- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Co-immunofluorescence of MRPL12 and Nrf2 in HK2 Cells
Published: Vol 11, Iss 20, Oct 20, 2021 DOI: 10.21769/BioProtoc.4191 Views: 3898
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

ZnCl2 Precipitation-Assisted Sample Preparation for Proteomic Analysis
Qiqing He [...] Fuchu He
Jul 20, 2025 2726 Views

Fluorescence Polarization-Based High-Throughput Screening Assay for Inhibitors Targeting Cathepsin L
Keyu Guo [...] Shuyi Si
Jul 20, 2025 2277 Views

SiMPull-POP: Quantification of Membrane Protein Assembly via Single Molecule Photobleaching
Ryan J. Schuck [...] Rajan Lamichhane
Jan 5, 2026 287 Views
Abstract
Immunofluorescence is a technique to visualize the localization of specific molecule targets within cells using the specificity of antibodies. Here, we describe a protocol to detect two different protein components in a cell simultaneously. Antibody concentrations to be used vary from cell to cell and should be optimized for different cell types. In this protocol, we perform co-immunofluorescence of mitochondrial ribosomal protein L7/L12 (MRPL12) and nuclear factor erythroid 2-related factor 2 (Nrf2), a potential transcription factor of MRPL12, in HK-2 cells, as an example. Taking advantage of the diverse set of antibodies raised in different species, we are able to analyze the colocalization and expression of these proteins.
Keywords: MRPL12Background
The mitochondrial ribosomal protein L7/L12 (MRPL12), a member of the mitochondrial ribosomal protein (MRP) family, plays pivotal roles in mitochondrial function, including mitochondrial ribosome assembling (Surovtsevaa et al., 2011; Kühl et al., 2016), mtDNA transcription (Surovtsevaa et al., 2011; Nouws et al., 2016), mitochondrial ATP production (Marty and Fort, 1996), and mitochondrial oxidative phosphorylation (OXPHOS) (Ma et al., 2020). As a canonical molecule coordinating the expression of antioxidant genes (Hayes and Dinkova-Kostova, 2014; Dinkova-Kostova and Abramov, 2015), nuclear factor erythroid 2-related factor 2 (Nrf2) is predicted to be a potential transcription factor for MRPL12 (Gu et al., 2021). To examine the regulation of Nrf2 on the expression of MRPL12, co-immunofluorescence was applied to identify the abundance and colocalization of the two proteins. Here, we performed an indirect immunofluorescence technique where the sample cells were incubated with primary antibodies raised in different species, which were detected by fluorophore-conjugated secondary antibodies. Using this co-immunofluorescence technique, we can simultaneously analyze the distribution of the proteins and perform a semi-quantitative analysis to elucidate the interaction between the two molecules indirectly.
Materials and Reagents
24-well cell culture plate (Corning, Costar®, catalog number:3524)
Cell culture flask, T25 (Corning, Costar®, catalog number:430639)
Round coverslip, 9 mm (Biosharp, catalog number: BS-09-RC), place the coverslips in a 75% ethanol-containing centrifuge cube
Adhesion microscope slides (CITOTEST, catalog number: 80312-3161)
Parafilm (Bemis, catalog number:4157)
For cell preparation:
Dulbecco’s Modified Eagle Medium (DMEM), 1g/L D-Glucose, with L-Glutamine (Gibco, catalog number: C11885500BT). Store at 2-8°C for 1 year
Fetal Bovine Serum (FBS) (Gibco, catalog number:10099141). Aliquot and store at -20°C
Penicillin-Streptomycin (10,000U/ml) (Gibco, catalog number:15140122). Store at -20~-5°C for 1 year
Phosphate Buffered Saline (PBS), sterile-filtered (Servicebio, catalog number: G4202). Store at 2-30°C for 18 months
0.25% Trypsin-EDTA, (Gibco, catalog number: 25200-072). Store at -20~-5°C for 24 months
Phosphate buffered saline (PBS) powder (Absin, catalog number: abs9266). Store at room temperature (RT) for 2 years
4% paraformaldehyde (PFA) (Solarbio, catalog number: P1110). Aliquot and store at -20°C for 1 year, and avoid repeated freezing and thawing. Store at 4°C for up to a month
Triton X-100 (Solarbio, catalog number: T8022). Store at RT for 2 years
Normal goat serum (Solarbio, SL038). Store at -20°C for 5 years
Primary antibodies:
MRPL12 antibody (Novas, catalog number: 3B12-1A3). Aliquot and store at -20°C. Avoid freeze-thaw cycles
Nrf2 antibody (Abcam, catalog number: ab62352). Aliquot and store at -20°C. Avoid freeze-thaw cycles
Secondary antibodies:
Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 488 (Thermo Fisher Scientific, Invitrogen, catalog number: A32723). Store at 4°C in the dark
Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 594 (Thermo Fisher Scientific, Invitrogen, catalog number: A32754). Store at 4°C in the dark
Hoechst 33342 (Life technologies, catalog number: HH-H3570). Aliquot and store at -20°C in the dark
Mounting medium, antifading (Solarbio, catalog number: S2100). Store at 4°C for 1 year
Cell culture medium (see Recipes)
PBS (see Recipes)
Permeabilization solution (see Recipes)
Blocking solution (see Recipes)
Equipment
Tweezers
Note: The tweezers should be sterile when conducting cell preparations. The subsequent steps do not require sterile conditions.
Paper towel
(Optional) Wet chamber
(Optional) Refrigerator (Haier, model: BCD-328WDPT)
Constant temperature incubator (Panasonic, model: MIR-262-PC)
Nikon inverted fluorescence microscope (Nikon, model: Ti-S)
Clean bench (Airtech, model: SW-CJ-1FD)
Directly heated CO2 incubator (Thermo Fisher Scientific, model: 240i)
Software
ImageJ
Prism GraphPad
Procedure
Cell preparation
Place coverslips in 24-well plate
Take out coverslips from the 75% ethanol-containing centrifuge cube using sterile tweezers and place them on a sterile plate. Expose them to ultraviolet (UV) light for approximately 30 min with the other required material, including PBS, trypsin, culture medium (DMEM medium with 10% FBS and 1% penicillin-streptomycin, see Recipe 1), and 24-well cell culture plate.
Ensure that both sides of the coverslips are dry to avoid cells being killed by the remaining ethanol.
Drop approximately 5 μl cell culture medium on each well. Place dry coverslips on the drop with sterile tweezers. Press the coverslips gently and remove the excess fluid to ensure they are firmly stuck to the bottom of the well.
Cell seeding and culture
Remove the medium from the cell culture flask (T25) containing the cells and wash with 2 ml PBS. Remove the PBS and add 500 μl 0.05% trypsin for 2 min to detach cells from the cell culture flask wall.
Seed a number of cells according to the experimental requirements, e.g., transfecting with siRNAs or overexpression plasmids (see Note 1). Shake the plate crosswise and lengthwise to prevent the local growth of cells from being too dense. Then allow cells to grow at 37°C in an incubator with 5% CO2. Remember to add control groups when performing the colocalization (see Note 8).
Treat cells in accordance with the experimental requirements. Aim at cell densities resulting in 70-80% confluency to avoid cells overlapping.
Immunofluorescence staining
When cells grow to 70-80% confluency, remove the medium and rinse once with 500 μl PBS for 5 min. The following steps do not need to be performed under sterile conditions.
Unfreeze the 4% PFA in advance. Add 300 μl of 4% PFA to each well for 20 min at RT.
Remove the fixative and rinse cells with PBS three times for 5 min (see Note 4).
Remove the PBS and add 300 μl of permeabilization solution to each well for 5 min at RT.
Remove the permeabilization solution and rinse with PBS three times for 5 min.
Block with 300 μl of blocking solution for 1 h at 37°C.
Remove the solution and rinse with PBS three times for 5 min.
Primary antibody incubation:
Dilute antibody A (MRPL12, 1:200) and antibody B (Nrf2, 1:300) with PBS (see Notes 5, 6, 7).
Add several drops of PBS on adhesion microscope slides, and then place a piece of parafilm on each slide. Expel bubbles to ensure the film tightly attaches to the slide.
Clamp out coverslips from the plate with tweezers and touch the edge of the paper towel to remove the remaining solution.
Put coverslips on the parafilm with cell-facing upward and add approximately 50 μl diluted antibody to cover it fully.
To avoid loss of the antibody by volatilization, incubate the slides in a wet chamber overnight at 4°C.
Optional step: After overnight incubation, keep slides at room temperature for approximately 30 min.
Lift up coverslips and put them back into wells with the cell-facing surface facing up. Rinse with PBS three times for 5 min.
Secondary antibody incubation:
Dilute the secondary antibody Alexa Fluor®-488 (1:1,000) and Alexa Fluor®-594 (1:1,000) with PBS (see Notes 5, 6).
Add 300 μl secondary antibody solution to each well.
Incubate the samples at 37°C for 1 h in the dark.
After secondary antibody incubation, protect the cells from light exposure. Remove the secondary antibody and rinse with PBS three times for 5 min.
To visualize nuclei, add the PBS-diluted Hoechst 33342 (1:1,000) to the coverslips for 10 min at RT.
Remove the solution. Rinse with PBS three times for 5 min.
Mounting, storage, and imaging
Label microscope slides with the sample information. Drop approximately 5 μl antifade mounting medium for each coverslip on the glass slide. Then, place the coverslips on the mounting medium with the surface containing the cells facing down and ensure that there are no bubbles. Remove the excess fluid if needed.
Samples can be stored for approximately 1 week at 4°C or 2 months at -20°C in the dark. Longer storage is not recommended.
Use a Nikon inverted fluorescence microscope to capture images from five different fields on each sample; select regions randomly (Figure 1). The excitation wavelengths of Hoechst 33258, Alexa Fluor 488, and Alexa Fluor 594 are 345 nm, 499 nm, and 591 nm, respectively. Record fluorescence from individual channels separately.
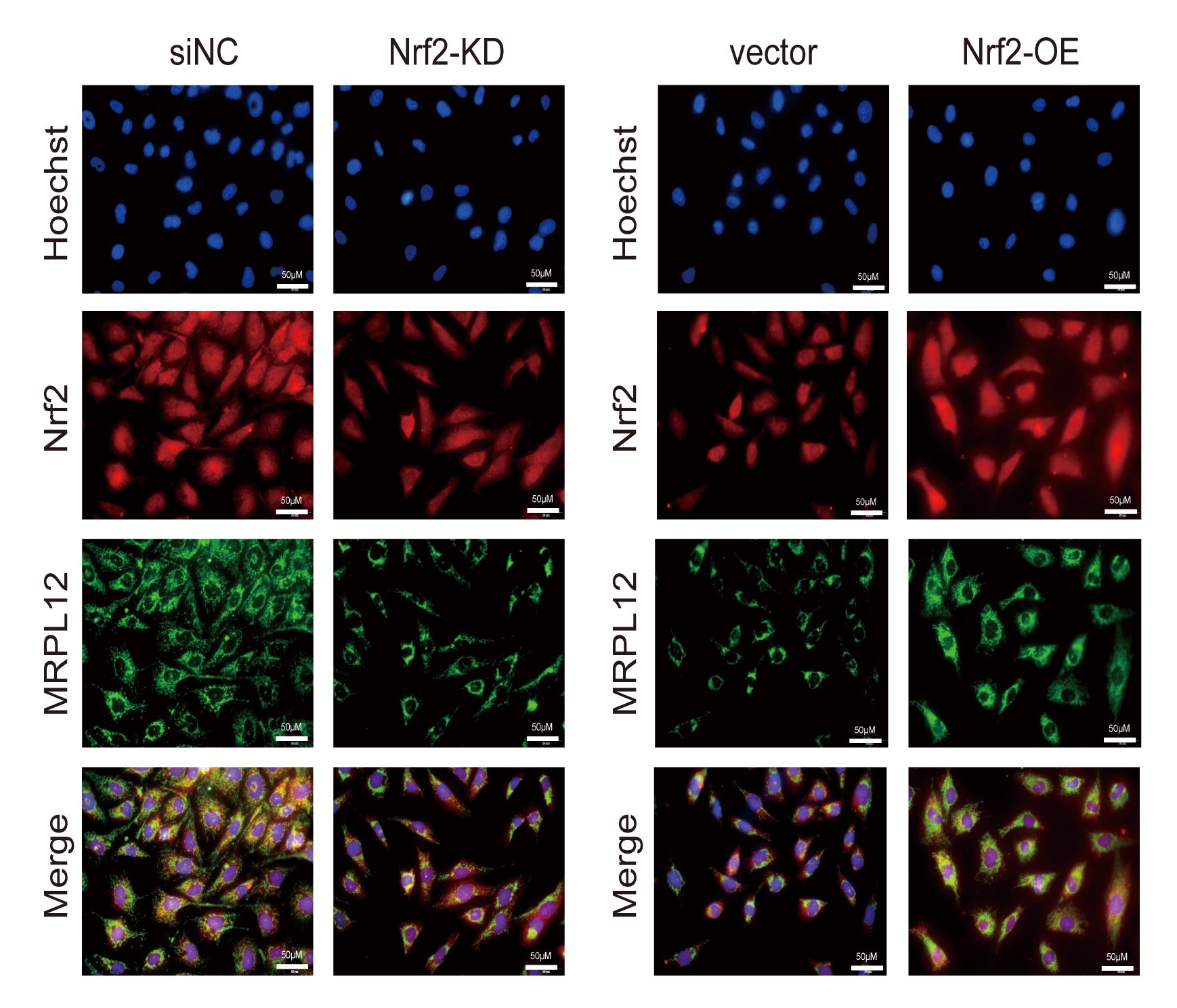
Figure 1. Immunofluorescent staining in Nrf2 overexpressed or knocked down HK-2 cells. HK-2 cells were fixed with methanol. Nrf2 (red), MRPL12 (green), and nuclei (blue) were stained as described in the protocol. Images were captured and merged using a Nikon inverted fluorescence microscope. Scale bar is 50 μm. Figure 1 reprinted with permission from Gu et al. (2021) .
Data analysis
In our published studies, we quantitatively analyzed the fluorescence intensity of HK2 cells among compared groups.
Fluorescence intensity determination
The fluorescence in stained HK2 cells is estimated using ImageJ. To quantify the fluorescence, first open the image in ImageJ, then choose Image > type > 16-bit to convert the image to grayscale. Afterward, go to Analyze > Set Measurements and choose “Area,” “Mean gray value (Mean),” and “Integrated intensity (IntDen)” as measurement parameters. Next, choose Analyze > Tools > ROI Manager. Randomly select a stained cell followed by Add[t] > Rename. Then mark another two cells and three spots of fluorescence background through the above method. Select all items and click “Measure” (Figure 2).
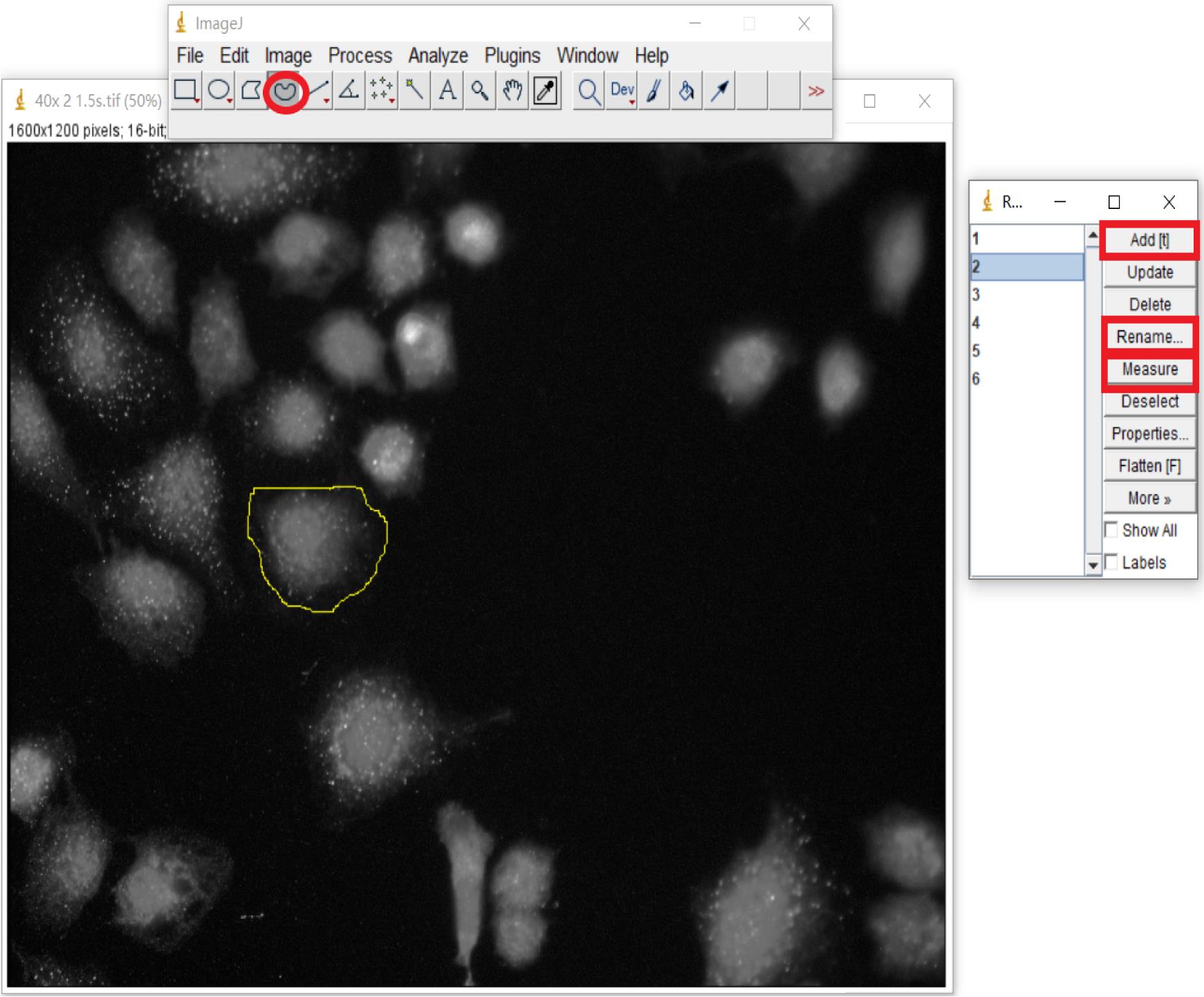
Figure 2. Fluorescence intensity determination using ImageJ. Select the measuring object and calculate with ROI Manager. Circle a cell randomly (oval highlighted) followed by Add[t] (rectangle highlighted) > Rename (rectangle highlighted). Three cells and three spots of fluorescence background are totally marked. Select all items, then choose “Measure” (rectangle highlighted).Statistical analysis
Use the following formula to determine the corrected integrated intensity values for each cell: Corrected IntDen (cell) = IntDen (cell) - Area (cell) × Mean (background).
Plot the corrected integrated intensity values of HK-2 cells into column graphs and assess statistical significance with the Student’s t-test using Prism GraphPad (Figure 3).

Figure 3. Statistical analysis for fluorescence intensity of Nrf2 and MRPL12 expression in Nrf2 overexpressed or knocked down HK-2 cells. Data are presented as the mean ± S.D. (n = 3). *P < 0.05, **P < 0.01, ***P< 0.001. Figure 3 reprinted with permission from Gu et al. (2021) .
Notes
It is also feasible to transfect siRNAs or overexpression plasmids in 6-well plates and passage the transfected cells to a 24-well plate.
Protect the cover glass from drying out throughout the entire procedure. For example, change solution quickly and ensure that solutions perfectly cover the cells.
To ensure cells will not be rinsed out during the whole protocol, place the pipette tip at the edge of the well to draw up the fluid and gently add the new solution to the sidewall of the well.
Pause point: There could be a pause after rinsing the fixative. The fixed samples can be stored at 4°C for approximately 1 week if required. Nevertheless, we suggest continuing with the next procedures immediately. Longer storage is not recommended.
The two antibodies must have different species origins, and the secondary antibodies must have different exciting light wavelengths.
The concentration of the antibody may be determined based on antibody instructions or on data from a concentration gradient test done as a preliminary experiment to find the optimum dosage.
To reduce antibody use, it is advisable to put coverslips on the parafilm to incubate the antibody. You can directly add the diluted antibody to the well without putting the cover glass on the parafilm.
To control for nonspecific staining, “no primary antibody” and “no secondary antibody” controls should be performed for each new antibody. In addition, at least a “no primary antibody control” should be performed for each colocalization experiment.
Recipes
Cell culture medium
Low glucose (1g/L) DMEM medium containing:
10% FBS
1% penicillin-streptomycin
PBS
Dissolve the PBS powder in double distilled water, adjusting the pH to 7.2-7.4.
Permeabilization solution
PBS containing:
0.5% Triton X-100
Note: It should be freshly prepared.
Blocking solution
PBS containing:
1% normal goat serum
Note: It should be freshly prepared.
Acknowledgments
This protocol was adapted from the previously published paper (Gu et al., 2021). This work was support by funds from the National Natural Science Foundation of China (grants numbers 81770729, 91749111, 82070756); Shandong Province Taishan Scholar Project (grants numbers tsqn20161073).
Competing interests
The authors declare no financial or non-financial conflicts of interest within this work.
References
- Dinkova-Kostova, A. T. and Abramov, A. Y. (2015). The emerging role of Nrf2 in mitochondrial function. Free Radic Biol Med 88(Pt B): 179-188.
- Gu, X., Liu, Y., Wang, N., Zhen, J., Zhang, B., Hou, S., Cui, Z., Wan, Q. and Feng, H. (2021). Transcription of MRPL12 regulated by Nrf2 contributes to the mitochondrial dysfunction in diabetic kidney disease. Free Radic Biol Med 164: 329-340.
- Hayes, J. D. and Dinkova-Kostova, A. T. (2014). The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci 39(4): 199-218.
- Kühl, I., Miranda, M., Posse, V., Milenkovic, D., Mourier, A., Siira, S. J., Bonekamp, N. A., Neumann, U., Filipovska, A., Polosa, P. L., Gustafsson, C. M. and Larsson, N. G. (2016). POLRMT regulates the switch between replication primer formation and gene expression of mammalian mtDNA. Sci Adv 2(8): e1600963.
- Marty, L. and Fort, P. (1996). A delayed-early response nuclear gene encoding MRPL12, the mitochondrial homologue to the bacterial translational regulator L7/L12 protein. J Biol Chem 271(19):11468-76.
- Ma, Y., Zhu, S., Lv, T., Gu, X., Feng, H., Zhen, J., Xin, W. and Wan, Q. (2020). SQSTM1/p62 controls mtDNA expression and participates in mitochondrial energetic adaption via MRPL12. iScience 23(8): 101428.
- Nouws, J., Goswami, A. V., Bestwick, M., McCann, B. J., Surovtseva, Y. V. and Shadel, G. S. (2016). Mitochondrial ribosomal protein L12 is required for POLRMT stability and exists as two forms generated by alternative proteolysis during import. J Biol Chem 291(2): 989-997.
- Surovtseva,. Y. V., Shutt, T. E., Cotney, J., Cimen, H., Chen, S. Y., Koc, E. C. and Shadel, G. S. (2011). Mitochondrial ribosomal protein L12 selectively associates with human mitochondrial RNA polymerase to activate transcription. Proc Natl Acad Sci USA 108(44): 17921-17926.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Lu, Y., Gu, X., Wan, Q., Feng, H. and Liu, Y. (2021). Co-immunofluorescence of MRPL12 and Nrf2 in HK2 Cells. Bio-protocol 11(20): e4191. DOI: 10.21769/BioProtoc.4191.
Category
Biochemistry > Protein > Fluorescence
Molecular Biology > Protein > Expression
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








