- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Electrophysiological Recordings of the Polycystin Complex in the Primary Cilium of Cultured Mouse IMCD-3 Cell Line
Published: Vol 11, Iss 20, Oct 20, 2021 DOI: 10.21769/BioProtoc.4196 Views: 3037
Reviewed by: Chiara AmbrogioJohn W PetersonAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Capacitance Measurements of Exocytosis From AII Amacrine Cells in Retinal Slices
Espen Hartveit and Margaret L. Veruki
Jan 5, 2025 2319 Views

Voltage Clamp Fluorometry in Xenopus laevis Oocytes to Study the Voltage-sensing Phosphatase
Victoria C. Young [...] Susy C. Kohout
Feb 20, 2025 2003 Views

Visualization of Gap Junction–Mediated Astrocyte Coupling in Acute Mouse Brain Slices
Nine F. Kompier [...] Fritz G. Rathjen
Feb 20, 2025 2258 Views
Abstract
PC-1 and PC-2 form an ion channel complex called the polycystin complex, which predominantly localizes to a small hair-like organelle called the primary cilium. The polycystin complex permeates cations, K+, Na+, and Ca2+, and has an unusual 1:3 stoichiometry that combines one PC-1 subunit with three PC-2 subunits. However, the small size and shape of primary cilia impose technical challenges to study the polycystin complex in its native environment. In this paper, we describe the methodology to directly record ion channel activity in primary cilia. This method will allow a detailed functional characterization of how mutations within the polycystin complex cause Autosomal Dominant Polycystic Kidney Disease (ADPKD), essential to develop novel therapeutics for this ciliopathy.
Keywords: Polycystin complexBackground
Autosomal Dominant Polycystic Kidney Disease (ADPKD) is the most common monogenic ciliopathy in humans, responsible for 6-10% of end-stage renal disease (ESRD) (Harris and Torres, 2009). Mutations in the polycystin proteins PC-1 or PC-2 are the major cause of ADPKD, resulting in renal cysts and numerous extra-renal manifestations (Torres et al., 2007). The polycystin complex is a heteromeric channel complex consisting of PC-1 and PC-2 subunits in a 1:3 ratio (Su et al., 2018). Direct recordings from primary cilia (ciliary patch clamp) of mouse Inner Medullary Collecting Duct-3 (IMCD-3) cells have shown that the polycystin complex is the dominant ion channel complex to permeate cations (Icilium) across the ciliary membrane (Kleene and Kleene, 2017; Liu et al., 2018). We and others recently developed novel electrophysiological methods to measure ion channel activity in this tiny, previously inaccessible organelle (Kleene and Kleene, 2012; DeCaen et al., 2013; Ha et al., 2020). The volume ratio of cilioplasm to cytoplasm is approximately 1:30,000, suggesting that a much smaller pipette tip is required to make a tight seal on the ciliary membrane (Delling et al., 2013). In this manuscript, we provide useful tips to perform the ciliary patch clamp technique for students and scientists who want to investigate ciliary ion channels.
Materials and Reagents
12 mm diameter cover slip (Chemglass Life Science, catalog number: CLS-1760-012)
24-well culture plate (Geiner Bio-One, Cell Star®, catalog number: 662-160)
Pipette tips
Borosilicate glass pipettes with filament (Sutter Instrument, catalog number: BF150-75-10)
Lipofectamine LTX (Thermo Fisher Scientific, catalog number: 15338100)
Dulbecco’s modified Eagle’s medium (DMEM) (Life Technologies, Gibco®, catalog number: 11965-092)
Penicillin-streptavidin (Life Technologies, Gibco®, catalog number: 15140-122)
Fetal bovine serum (GEMINI, catalog number: 900-108)
OPTI-MEM (Life Technologies, Gibco®, catalog number: 31985-070)
0.25% trypsin-EDTA (1×) (Life Technologies, Gibco®, catalog number: 25200-056)
Sodium chloride (NaCl) (Millipore Sigma, catalog number: S9888-1KG)
Potassium chloride (KCl) (Millipore Sigma, catalog number: P9333)
Calcium chloride (CaCl2) (Millipore Sigma, catalog number: C8106)
Magnesium chloride (MgCl2) (Millipore Sigma, catalog number: M1028)
HEPES (Millipore Sigma, catalog number: H3325-1KG)
Sodium methanesulfonate (NaMES) (Millipore Sigma, catalog number: 302406-100G)
EGTA (Millipore Sigma, catalog number: E3889)
D-mannitol (Millipore Sigma, catalog number: M4125-1KG)
NaOH (Millipore Sigma, catalog number: 655104-25G)
Extracellular Solution for patch clamp (see Recipes)
Intracellular Solution for patch clamp (see Recipes)
Equipment
Inverted microscope (Carl Zeiss, model: Axiovert 200M)
Patch clamp chamber (Warner, PI/PH1)
Capacitor feedback headstage CV203BU (Molecular Devices)
Micropipette puller (Sutter Instruments, model: SU-P1000)
Micro forge (NARISHIGE Japan, model: MF-830)
63× 1.2 NA Water Lens ∞/0.14-0.19 (Carl Zeiss, C-Apochromat)
Amplifier Axopatch 200B (Molecular Devices)
Digidata 1550B (Molecular Devices)
Manipulator (Sutter Instruments, model: MPC-200)
Vapor pressure osmometer (Wescor, model: 5520)
Cover slip (Fisher Scientific, catalog number: 19804)
CO2 incubator (Heraeus, model: 240)
Software
Origin8 (OriginLab, https://www.originlab.com)
Clampfit (Molecular Devices, https://www.moleculardevices.com)
Prism 10.0 (GraphPad, https://www.graphpad.com/scientific-software/prism/)
Clampex (Molecular Devices, https://www.moleculardevices.com)
Procedure
Pipette fabrication for ciliary patch clamp
Run a ramp test to determine the level of heat required to melt the individual batch of glass pipettes. To run a ramp test, select the option of “ramp test” on the screen of the puller. The result of the ramp test is automatically determined when the glass pipette is melted and the puller bars drift apart.
Customize program to fabricate pipettes with bath resistance 18-26 MΩ. We designed the program (Table 1) and pulled the pipette using a Sutter micropipette puller (given that ramp is 584).
Table 1. Micropipette program using BF150-75-10 borosilicate glass. The number of each setting can be re-calculated based on ramp test of the pipette.
HEAT PULL VELOCITY DELAY PRESSURE 600 0 25 1 500 After pulling, carefully polish pipette tip using microforge.
After polishing the pipette, place the pipette into the bath solution of the chamber.
Confirm the bath resistance of pipette (18-26 MΩ). The bath resistance of the pipette is automatically shown in the Clampex program (Figure 1).
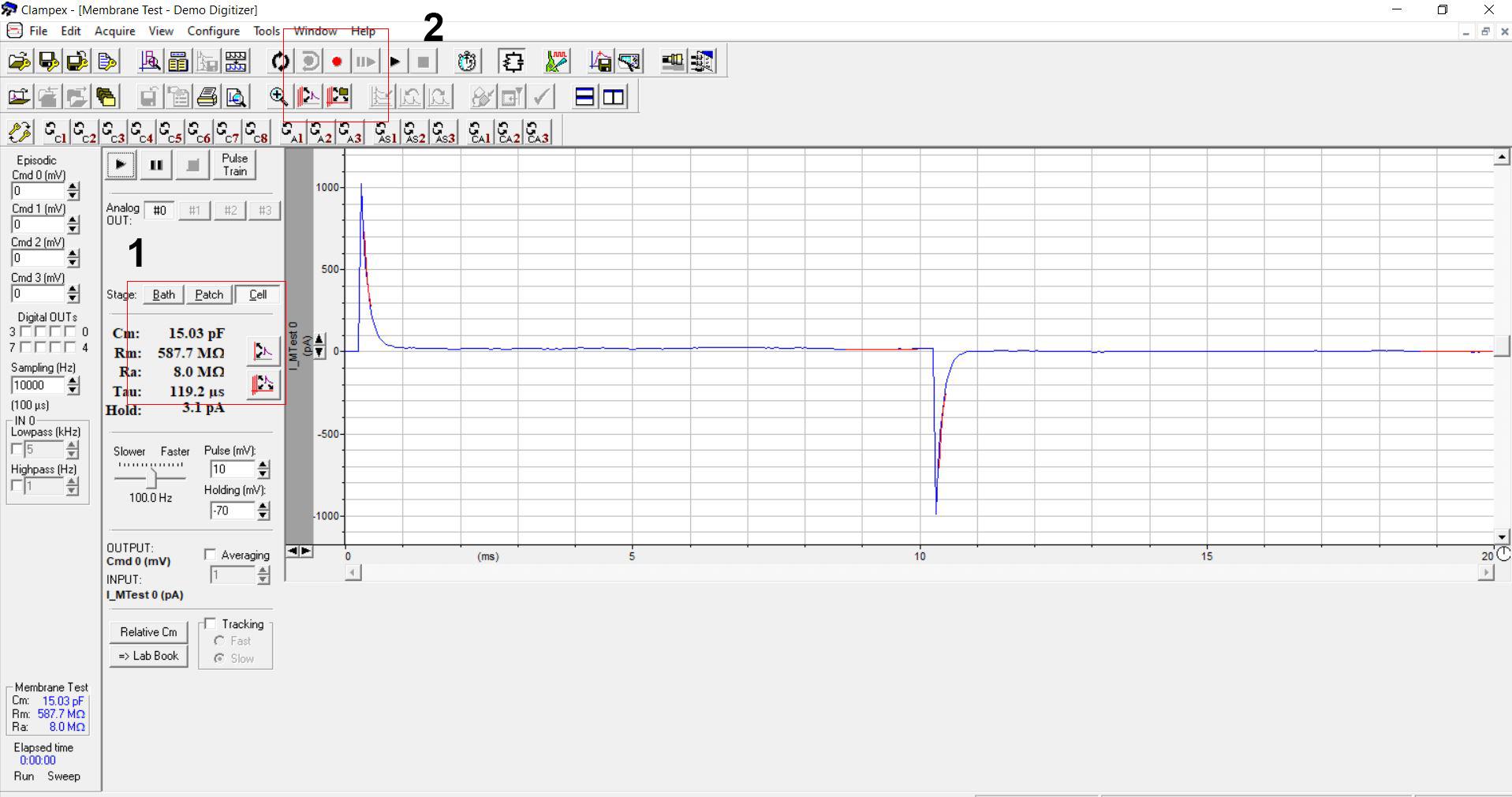
Figure 1. Clampex software for patch clamp recording. Red box 1 shows the resistance of the pipette in the bath mode (bath resistance). Red box 2 indicates the record button that can be selected for electrophysiology recording. In this figure, the Clampex software is run in the demo mode.
Ciliary patch clamp recordings
Seed IMCD-3 cells (number of cells: 0.05 × 106) on cover slips (12 mm diameter) in 24-well culture plate and add 1 ml of DMEM media.
Transfect with the ARL 13B gene encoding ADP-ribosylation factor-like protein 13B-Enhanced green fluorescent protein (Caspary et al., 2007) as a ciliary marker when cell confluency is 70-90% with Lipofectamine LTX following the manufacturer’s instruction.
Incubate overnight in a CO2 incubator.
The next day, replace medium with OPTI-MEM and incubate for 1-2 days until the number of cells reaches >0.24 × 106 to induce primary cilium formation.
Transfer cover slip to patch clamp chamber and confirm that cells have formed a primary cilium by detecting green fluorescent labeled ARL-13B in the microscope (Figure 2B).
Place the borosilicate pipette into the chamber using the manipulator (Figure 2B) and carefully position it near the primary cilium (Figure 2B).
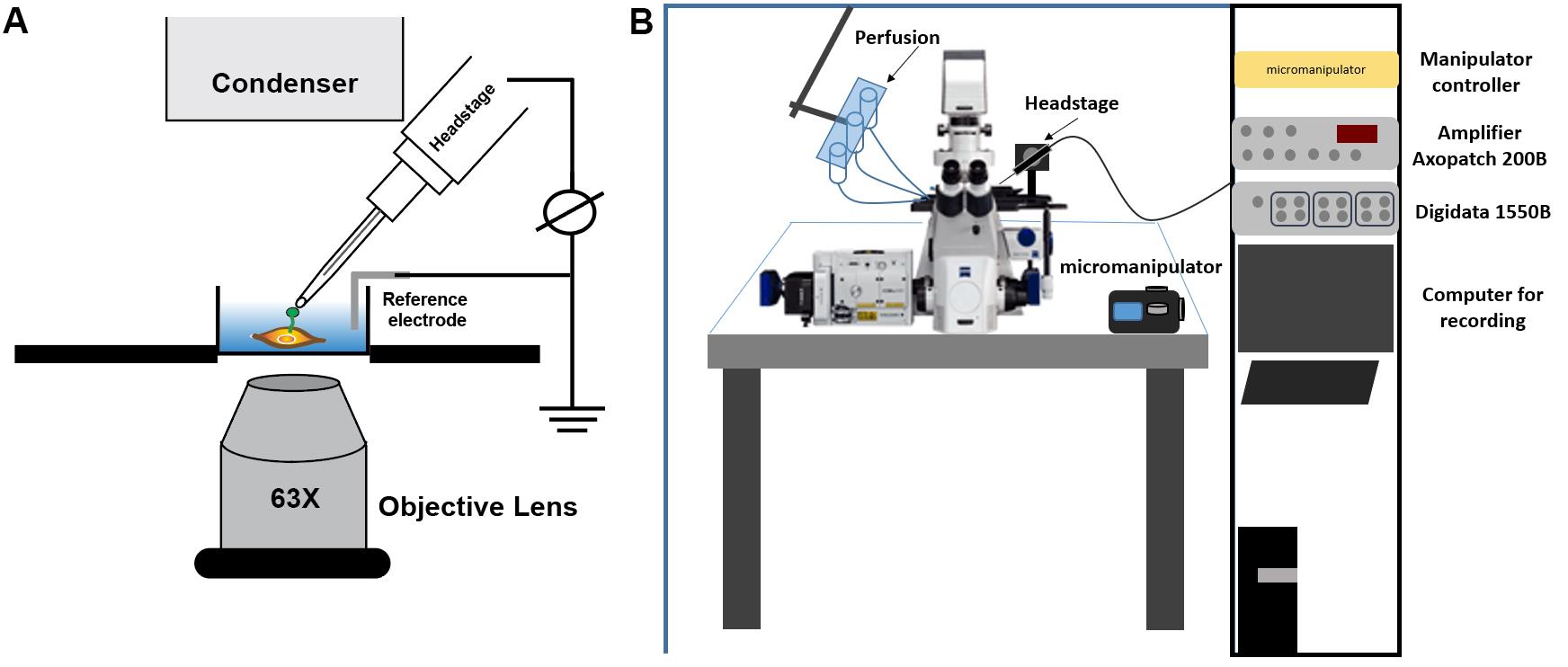
Figure 2. Schematic illustration for ciliary patch clamp set-up. A. Schematic drawing for single channel patch clamp recordings of ciliary membrane. B. Full view of patch clamp set-up.Position pipette at ciliary tip and apply negative pressure by suctioning the tube connected to the electrode to form a gigaseal that should be between 8.0-32.0 Giga Ohms (GΩ). The value of a giga-seal is automatically recorded in the Clampex software.
Record currents by selecting the record option in the Clampex and discard data obtained at less than 8.0 Giga Ohms (GΩ) seal (Figure 1).
Data analysis
Single channel analysis (pClamp 10.2 and Clampfit 10.0)
Conductance: Select the recording obtained from each voltage potential and plot the data using a conventional histogram. The conventional histogram data are fitted with the Gaussian function below for single channel conductance analysis.
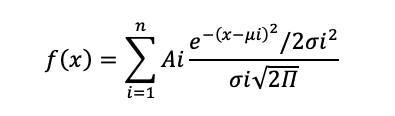
where n is the components, A is the amplitudes, µ is the gaussian mean, and σ is the Gaussian standard deviation. After fitting the Gaussian fitting, subtract µ1 from µ2 and plot the value on each voltage potential.
Open probability of single channel activity: Select the recording section and calculate the open probability using the equation below:
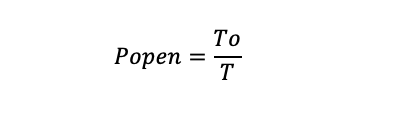
where To is the total time that the channel presented in the open state, and T is the total observation time. If a patch contains more than one of the same type of channel, Popen was computed by:
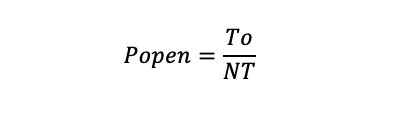
where N indicates the number of channels in the patch. The following equation is used to populate data.
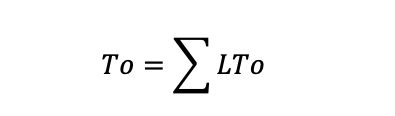
where L indicates the level of the channel opening. The level of the channel opening can be set at the unitary amplitudes. In Figure 3C, the top red dotted line indicates the level of the channel opening. The absolute probability of the channel being open NPo is computed by:
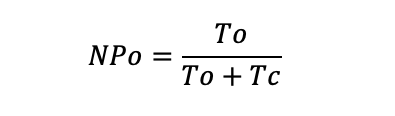
where Tc indicates the total close.
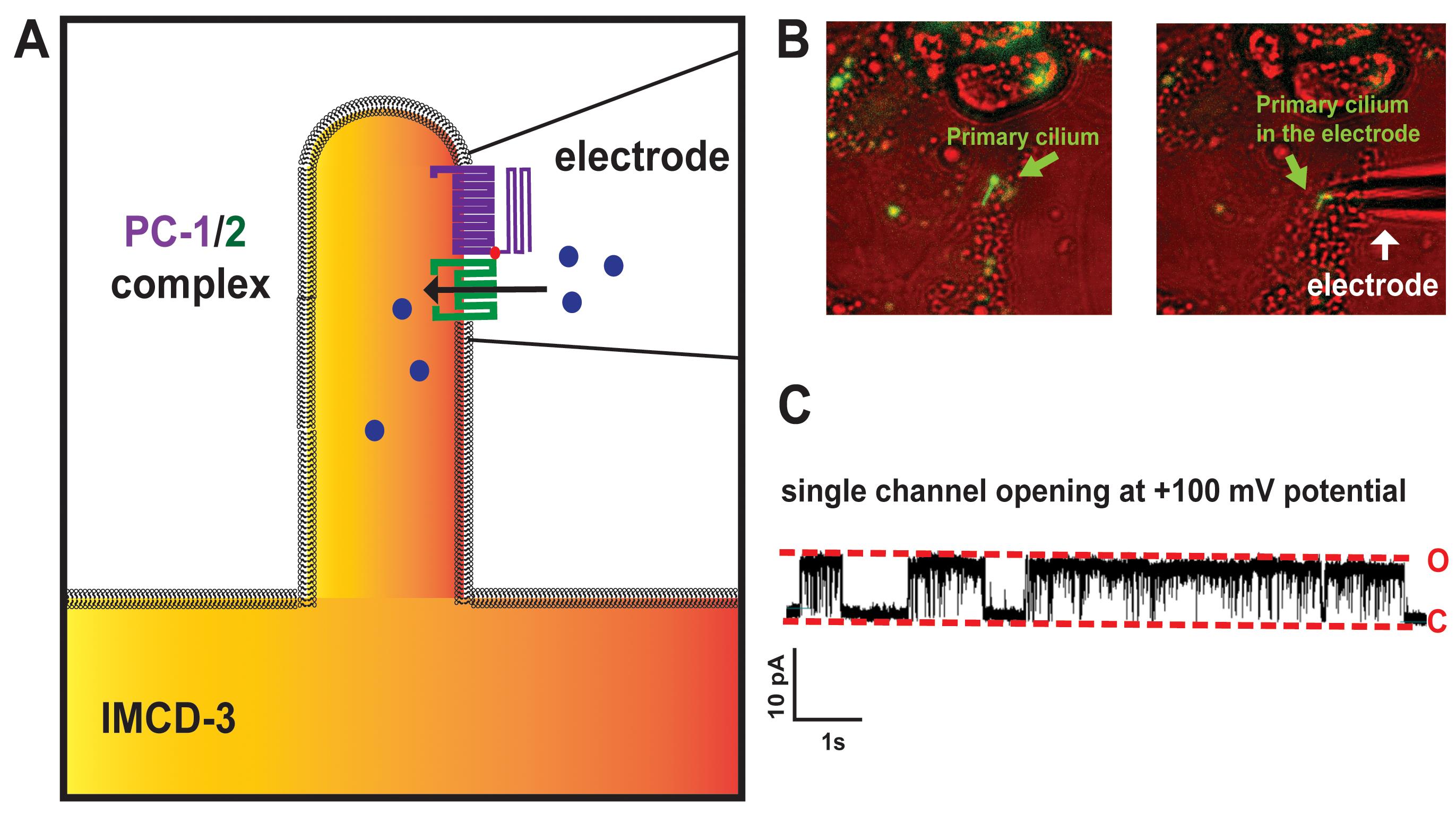
Figure 3. Ciliary patch clamp recording in IMCD-3 cells. A. Illustration of PC-1/2 single channels within the electrode. B. Image of ciliary patch clamp recording in IMCD-3 cell. C. Representative single channel recordings obtained from the primary cilium of IMCD-3 cells. Red dotted lines indicate the open (O) and close (C) states of ciliary ion channel.
Recipes
Extracellular Solution for patch clamp
145 mM sodium chloride (NaCl)
5 mM potassium chloride (KCl)
2 mM calcium chloride (CaCl2)
1 mM magnesium chloride (MgCl2)
10 mM HEPES
Intracellular Solution for patch clamp
90 mM sodium methanesulfonate (NaMES)
10 mM sodium chloride (NaCl)
10 mM HEPES
5 mM EGTA
2 mM magnesium chloride (MgCl2)
100 nM free calcium
Osmolarity was adjusted to 290 mOsm/Kg using D-mannitol
pH was adjusted to 7.4 using NaOH
Acknowledgments
This work was supported by the National Institute of Health Grant R01GM130908 (MD) and the National Research Foundation of Korea (NTF) grant funded by the Korean government (MSIT) (No.2019R1A6A3A03033302) (KH). The protocol was used in the publication by Ha et al. (2020; DOI: 10.7554/eLife.60684).
Competing interests
Authors declare that no competing interests exist.
References
- Caspary, T., Larkins, C. E. and Anderson, K. V. (2007). The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell 12(5): 767-778.
- DeCaen, P. G., Delling, M., Vien, T. N. and Clapham, D. E. (2013). Direct recording and molecular identification of the calcium channel of primary cilia. Nature 504(7479): 315-318.
- Delling, M., DeCaen, P. G., Doerner, J. F., Febvay, S. and Clapham, D. E. (2013). Primary cilia are specialized calcium signalling organelles. Nature 504(7479): 311-314.
- Ha,K., Nobuhara, M., Wang, Q., Walker, R. V., Qian, F., Schartner, C., Cao, E. and Delling, M. (2020). The heteromeric PC-1/PC-2 polycystin complex is activated by the PC-1 N-terminus. Elife 9: e60684.
- Harris, P. C. and Torres, V. E. (2009). Polycystic kidney disease. Annu Rev Med 60: 321-337.
- Kleene, S. J. and Kleene, N. K. (2017). The native TRPP2-dependent channel of murine renal primary cilia. Am J Physiol Renal Physiol 312(1): F96-f108.
- Liu, X., Vien, T., Duan, J., Sheu, S. H., DeCaen, P. G. and Clapham, D. E. (2018). Polycystin-2 is an essential ion channel subunit in the primary cilium of the renal collecting duct epithelium. Elife 7: e33183.
- Su, Q., Hu, F., Ge, X., Lei, J., Yu, S., Wang, T., Zhou, Q., Mei, C. and Shi, Y. (2018). Structure of the human PKD1-PKD2 complex. Science 361(6406): eaat9819.
- Torres, V. E., Harris, P. C. and Pirson, Y. (2007). Autosomal dominant polycystic kidney disease. Lancet 369(9569): 1287-1301.
Article Information
Copyright
Ha and Delling. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Ha, K. and Delling, M. (2021). Electrophysiological Recordings of the Polycystin Complex in the Primary Cilium of Cultured Mouse IMCD-3 Cell Line. Bio-protocol 11(20): e4196. DOI: 10.21769/BioProtoc.4196.
- Ha,K., Nobuhara, M., Wang, Q., Walker, R. V., Qian, F., Schartner, C., Cao, E. and Delling, M. (2020). The heteromeric PC-1/PC-2 polycystin complex is activated by the PC-1 N-terminus. Elife 9: e60684.
Category
Biophysics > Electrophysiology > Patch-clamp technique
Developmental Biology > Cell signaling > Electrical response
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








