in trans by transposable element small interfering RNAs. RNA Biol 10(8): 1379-95.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Lei, Z., Kim, E. Y. and Cho, J. (2021). Enrichment of Cytoplasmic RNA Granules from Arabidopsis Seedlings. Bio-protocol 11(21): e4212. DOI: 10.21769/BioProtoc.4212.
Category
Molecular Biology > RNA > mRNA translation
Plant Science > Plant cell biology > Organelle isolation
Plant Science > Plant molecular biology > RNA
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








