- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Glucose Starvation, Magnesium Ion Starvation, and Bile Stress Assays
Published: Vol 11, Iss 18, Sep 20, 2021 DOI: 10.21769/BioProtoc.4157 Views: 2961
Reviewed by: Alexandros AlexandratosElizabeth LibbyAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

ODELAM: Rapid Sequence-independent Detection of Drug Resistance in Mycobacterium tuberculosis Isolates
Thurston Herricks [...] John D. Aitchison
May 20, 2021 4732 Views

Analysis of Heterocyst and Akinete Specific Glycolipids in Cyanobacteria Using Thin-layer Chromatography
Ritu Garg [...] Iris Maldener
Mar 20, 2022 2451 Views

Detecting Photoactivatable Cre-mediated Gene Deletion Efficiency in Escherichia coli
Yuta Koganezawa [...] Miki Umetani
Jun 5, 2023 1932 Views
Abstract
Salmonella enterica serovar Enteritidis (S. Enteritidis) is a leading causative pathogen for food-borne gastroenteritis. During its course of infection, it confronts myriads of physiological barriers inside the host, such as nutrient deprivation, low micronutrient availability, and toxicity from bile salts, to promote bacterial survival and infection inside the host. The ability of the pathogen to overcome these stressful conditions determines the degree of virulence in the host. Therefore, assessment of the survival of a pathogen during different stress conditions, like glucose starvation, magnesium starvation, and bile stress, are important parameters to assess the virulence of the pathogen. Here, we describe protocols for estimating the survival of the pathogen during the above-mentioned stress conditions. We culture S. Enteritidis in an appropriate growth medium to a required O.D.600 and treat it with glucose starvation (M9 minimal culture medium containing 0.03% glucose), magnesium starvation (M9 minimal culture medium containing 20 µM MgSO4), and bile stress (bacterial cells treated with 15% bile salts in Luria Bertani (LB) culture medium) conditions. The number of surviving bacteria is obtained after the treatment by calculating the colony-forming units (CFU) of the surviving pathogen obtained on LB agar plates at relevant time intervals. The experiments are performed in biological replicates, and statistical analysis is performed to validate the experimental findings. The methodology of these stress response assays is simple and can be adapted to study the pathogenesis and stress response in other relevant and culturable enteric pathogens.
Keywords: Food-borne gastroenteritisBackground
Salmonella enterica serovar Enteritidis (S. Enteritidis) is one of the four leading enteric pathogens that cause food-borne gastroenteritis. About 93 million infections are reported every year caused as a result of non-typhoidal salmonellosis (Balasubramanian et al., 2018). It poses a severe clinical and economic burden, especially in developing countries, due to poor sanitation, hygiene, and improper food sterilization practices. Approximately 155,000 deaths are reported worldwide every year arising from non-typhoidal salmonellosis (Balasubramanian et al., 2018). During infection by S. Enteritidis, the pathogen confronts a wide range of physiological barriers or stress factors in the host system, such as temperature, antimicrobial peptides, nutrient-starved conditions (e.g., glucose limitation in the gut), micronutrients (e.g., Mg2+, Mn2+, Ca2+, and Fe3+) starvation, high osmolarity and hypoxia, bile salts, and oxidative and nitrosative stresses (Fang et al., 2016). The pathogen employs a set of stress responses and virulence genes to survive during these hostile conditions that indirectly determine its resilience during host-pathogen interactions (Fang et al., 2016). In particular, stress factors like glucose starvation are encountered by the pathogen in the anaerobic environment of the gut (Bowden et al., 2009; Arunima et al., 2020a). Mg2+ starvation conditions are encountered within the phagocytic environment (Connor et al., 2009; Choi et al., 2019; Arunima et al., 2020a and 2020b), and Bile salts are encountered as the pathogen transits through the liver and small intestine during infection (Spector et al., 1999; Kenyon et al., 2002 and 2005; Rychlik and Barrow, 2005; Álvarez-Ordóñez et al., 2011; Hernández et al., 2012; Arunima et al., 2020a). While these host factors exert antimicrobial effects on pathogen survival during infection (Fang et al., 2016), Salmonella serovars are known to overcome these stressful conditions (Rychlik and Barrow, 2005). The ability of the pathogen to survive these factors has been previously documented to determine the degree of virulence within the host system (GarcíaVéscov et al., 1996; Merritt and Donaldson, 2009; Waligora et al., 2014; Arunima et al., 2020a and 2020b). Hence, assessing survival during glucose and Mg2+ starvation and bile stress is critical to studying the pathogenesis of Salmonella infection.
In this article, we have described the protocols for assessing the survival of S. Enteritidis and its derivative strains during glucose starvation, Mg2+ starvation, and bile stress, to phenotypically understand the role of genetic factors that can contribute to its stress responses. To note, the protocols have been widely adopted in our previously published work (Arunima et al., 2020a and 2020b), and similar techniques were implemented in work by others (GarcíaVéscov et al., 1996; Soncini et al., 1996; Groisman et al., 1997; Spector et al., 1999; Kenyon et al., 2002 and 2005; Prouty et al., 2004; Abdallah et al., 2007; Merritt and Donaldson, 2009; Hernández et al., 2012; Moreira et al., 2013; Lofton et al., 2014). Here, we provide the protocols to estimate the survival of S. Enteritidis following treatment with these stresses in vitro to facilitate their adoption by the scientific community. The methodology of these stress response assays is simple and can be expanded to study the stress response in other relevant and culturable enteric pathogens.
Materials and Reagents
0.22 µm pore Size, PVDF Sterile hydrophilic syringe membrane filter (Merck, Millex® 33 mm PVDF 0.22 µm Sterile RUO, catalog number: SLGVR33RS)
15 ml Sterile Falcon tubes (Tarsons, Sterile Centrifuge Tube Conical Bottom, catalog number: 546021)
50 ml Sterile Falcon tubes (Tarsons, Sterile Centrifuge Tube Conical Bottom, catalog number: 546041)
1.5 ml Microcentrifuge tubes (Tarsons, Polypropylene Spinwin Micro Centrifuge Tube 1.5 ml, catalog number: 500010)
1.5 ml Microcentrifuge tubes rack (Scientific Research & Instruments, 1.5 ml Microcentrifuge Tubes Places 24 (6 × 4 Array), catalog number: P20201)
Pipette Tips (Tarsons, Micro Tips, catalog numbers: 521000, 521010, 521020)
90 mM Disposable Petri Dishes (Tarsons, Petri dish Radiation Sterile 90 mm, catalog number: 460095)
Borosilicate glass rim culture tubes (Borosil, 18 × 150 mm Culture Tubes with Rim, catalog number: 9800U06)
Salmonella enterica serovar Enteritidis str. P125109
Ethanol pure ≥99.5% (Himedia, catalog number: MB106)
Agar Agar (Himedia, Agar Agar, Type I, catalog number: GRM666)
Streptomycin Sulfate (Himedia, catalog number: CMS220)
Casamino acids (Himedia, Casein Enzyme Hydrolysate, catalog number: CR014)
Glucose (Himedia, D-(+)-Glucose anhydrous, catalog number: MB037)
CaCl2·2H2O (Sigma-Aldrich, catalog number: C3306)
MgSO4·7H2O (Sigma-Aldrich, catalog number: M2773)
Bile salts (Himedia, catalog number: RM009)
Na2HPO4·7H2O (Himedia, catalog number: GRM1417)
KH2PO4 (Sisco Research Laboratories Pvt. Ltd.(SRL), catalog number: 52403)
NaCl (Fisher Scientific, catalog number: 27605)
NH4Cl (Sisco Research Laboratories Pvt. Ltd.(SRL), catalog number: 96452)
KCl (Fisher Scientific, catalog number: 19255)
Sterile 1 M MgSO4 solution (see Recipes)
Sterile 1 M CaCl2 solution (see Recipes)
Sterile 20 % Glucose (see Recipes)
Sterile 20 % Casamino acids (see Recipes)
Sterile 5× M9 salt see Recipes)
Sterile M9 minimal medium (see Recipes)
Sterile M9 minimal medium containing 0.03% glucose (see Recipes)
Sterile M9 minimal medium containing 20 µM MgSO4 (see Recipes)
50 mg/ml Streptomycin Sulfate solution (see Recipes)
Sterile 1× Phosphate-buffered saline (PBS) buffer (see Recipes)
30% Bile Salts (see Recipes)
Luria Bertani (LB) medium (Himedia, Luria Bertani Broth, Miller, catalog number: M1245) (see Recipes)
LB agar (see Recipes)
Equipment
Hand Tally Counter (Humboldt, catalog number: H-9700)
Pipettes EP-2.5, EP-20, EP-200, EP-1000 (Eppendorf, Research® 2100 Series single channel adjustable volume pipettes, model: 0.1-2.5 µl, 2-20 µl, 20-200 µl, 100-1,000 µl)
-20°C refrigerator (CelFrost, catalog number: BFS-150) and 4°C refrigerator (LG, catalog number: GL-T402LPZU)
250 ml borosilicate glass conical flask with screw caps (Borosil, Conical With Screw Cap Flasks Capacity 250 ml, catalog number: 5021021)
500 ml and 250 ml borosilicate glass reagent bottles (Borosil, catalog numbers: 1501024, 1501021)
500 ml borosilicate glass beaker (Borosil, catalog number: BRL_1000D24)
Autoclavable beaker (Tarsons, catalog number: 422040)
500 ml and 100 ml measuring cylinder (Tarsons, catalog numbers: 345060, 345040)
Stirring hotplate
Autoclave (Osworld Scientific Equipments Pvt. Ltd, Osworld, Autoclave Vertical Top Opening Steam Sterilizer, OAT G – 125)
Burner
Biosafety cabinet (Nuaire, Class II Biosafety Cabinet Sliding Window, model: NU-425-600S)
Incubator for bacterial culture at stable temperature (Eppendorf, New BrunswickTM Innova® 42 R Incubator, and Refrigerated Shaker, catalog number: M1335-0016)
Centrifuge (Eppendorf, model: 5424 R)
Spectrophotometer (Agilent Cary 60 UV-visible light (UV-Vis) spectrophotometer)
Inoculating turntable (ISOLAB Laborgeräte GmbH, catalog number: 6286850)
Metal rod cell spreader (Fisher Scientific, catalog number: 08-769-2B)
Inoculation loop (Himedia, model: LA023)
Software
GraphPad Prism v. 7.0
Microsoft® Excel
Procedure
Note: All the bacteriological growth and survival assays should be carried out in an aseptic environment, inside a Class II-Biosafety Cabinet. Use sterile micro tips, culture medium, flasks, and Petri dishes in all experiments.
Storage of bacterial strains (S. Enteritidis)
Grow the Salmonella strains in nutrient-rich LB medium at 37°C and 150 rpm until the logarithmic (log)-phase is achieved.
Store bacteria in 50% glycerol. Add 300 µl of (log)-phase culture to 700 µl of 50% glycerol. Mix it gently by inverting the cryo-chill tubes.
Streak S. Enteritidis and its derivative strains before initiating experiments on LB agar plates containing 50 µg/ml Streptomycin (see Recipes). You can streak with micro-tips or a sterile inoculation loop.
Grow the strains by placing the streaked LB plates in the incubator at 37°C overnight.
Note: The time required to reach a particular O.D. may vary depending on the incubator, temperature, and rpm used to culture the bacterial strains. Thus, it is necessary to determine the growth curve pattern of the strains before conducting stress survival assays, especially if the role of the target genes during stress is growth phase-dependent.
Growth curve analysis of bacterial strains
Pick one bacterial colony from the bacterial LB agar plates with sterile 200 µl micro-tips [For example, S. Enteritidis (WT) and S. Enteritidis (mutant X) will serve as the representative strains for the protocols in this article].
Inoculate the bacterial strains (one colony each) in 5 ml of growth medium (LB or M9 minimal medium) in culture test tubes.
Culture the strains overnight (12-16) h at 37°C and150 rpm.
Sub-culture from the overnight bacterial culture in sterile conical flasks containing 100 ml of growth medium at 1:100 ratio (1 ml of overnight bacterial culture in 100 ml of LB or M9 minimal medium).
Collect 1 ml of the culture of each bacterial strain in 1.5 ml microcentrifuge tubes at 0 h (just at the time of sub-culturing), 1 h, 2 h, and up to 24 h.
Measure the O.D. of the cultures at an hourly interval (0 h, 1 h, and up to 24 h) in a UV-Spectrophotometer at λmax of 600 nm.
Take note of the O.D. at different time points in an excel sheet.
Repeat the experiment in biological replicates on different days. Acquire the data and save the raw data in an excel sheet.
Plot the O.D.600 of the strains versus the time (in hours) using GraphPad Prism (see Figure 1 and Figure 2).
Take note of the time and the culture growth conditions required to reach a particular desired O.D. to carry out the subsequent experiments.
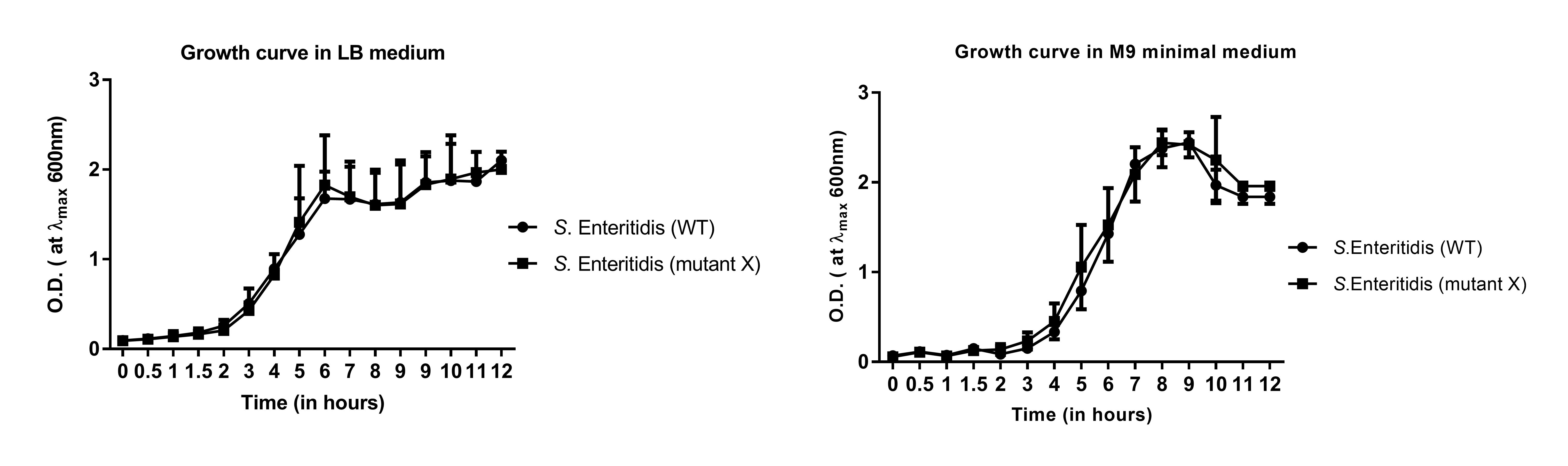
Figure 1. Representative growth curve analysis of S. Enteritidis strains in LB medium (A) and M9 minimal medium (B). The graph represents the O.D. of the bacterial strains (representative S. Enteritidis WT and mutant strains) across different time points up to 12 h.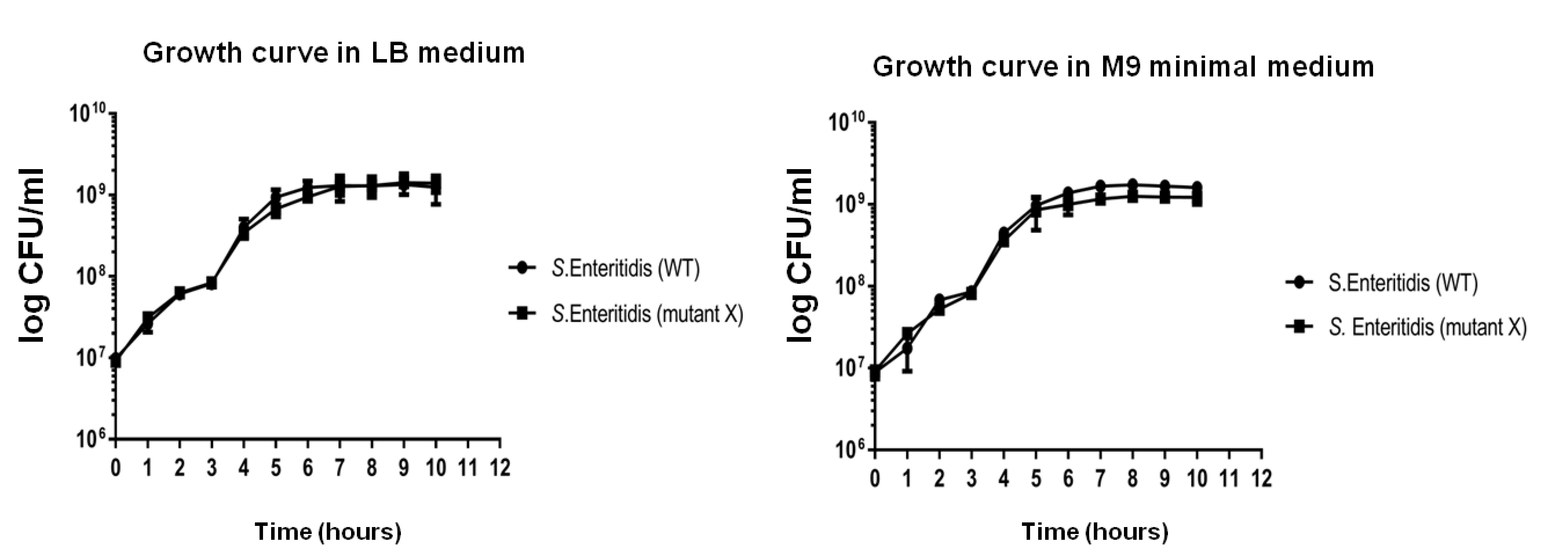
Figure 2. Representative growth curve analysis of S. Enteritidis strains in LB medium (A) and M9 minimal medium (B). The graph represents the logCFU/mL of the bacterial strains (representative S. Enteritidis WT and mutant strains) across different time points up to 12 h. The approximate doubling time of WT is 22.2 min in LB medium and 20.3 min in M9 minimal medium.You can collect 200 µl of culture at the indicated time points (above) for assessing the CFU of the strains at the corresponding O.D. and time points.
Serially dilute all the cultures collected with 1× PBS at 10-1, 10-2, 10-3, 10-4, and 10-5 dilution (100 µl culture+ 900 µl 1× PBS).
Place the LB agar plate containing Streptomycin (50 µg/ml) on the turntable. Spread plate 50 µl of each dilution onto the LB agar plates with a sterile cell spreader. Label the bottom of each LB agar plate with the organism name, time, and the dilution factor.
Note: Dry the plates completely to avoid condensation on the lid of the Petri dish. Bring the plates to room temperature before spread-plating.
Incubate the plates at 37°C overnight to obtain the bacterial colonies.
Glucose Starvation Assay
Pick one bacterial colony from the bacterial LB agar plates with sterile 200 µl micro-tips.
Inoculate the bacterial strains (one colony each) in 5 ml of M9 minimal medium (0.4% glucose) in culture test tubes (see Recipes).
Culture the strains overnight (12-16) h at 37°C and 150 rpm.
Sub-culture from the overnight bacterial culture in two sets of sterile conical flasks containing 100 ml of M9 minimal medium (0.4% glucose) at a 1:100 ratio (1 ml of overnight bacterial culture into 100 ml of M9 minimal medium) at 37°C and 150 rpm.
Monitor the O.D.600 of strains intermittently, until an O.D.600 of 0.4 (early log phase) is reached.
Note: Most stress response genes are growth phase-dependent. Depending on your study requirement, choose your growth phase to carry stress response survival assays. In this protocol, we have conducted stress response assays in early log phase culture, which is denoted by O.D.600 of 0.4.
Culture one set of flasks with strains in 100 ml of M9 minimal medium (0.4% glucose) at a 1:100 ratio, at 37°C and 150 rpm for the entire experiment. It will serve as an experimental control to monitor the growth rate of the pathogen, which should be similar (not growth defective) under normal culture conditions.
To achieve this, transfer one set of log-phase bacterial cultures obtained from step 4 into 50 ml Falcon tubes. Recover all the bacterial cells from the culture by centrifuging at 3,000 rpm for 10 min at room temperature. Remove the media supernatant.
Wash the cells twice with M9 buffer (M9 minimal medium without casamino acids and glucose) by resuspending the bacterial pellet in 20-30 ml of M9 buffer. Centrifuge at 3,000 rpm for 10 min at room temperature. Remove the M9 buffer supernatant. This step is done to remove M9 minimal medium traces from the bacterial cells.
Resuspend the washed pellet in 1 ml of M9 buffer. Transfer the culture into a sterile conical flask with M9 minimal medium containing 0.4% glucose. At this point, collect the culture for serial dilution and plating on LB agar immediately. This time point will serve as 0 h.
The growth will be monitored by collecting the cultures at 1 h, 2 h, 3 h, and 4 h of growth in normal M9 minimal medium. Serially dilute them and plate them on LB agar containing Streptomycin (50 µg/ml). The CFU at these time points will serve as control compared to the CFU of strains grown in glucose-starved conditions.
To create glucose-starved medium, transfer the second set of log-phase cultures obtained from step 4 into 50 ml Falcon tubes. Recover all the bacterial cells from the culture by centrifuging at 3,000 rpm for 10 min at room temperature. Remove the media supernatant.
Wash the cells twice with M9 buffer (M9 minimal medium without casamino acids and glucose) by resuspending the bacterial pellet in 20-30 ml of M9 buffer. Centrifuge at 3,000 rpm for 10 min at room temperature. Remove the M9 buffer supernatant. This step is done to remove M9 minimal medium traces from the bacterial cells.
Resuspend the washed pellet in 1 ml of M9 buffer. Transfer the culture into a sterile conical flask with M9 minimal medium containing 0.03% glucose (see Recipes). This medium will serve as the glucose-starved medium for the strains. Collect the culture for 0 h time point.
Culture the strains in glucose-starved medium at 37°C and 150 rpm. Collect the cultures after 1 h, 2 h, 3 h, and 4 h of exposure to get the glucose starvation data.
Note: You can collect for longer time points if the bacterial cells are surviving beyond 4 h. The time of analysis is subjected to the survival of your bacterial strain post-exposure to stress.
Serially dilute all the cultures collected at different time points with 1× PBS at 10-1, 10-2, 10-3, 10-4, and 10-5 dilution (100 µl culture+ 900 µl 1× PBS).
Place the LB agar plate containing Streptomycin (50 µg/ml) on the turntable. Spread plate 50 µl of each dilution onto the LB agar plates with a sterile cell spreader. Label the bottom of each LB agar plate with the organism name, time following treatment with stress, and the dilution factor.
Note: Dry the plates completely to avoid condensation on the lid of the Petri dish. Bring the plates to room temperature before spread-plating.
Incubate the plates at 37 °C overnight to recover the surviving bacterial colonies.
Magnesium (Mg2+) Starvation Assay
Pick one bacterial colony from the bacterial LB agar plates with sterile 200 µl micro-tips.
Inoculate the bacterial strains (one colony each) in 5 ml of M9 minimal medium (Optimal working concentration of MgSO4: 200 µM) in culture test tubes (see Recipes).
Culture the strains overnight (12-16) h at 37°C and 150 rpm.
Sub-culture from the overnight bacterial culture in two sets of sterile conical flasks containing 100 ml of M9 minimal medium (200 µM MgSO4) at 1:100 ratio (1 ml of overnight bacterial culture into 100 ml of M9 minimal medium) at 37°C and 150 rpm.
Monitor the O.D.600 of strains intermittently, until an O.D.600 of 0.4 (early log phase) is reached.
Culture one set of flasks with strains in 100 ml of M9 minimal medium (200 µM MgSO4) at a 1:100 ratio (1 ml of overnight bacterial culture into 100 ml of medium) at 37°C and 150 rpm for the entire experiment. It will serve as an experimental control to monitor the growth rate of the pathogen, which should be similar (not growth defective) under normal culture conditions.
To achieve this, transfer one set of log-phase bacterial cultures obtained from step 4 into 50 ml Falcon tubes. Recover all the bacterial cells from the culture by centrifuging at 3,000 rpm for 10 min at room temperature. Remove the media supernatant.
Wash the cells twice with M9 buffer (M9 minimal medium without casamino acids, glucose, and MgSO4) by resuspending the bacterial pellet in 20-30 ml of M9 buffer. Centrifuge at 3,000 rpm for 10 min at room temperature. Remove the M9 buffer supernatant. This step is done to remove M9 minimal medium traces from the bacterial cells.
Resuspend the washed pellet in 1 ml of M9 buffer. Transfer the culture into a sterile conical flask with M9 minimal medium containing 200 µM MgSO4. At this point, collect the culture for serial dilution and plating on LB agar immediately. This time point will serve as 0 h.
Growth will be monitored by collecting cultures at 1 h, 2 h, 3 h, and 4 h of growth in normal M9 minimal medium. Serially dilute them and plate them on LB agar containing Streptomycin (50 µg/ml). The CFU at these time points will serve as a control compared to the CFU of strains grown in Mg2+ starved conditions.
To create Mg2+ starved medium, transfer the second set of log-phase cultures obtained from step 4 into 50 ml Falcon tubes. Recover all the bacterial cells from the culture by centrifuging at 3,000 rpm for 10 min at room temperature. Remove the media supernatant.
Wash the cells twice with M9 buffer (M9 minimal medium without casamino acids, glucose, and MgSO4) by resuspending the bacterial pellet in 20-30 ml of M9 buffer. Centrifuge at 3,000 rpm for 10 min at room temperature. Remove the M9 buffer supernatant. This step is done to remove M9 minimal medium traces from the bacterial cells.
Resuspend the washed pellet in 1ml of M9 buffer. Transfer the culture into a sterile conical flask with M9 minimal medium containing 20 µM MgSO4 (see Recipes). This medium will serve as the Mg2+ starved medium for the strains. Collect culture for 0 h time point.
Note: You change MgSO4 of concentration (2 to ≤20 µM) in M9 minimal medium to mimic the Mg2+ starved environment in vitro.
Culture the strains in Mg2+ starved medium at 37°C and 150 rpm. Collect the cultures after 1 h, 2 h, 3 h, and 4 h of exposure to Mg2+ starvation.
Note: You can collect for longer time points. The time of analysis is subjected to the survival of your bacterial strain post-exposure to stress.
Serially dilute the cultures collected at different time points with 1× PBS at 10-1, 10-2, 10-3, 10-4, and 10-5 dilution (100 µl culture+ 900 µl 1× PBS).
Place the LB agar plate containing Streptomycin (50 µg/ml) on the turntable. Spread plate 50 µl of each dilution onto the LB agar plates with a sterile cell spreader. Label the bottom of each LB agar plate with the organism name, time following treatment with stress, and the dilution factor.
Note: Dry the plates completely to avoid condensation on the lid of the Petri dish. Bring the plates to room temperature before spread-plating.
Incubate the plates at 37°C overnight to recover the surviving bacterial colonies.
Bile Stress Assay
Note: The sensitivity to bile salts may vary depending on the bacterial strain and the commercial source. Before conducting assays with compounds that exhibit bactericidal/toxicity against the bacteria (like bile salts), determine the minimal inhibitory concentration (MIC) of the compound and use a concentration below it for studying the stress response of target strains against the reference strain. Reference the research articles by Hernández et al. (2012) and Wiegand et al. (2008) to calculate the MIC of bile salts for S. Enteritidis and other culturable (aerobic) micro-organisms. We calculated the MIC of bile salts for S. Enteritidis from 0.4 OD culture to be 30% (data not shown). Therefore, we took a Bile salt concentration of 15% for our experiment.
Pick one bacterial colony from the bacterial LB agar plates with sterile 200 µl micro-tips.
Inoculate the bacterial strains (one colony each) in 5 ml of LB medium in culture test tubes (see Recipes).
Culture the strains overnight (12-16) h at 37°C and 150 rpm.
Sub-culture from the overnight bacterial culture in sterile conical flasks containing 100 ml LB medium at 1:100 ratio (1 ml of overnight bacterial culture into 100 ml of LB medium) at 37°C and 150 rpm.
Monitor the O.D.600 of strains intermittently, until an O.D.600 of 0.4 (early log phase) is reached. At this point, collect 100 µl of culture. Serially dilute the culture in 1× PBS at 10-1, 10-2, 10-3, 10-4, and 10-5 dilution (Mix 100 µl culture+ 900 µl 1× PBS). Plate 50µl of each dilution onto LB agar plate containing Streptomycin (50 µg/ml). This will serve as the reference CFU of culture before treatment with bile stress (control, 0 h time point).
Collect 1 ml of log-phase culture for each strain in three individual 2 ml micro-centrifuge tubes. Add Bile salts to a final concentration of 15% from the 30% bile salts stock solution [1 ml of bile salts (30%) + 1 ml of bacterial culture].
Incubate the strains at 37°C for 1 h, 2 h, 3 h, and 4 h following treatment with bile salts.
Note: You can collect for longer time points. The time of analysis is subjected to the survival of your bacterial strain post-exposure to stress.
Serially dilute the cultures following treatment at the indicated time points with 1× PBS at 10-1, 10-2, 10-3, 10-4, and 10-5 dilution (100 µl culture+ 900 µl 1× PBS). Plate 50µl of each dilution onto LB agar plate containing Streptomycin (50 µg/ml).
Place the LB agar plate containing Streptomycin (50 µg/ml) on the turntable. Spread plate 50 µl of each dilution onto the LB agar plates with a sterile cell spreader. Label the bottom of each LB agar plate with the organism name, time following treatment with stress, and the dilution factor.
Incubate the plates at 37°C overnight to recover the surviving bacterial colonies.
Estimation of bacterial survival following stress treatments by of CFU/ ml calculation
The next day after the stress assay, you should have isolated colonies for each bacterial strain and the control strain (surviving colonies) on the LB agar plates at an appropriate dilution factor
Count the number of colonies using a Hand Tally Counter (Humboldt). Digital counters or image-based counters can also be used.
Make an entry of the number of colonies recovered before and after treatment with stress in an excel sheet. Save the raw data for each technical replicate and biological replicate of the experiment.
The number of surviving bacteria is calculated as described below.
If “X" is the number of bacterial colonies obtained on an LB agar plate at the dilution factor “Y” after plating 50 µl on an LB agar plate, then
CFU per 50 µl = X * 10Y
CFU per 1 µl= X * 10Y/ 50 µl
CFU per ml = (X * 10Y/ 50 µl) * 1000 µl
= (20 * X * 10Y) CFU/ml
Note: For a representative image, please see Figure 1B (Glucose starvation assay), 1C (Mg2+ starvation assay), and 1D (Bile stress assay) from Arunima et al. (2020a).
Data analysis
Each experiment should be conducted in three replicates made on different days, and using different bacterial stocks, different media stocks, and, if possible, different analysts.
The final data corresponds to the average CFU/ ml of the bacterial strains obtained from three biological experiments.
Plot the average CFU/ml of bacteria before treatment with stress (as the control) and average the CFU/ml of bacterial strains after treatment with different stress conditions at the other indicated time points.
We have used GraphPad Prism v. 7.0. for analysis, but you can use other software. Enter the CFU/ml of surviving bacterial strains (average of one biological replicate) into each Group column of a Grouped analysis, and enter the time points in the Row Title column.
The graph generated will be a line graph depicting the CFU/ml of different bacterial strains compared to the reference strain (Y-axis) versus the Time (X-Axis). You can also plot the graph as Bar±SD graph (survival) versus Time (in hours on X-Axis) (see Figure 1B, 1C, and 1D of Arunima et al., 2020a).
Use two-way ANOVA analysis to conduct the statistical analysis for the stress conditions. P-value < 0.05, P < 0.01 and P < 0.001 are considered statistically significant; P ≥ 0.05 is not significant.
Notes
Run a negative control in every experiment to check for contamination. A negative control will be the growth medium only (LB and M9 minimal medium without bacteria) that is processed in the same manner as the cultures in the experiment.
Carry out the experiments in aseptic conditions.
Recipes
LB medium
Weigh 25 g of LB medium (Himedia).
Suspend it in 1,000 ml double distilled water.
Stir mix on Stir plate.
Dispense in tubes or flasks as required.
Autoclave at 15 lbs pressure (121°C) for 20 min.
Once cooled, supplement necessary antibiotics for culturing of microbes.
Note: The formulation of the LB medium used for the experiment is the LB-Lennox medium.
LB Agar
Prepare LB medium as described above.
Add 7.5 g agar to 500 ml of LB medium, swirl it gently.
Autoclave at 15 lbs pressure (121°C) for 20 min.
Cool it to 55°C, and add Streptomycin (50 µg/ml) to the LB agar.
Swirl gently to avoid bubbles.
If using more than one antibiotic, add and label the plates accordingly.
Measure 30-35 ml of LB agar in a sterile 50 ml falcon tube. Then pour it into each sterile 90 mm Petri dish.
Note: The volume of LB agar medium should be maintained uniformly for each set of experiments to minimize variability from growth differences in CFU and survival assays. LB containing Streptomycin is used in the experiment because S. Enteritidis is naturally resistant to Streptomycin.
After the medium solidifies, invert the Petri dishes and store them at 4°C until required.
Remove the plates from storage before 2-3 h of using them.
Sterile 1 M CaCl2 Solution
Dissolve 11.1 gm of CaCl2·2H2O in 100 ml of Milli Q water.
Autoclave at 15 lbs pressure (121°C) for 20 min.
Aliquot into small volumes to avoid contamination of main stock.
Store it at room temperature.
Sterile 1 M MgSO4 Solution
Dissolve 24.65 g of MgSO4·7H2O in 100 ml of Milli Q water.
Autoclave at 15 lbs pressure (121°C) for 20 min.
Aliquot into small volumes to avoid contamination of main stock.
Store it at room temperature.
20% glucose
Dissolve 20 g of glucose in 100 ml of autoclaved Milli Q water.
Syringe Filter it through 0.22 µm PVDF membrane, do not autoclave.
Aliquot into small volumes to avoid contamination of main stock.
Store it at 4°C.
20% Casamino acids
Dissolve 20 g of Casein hydrolysate in 100 ml of autoclaved Milli Q water.
Syringe Filter it through 0.22 µm PVDF membrane, do not autoclave.
Aliquot into small volumes to avoid contamination of main stock.
Store it at 4°C.
50mg/ml Streptomycin Solution
Dissolve 50 mg of Streptomycin Sulfate in 1 ml of autoclaved Milli Q water.
Syringe Filter it through 0.22 µm PVDF membrane, do not autoclave.
Store it at -20°C.
Sterile 1× PBS buffer
Dissolve 8 g of NaCl, 0.2 g of KCl, 1.44 g of Na2HPO4, and 0.24 g of KH2PO4 in 500 ml of Milli Q water.
Adjust the pH to 7.4 with HCl.
Add Milli Q water to a total volume of 1 L.
Autoclave at 15 lbs pressure (121°C) for 20 min.
Store it at room temperature.
You can also make 10× PBS buffer and dilute it to 1× PBS buffer for use. Please follow Cold Spring Harbor Protocol (http://cshprotocols.cshlp.org/content/2006/1/pdb.rec8247).
Sterile 5× M9 Salts
Dissolve 64 g of Na2HPO4·7H2O, 15 g of KH2PO4, 2.5 g of NaCl, and 5 g of NH4Cl in 800 ml of Milli Q water.
Adjust the volume to 1,000 ml by adding water.
Autoclave at 15 lbs pressure (121°C) for 20 min.
Store it at room temperature.
Sterile M9 minimal medium
Add 20 ml of 5× M9 salt.
Add 200 µl of 1 M MgSO4 (Working concentration: 200 µM).
Add 10 µl of 1 M CaCl2 (Working concentration: 100 µM).
Add 2 ml of 20% glucose (Working Concentration: 0.4%).
Add 500 µl of 20% Casamino acids (Working Concentration: 0.1%).
Add all the above components and adjust the volume to 100 ml of autoclaved double distilled water. Use an autoclaved measuring cylinder for quantifying all ingredients.
Aliquot them to autoclaved 5 ml culture test tubes and 250 ml conical flasks for bacterial culture and experiments.
To note, add glucose and casamino acids freshly before use. M9 minimal medium without glucose and casamino acid (M9 buffer) can be stored at room temperature.
Sterile M9 minimal medium containing 0.03% glucose
Add 20 ml of 5× M9 salt.
Add 200 µl of 1 M MgSO4 ( Working concentration: 200 µM).
Add 10 µl of 1 M CaCl2 (Working concentration: 100 µM).
Add 150 µl of 20% glucose (Working Concentration: 0.03%).
Add 500 µl of 20% Casamino acids (Working Concentration: 0.1%).
Add all the above components and adjust the volume to 100 ml of autoclaved double distilled water.
Aliquot them to autoclaved 5ml culture test tubes and 250 ml conical flasks for bacterial culture and experiments.
Sterile M9 minimal medium containing 20 µM MgSO4
Add 20 ml of 5× M9 salt.
Add 20 µl of 1 M MgSO4 ( Working concentration: 20 µM).
Add 10 µl of 1 M CaCl2 (Working concentration: 100 µM).
Add 150 µl of 20% glucose (Working Concentration: 0.03%).
Add 500 µl of 20% Casamino acids (Working Concentration: 0.1%).
Add all the above components and adjust the volume to 100 ml of autoclaved double distilled water.
Aliquot them to autoclaved 5 ml culture test tubes and 250 ml conical flasks for bacterial culture and experiments.
30% Bile salts
Dissolve 30 g of Bile salts mixture to 100 ml of autoclaved double distilled water.
Syringe Filter it through 0.22 µm PVDF membrane. Do not autoclave.
Store it at room temperature.
Acknowledgments
The glucose starvation assay was derived and modified from previous work (Spector and Cubitt, 1992; O’Neal et al., 1994; Kenyon et al., 2005). The magnesium starvation assay was adapted from the work by Moreira et al. (2013). The bile stress assay was derived and modified from Hernández et al. (2012). These modified protocols were originally implemented and described in Arunima et al. (2020a).
Competing interests
The authors declare no financial and non-financial competing interests associated with this work.
References
- Abdallah, J., Caldas, T., Kthiri, F., Kern, R. and Richarme, G. (2007). YhbO protects cells against multiple stresses. J Bacteriol 189: 9140-9144.
- Álvarez-Ordóñez, A., Begley, M., Prieto, M., Messens, W., López, M., Bernardo, A. and Hill, C. (2011). Salmonella spp. survival strategies within the host gastrointestinal tract. Microbiology 157: 3268-3281.
- Arunima, A., Swain, S. K., Patra, S. D., Das, S., Mohakud, N. K., Misra, N. and Suar, M. (2020a). Role of OB-Fold protein YdeI in stress response and virulence of Salmonella enterica serovar enteritidis. J Bacteriol 203: 1-21.
- Arunima, A., Swain, S.K., Ray, S., Prusty, B.K. and Suar, M. (2020b). RpoS-regulated SEN1538 gene promotes resistance to stress and influences Salmonella enterica serovar enteritidis virulence. Virulence 11: 295-314.
- Balasubramanian, R., Im, J., Lee, J. S., Jeon, H. J., Mogeni, O. D., Kim, J. H., Rakotozandrindrainy, R., Baker, S. and Marks, F. (2018). The global burden and epidemiology of invasive non-typhoidal Salmonella infections. Hum Vaccin Immunother00: 1-6.
- Bowden, S. D., Rowley, G., Hinton, J. C. D. and Thompson, A. (2009). Glucose and glycolysis are required for the successful infection of macrophages and mice by Salmonella enterica serovar typhimurium. Infect Immun 77: 3117-3126.
- Choi, S., Choi, E., Cho, Y. J., Nam, D., Lee, J. and Lee, E. J. (2019). The Salmonella virulence protein MgtC promotes phosphate uptake inside macrophages. Nat Commun 10(1):3326.
- Connor, K.O., Fletcher, S.A. and Csonka, L.N. (2009). Increased expression of Mg2+ transport proteins enhances the survival of Salmonella enterica at high temperature. Proc Natl Acad Sci 106: 17522-17527.
- Fang, F.C., Frawley, E.R., Tapscott, T. and Vázquez-Torres, A. (2016). Bacterial stress responses during host infection. Cell Host Microbe 20: 133-143.
- GarcíaVéscov, E., Soncini, F.C. and Groisman, E.A. (1996). Mg2+ as an extracellular signal: Environmental Regulation of Salmonella Virulence. Cell 84: 165-174.
- Groisman, E.A., Kayser, J. and Soncini, F.C. (1997). Regulation of Polymyxin Resistance and Adaptation to Low-Mg2+ Environments. J Bacteriol 179: 7040-7045.
- Hernández, S.B., Cota, I., Ducret, A., Aussel, L. and Casadesús, J. (2012). Adaptation and Preadaptation of Salmonella enterica to Bile. PLoS Genet 8: 1-15.
- Kenyon, W.J., Sayers, D.G., Humphreys, S., Roberts, M. and Spector, M.P. (2002). The starvation-stress response of Salmonella enterica serovar Typhimurium requires σE-, but not CpxR-regulated extracytoplasmic functions. Microbiology 148: 113-122.
- Kenyon, W. J., Thomas, S. M., Johnson, E., Pallen, M. J. and Spector, M. P. (2005). Shifts from glucose to certain secondary carbon-sources result in activation of the extracytoplasmic function sigma factor σE in Salmonella enterica serovar Typhimurium. Microbiology 151: 2373-2383.
- Lofton, H., Anwar, N., Rhen, M. and Andersson, D. I. (2014). Fitness of Salmonella mutants resistant to antimicrobial peptides. J Antimicrob Chemother 70: 432-440.
- Merritt, M. E. and Donaldson, J. R. (2009). Effect of bile salts on the DNA and membrane integrity of enteric bacteria. J Med Microbiol 58: 1533-1541.
- Moreira, C. G., Herrera, C. M., Needham, B. D., Parker, C. T., Libby, S. J., Fang, F. C., Trent, M. S. and Sperandio, V. (2013). Virulence and stress-related periplasmic protein (VisP) in bacterial/host associations. Proc Natl Acad Sci 110: 1470-1475.
- O’Neal, C. R., Gabriel, W. M., Turk, A. K., Libby, S. J., Fang, F.C. and Spector, M. P. (1994). RpoS is necessary for both the positive and negative regulation of starvation survival genes during phosphate, carbon, and nitrogen starvation in Salmonella typhimurium. J Bacteriol 176: 4610-4616.
- Prouty, A. M., Brodsky, I. E., Manos, J., Belas, R., Falkow, S. and Gunn, J. S. (2004). Transcriptional regulation of Salmonella enterica serovar Typhimurium genes by bile. FEMS Immunol Med Microbiol 41: 177-185.
- Rychlik, I. and Barrow, P. A. (2005). Salmonella stress management and its relevance to behaviour during intestinal colonisation and infection. FEMS Microbiol Rev 29: 1021-1040.
- Soncini, F. C., Véscovi, E. G., Solomon, F. and Groisman, E. A. (1996). Molecular basis of the magnesium deprivation response in Salmonella typhimurium: Identification of PhoP-regulated genes. J Bacteriol 178: 5092-5099.
- Spector, M. P. and Cubitt, C. L. (1992). Starvation-inducible loci of Salmonella typhimurium: regulation and roles in starvation-survival. Mol Microbiol 6: 1467-1476.
- Spector, M. P., Del Portillo, F. G., Bearson, S. M. D., Mahmud, A., Magut, M., Finlay, B. B., Dougan, G., Foster, J. W. and Pallen, M. J. (1999). The rpoS-dependent starvation-stress response locus stiA encodes a nitrate reductase (narZYWV) required for carbon-starvation-inducible thermotolerance and acid tolerance in Salmonella typhimurium. Microbiology 145: 3035-3045.
- Waligora, E. A., Fisher, C. R., Hanovice, N.J., Rodou, A., Wyckoff, E. E. and Payne, S. M. (2014). Role of intracellular carbon metabolism pathways in Shigella flexneri virulence. Infect Immun 82: 2746-2755.
- Wiegand, I., Hilpert, K., and Hancock, R. E. W. (2008). Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3: 163-175.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Arunima, A. and Suar, M. (2021). Glucose Starvation, Magnesium Ion Starvation, and Bile Stress Assays. Bio-protocol 11(18): e4157. DOI: 10.21769/BioProtoc.4157.
Category
Microbiology > Microbial physiology > Adaptation
Environmental science > Bacterium
Biological Sciences > Microbiology
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









