- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
One-step White Blood Cell Extracellular Staining Method for Flow Cytometry
(*contributed equally to this work) Published: Vol 11, Iss 16, Aug 20, 2021 DOI: 10.21769/BioProtoc.4135 Views: 4124
Reviewed by: Alessandro DidonnaJidnyasa IngaleLaura CampisiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Detection of Autophagy in Human Peripheral Blood Mononuclear Cells Using Guava® Autophagy and Flow Cytometry
Melanie Scherer [...] Jörg Bergemann
Sep 20, 2025 1413 Views

Cluster FLISA—A Method to Compare Protein Expression Efficiency Between Cell Lines and Subunit Clustering of Proteins
Sabrina Brockmöller and Lara Maria Molitor
Nov 5, 2025 1207 Views

Protocol for the Isolation and Analysis of Extracellular Vesicles From Peripheral Blood: Red Cell, Endothelial, and Platelet-Derived Extracellular Vesicles
Bhawani Yasassri Alvitigala [...] Lallindra Viranjan Gooneratne
Nov 5, 2025 1442 Views
Abstract
Flow cytometry is a powerful analytical technique that is increasingly used in scientific investigations and healthcare; however, it requires time-consuming, multi-step sample procedures, which limits its use to specialized laboratories. In this study, we propose a new universal one-step method in which white blood cell staining and red blood cell lysis are carried out in a single step, using a gentle lysis solution mixed with fluorescent antibody conjugates or probes in a dry or liquid format. The blood sample may be obtained from a routine venipuncture or directly from a fingerprick, allowing for near-patient analysis. This procedure enables the analysis of common white blood cell markers as well as markers related to infections or sepsis. This simpler and faster protocol may help to democratize the use of flow cytometry in the research and medical fields.
Graphic abstract:

One-step White Blood Cell Extracellular Staining Method for Flow Cytometry.
Background
Flow cytometry is increasingly used in scientific investigations and healthcare; however, it requires a time-consuming, multi-step sample preparation procedure, which limits its use to specialized laboratories. Most whole blood sample preparation protocols for extracellular staining consist of at least three steps: using a venous blood sample collected in an anticoagulated sampling tube, 50-100 µl is pipetted into a reaction tube; as a second step, fluorescent antibodies are mixed and incubated with the blood to allow white blood cell staining; as a third step, the lysis solution is added to the reaction tube to allow for red blood cell removal; finally, an optional washing step is performed to reduce non-specific fluorescence.
Due to these multiple timed steps that need to be carried out in specialized laboratories, the use of flow cytometry is limited. It is, for example, not yet routinely used in the clinical field, even though the assessment of patient immune cells has proven helpful in the indication of potential disease activity (Brown and Wittwer, 2000). Some authors have tried to establish protocols with only one step, proposing to remove the lysis step; however, this method necessitates a dedicated flow cytometer, restricting its use(Petriz et al., 2018).
In this study, we propose a new universal method in which white blood cell staining and red blood cell lysis are carried out in a single step. The lysis solution is stable at room temperature (RT) and does not interfere with the staining owing to its composition, which comprises a substrate that is specifically cleaved by an enzyme uniquely present in RBC membranes (van Agthoven, 2007). White cells are maintained at neutral pH under isotonic conditions and are therefore not affected. The lysis solution can include low-dose formaldehyde (0.05%) to stabilize markers of interest (Bourgoin et al., 2020c).
The fine titration of the conjugates and their direct dilution in the lysis solution enable the final washing step to be eliminated while maintaining low background levels. Using the new Dried Unitized Reagent Assays (DURA) Innovations technology, the conjugates can be dried, which enables RT storage and removes the need for pipetting, thereby supporting better standardization. The blood sample may be of venous origin or from a fingerprick, allowing for less invasive sampling suitable for a point-of-care setting. This approach is possible thanks to an excess volume of lysis solution, which permits efficient RBC lysis in up to 50 µl of blood. The conjugate quantities have also been adjusted to stain white blood cells contained in 2-50 µl blood, with a small quantity of blood no longer being a limitation because 2 µl blood contains an average of 10,000 leukocytes. As a 100-cell subpopulation is usually considered statistically representative, using 2 µl of blood allows for analysis of subpopulations as low as 1%, which is sufficient for most routine applications.
Since management of patients with infections in the Emergency Department (ED) is challenging for practitioners, we developed a panel that enables rapid patient triage in the ED using flow cytometry. It has been shown that monitoring the expression of CD169 on monocytes (mCD169), CD64 on neutrophils (nCD64), and HLA-DR on monocytes (mHLA-DR) by flow cytometry can be indicative of viral or bacterial infection, or sepsis, respectively. We therefore established a panel consisting of antibodies targeting these three markers and evaluated the one-step method in subjects with infection and septic conditions by measuring the expression of the three infection-related markers (Bourgoin et al., 2019a, 2019b, 2020a, 2020b, 2020c, and 2021; Bedin et al., 2020; Michel et al., 2020).
Many other applications can be envisioned in fields where flow cytometry is routinely performed, such as measuring T, B, and NK cell proportions, detecting leukemias and lymphomas, and enumerating CD34+ stem cells for transplantation.
Materials and Reagents
Regular flow cytometry 5-ml test tubes, polypropylene or polystyrene (12 × 75 mm), or 1.4-ml microtubes (e.g., from Micronic), or deep-well plates (any vendor).
Lysis solution (Beckman Coulter, VersaLyseTM, catalog number: IM3648, store at 18-24°C, 2-year shelf-life)
Fixative solution (IOTest®3 10×; Beckman Coulter, catalog number: A07800, store at 2-8°C, 1-year shelf-life)
Antibody cocktail (IOTest Myeloid Activation CD169-PE (Phycoerythrin)/HLA-DR-APC (Allophycocyanin)/CD64-PB (Pacific-Blue) Antibody Cocktail from Beckman Coulter, catalog number: C63854, store at 2-8°C, 1-year shelf-life)
Easy batch preparation of the lysis buffer (see Recipes)
Batch preparation of the staining and lysis buffer for n tests (see Recipes)
Equipment
Pipettes (Gilson, PIPETMAN®, catalog numbers: FA10003M [2-20 µl] and FA10006M [100-1000 µl])
3-laser, 10-color cytometer (Beckman Coulter, Navios, catalog number: B47904) or 3-laser 13-color cytometer (Beckman Coulter, CytoFLEX, catalog number: B53000). Any other 3-laser cytometer can be used (e.g., Becton Dickinson FACSCanto, Cytek Aurora)
Software
Kaluza Software version 2.1 (Beckman Coulter, https://www.beckman.fr/flow-cytometry/software/kaluza)
Note: Any other flow cytometry software can be used (e.g., Becton Dickinson Flowjo and Cytek SpectroFlo).
Procedure
Prepare the lysis buffer (wear common Personal Protective Equipment).
Dilute 1:200 the fixative solution in the lysis solution under chemical hood protection and mix.
Perform the lysis–staining step.
In a tube containing 500 µl lysis buffer and 10 µl conjugate panel, add 2-50 µl blood sample (venous or capillary), mix, and incubate for 15 min in the dark at RT (18-25°C).
Acquire the data on a 3-laser cytometer (then dispose of the waste in containers dedicated to biohazard waste).
Data analysis
Acquiring data on the flow cytometer
Unstained cells are used to set the parameters of the flow cytometer.
Turn on the flow cytometer.
Open the acquisition software and create four dot plots:
Side Scatter (SSC) on the x-axis and Forward Scatter (FSC) on the y-axis.
Channel dedicated to PE (FL2 on Navios) detection on the x-axis and SSC on the y-axis.
Chanel dedicated to APC (FL6 on Navios) detection on the x-axis and SSC on the y-axis.
Chanel dedicated to PB (FL9 on Navios) detection on the x-axis and SSC on the y-axis.
Set the voltages/gains for the SSC-FSC plot such that most of the cell population is in the middle of the graph (Figure 1a). The discriminant/threshold should be set up in such a way that minimum debris is acquired.
Set all the compensations to 0.
Set the voltages for FL9, FL2, and FL6 such that the lymphocyte mean or median fluorescence intensity is around 0.3 (Figure 1b).
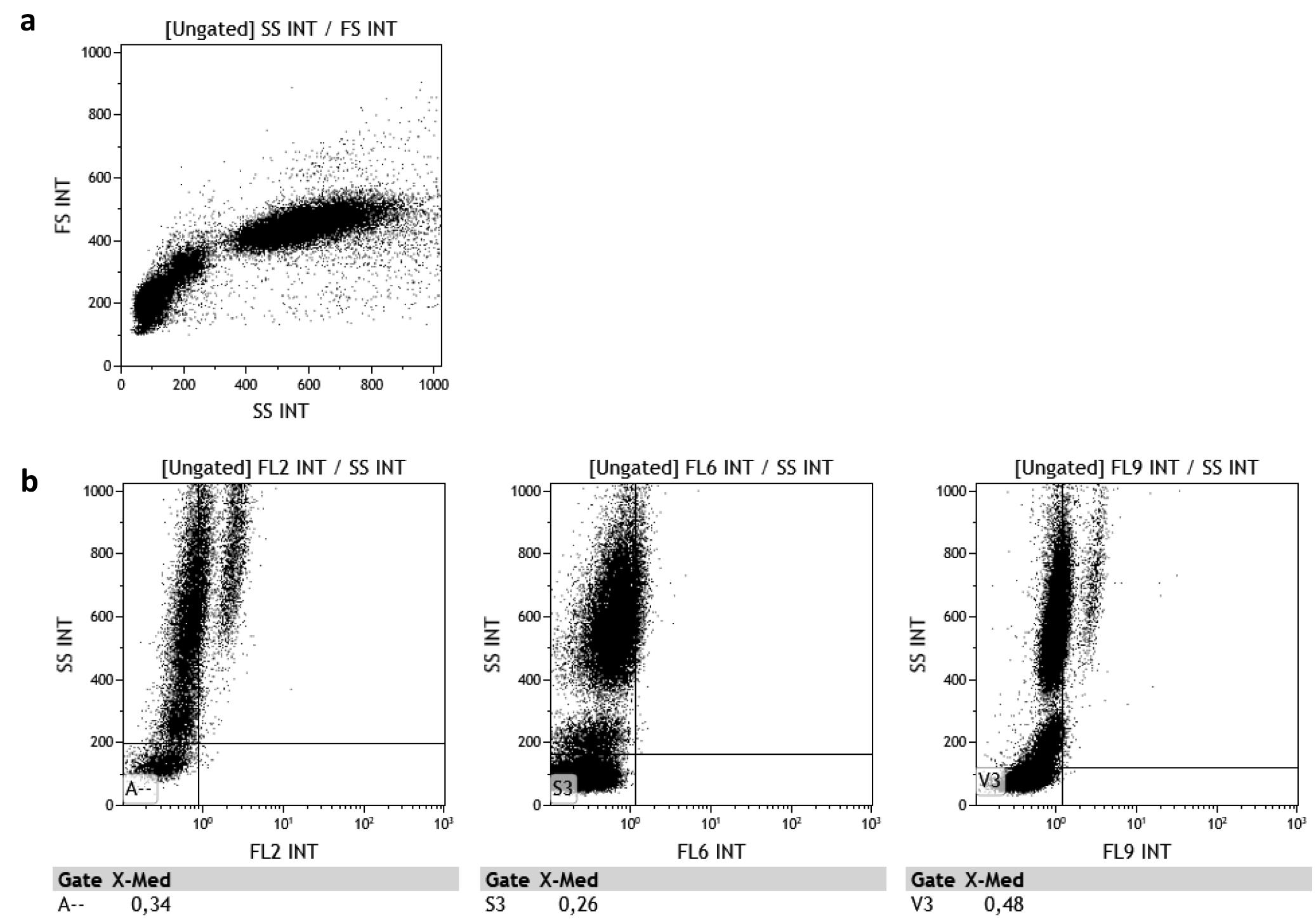
Figure 1. Flow cytometry parameters on (a) SSC-FSC plot and (b) FL2-, FL6-, and FL9-SSC plotsSave the protocol.
Acquire the first stained sample using a high flow rate (~1 µl/s) without any compensation. Make sure to acquire at least 500 monocytes for a robust analysis (usually 30 to 90 s).
On the SSC-FSC plot, draw a leukocyte gate, avoiding debris (Figure 2a).
On the SSC-FL9 (CD64-PB) plot, draw the lymphocyte, monocyte, and neutrophil gates (Figure 2b).
CD169 and HLA-DR expression levels should be assessed on monocytes. CD64 expression levels should be assessed on neutrophils (Figure 2b and 2c).
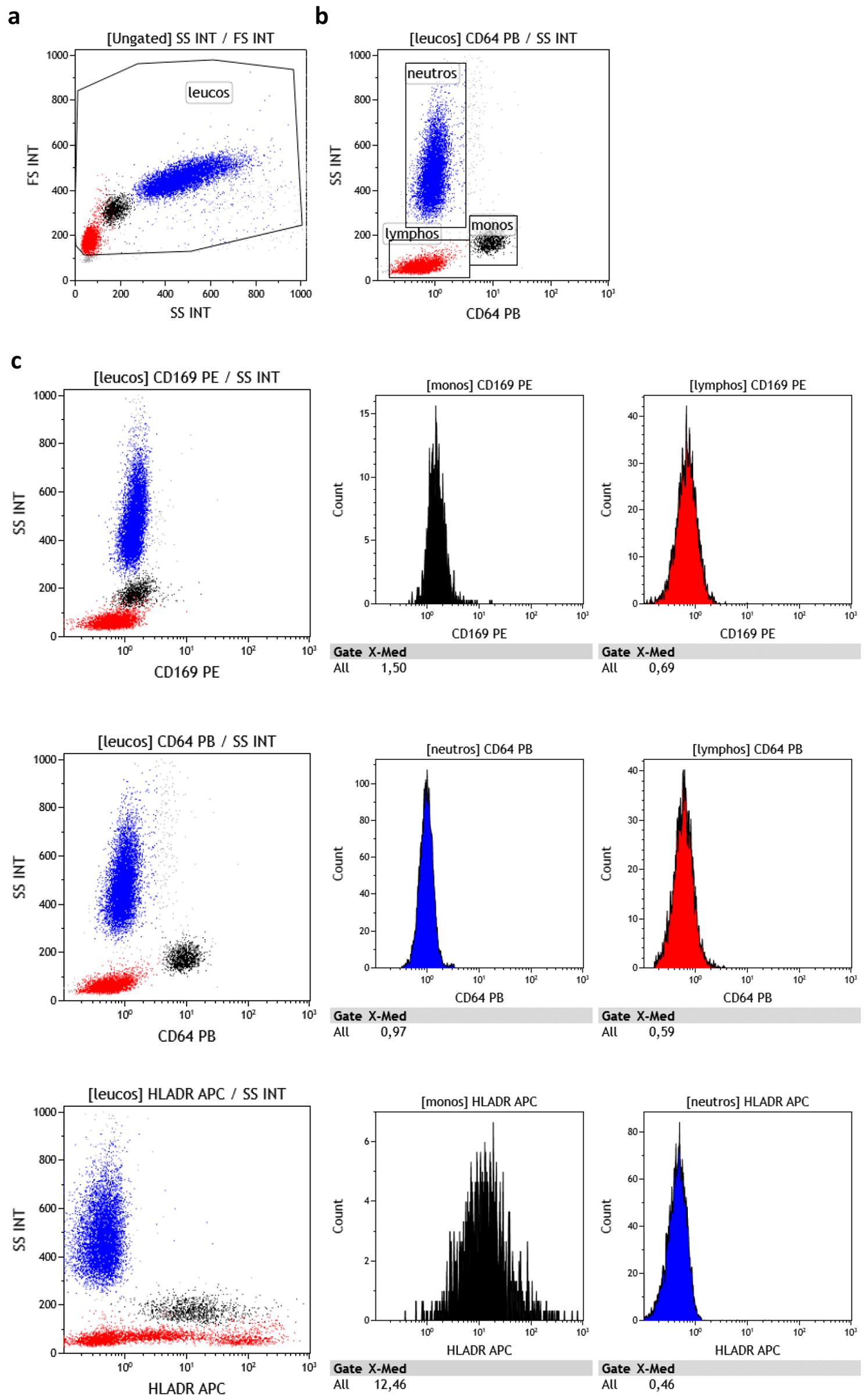
Figure 2. Data analysis procedure. (a and b) Gating strategy of lymphocytes, monocytes, and neutrophils. (c) CD169, CD64, and HLA-DR expression level assessment strategy.Marker levels should be expressed as the Mean of Fluorescence Intensity, Median of Fluorescence Intensity, or Signal-to-noise. Signal-to-noise is calculated as follows: CD169 expression levels on monocytes should be divided by CD169 expression levels on lymphocytes; CD64 expression levels on neutrophils should be divided by CD64 expression levels on lymphocytes; and HLA-DR expression levels on monocytes should be divided by HLA-DR expression levels on neutrophils (Figure 3).
Results are considered as follows: High mCD169 expression is indicative of viral stimuli, high nCD64 expression is indicative of bacterial stimuli, high mHLA-DR expression is indicative of infectious stimuli, and low mHLA-DR expression is indicative of immune exhaustion (Figure 3).
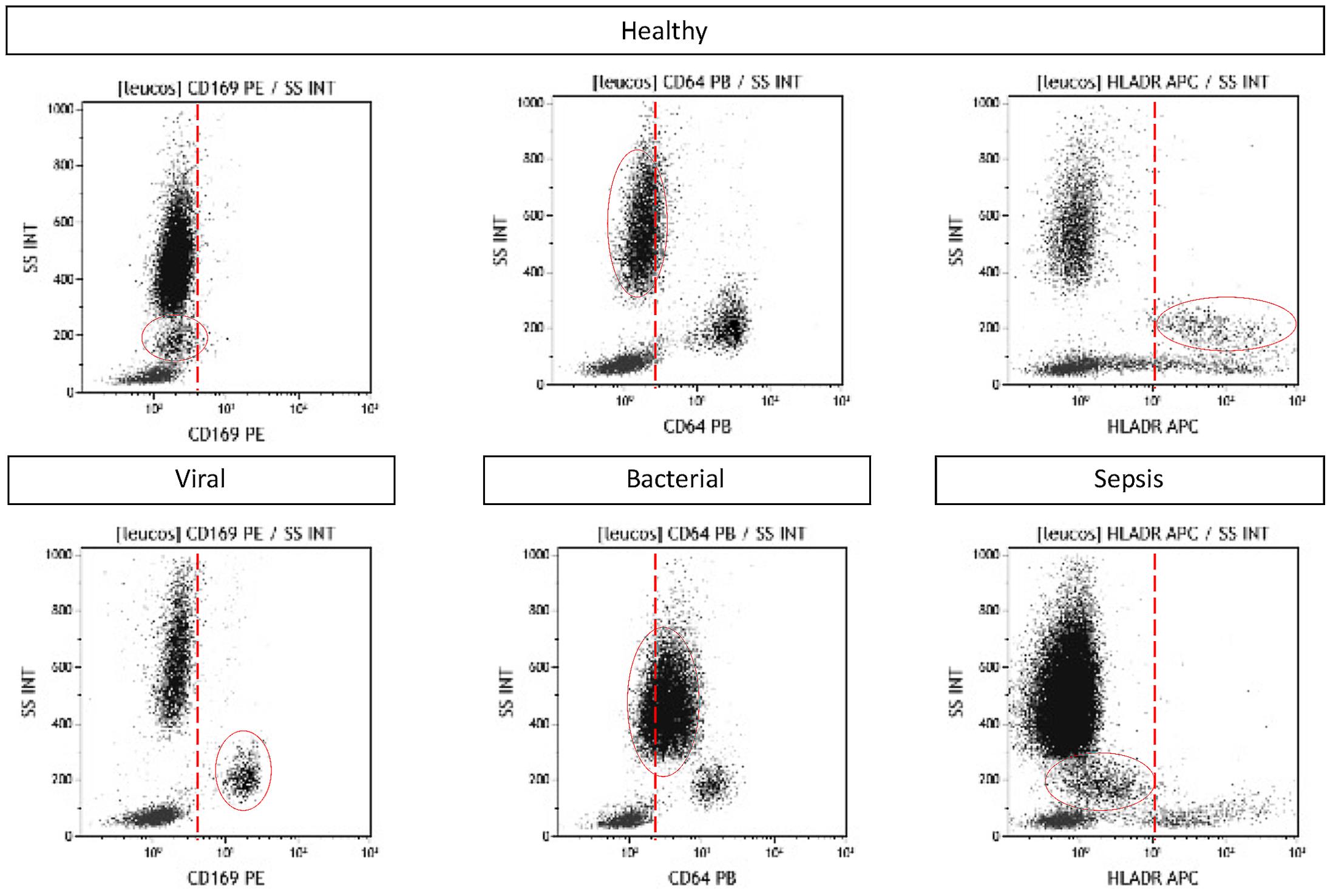
Figure 3. mCD169, nCD64, and mHLA-DR expression assessment in whole blood using the one-step method. Examples are given for one healthy volunteer, one virus‐infected patient, one bacteria‐infected patient, and one patient with sepsis.More research will be necessary to precisely set the thresholds in each laboratory setting, but as an indication, previous evaluations have shown that thresholds should be close to 3 for the CD169 and CD64 indexes.
Further analysis can be performed using software such as Kaluza (re-analysis, other gating and auto-gating, other histograms or dot plots, coloring, compensation adjustments, refined percentages and fluorescent values, overlay and merge data sets, batch analysis for higher throughput, etc.).
Notes
Depending on the target marker, application, and required stability, the method should be adapted in each laboratory setting. For example, and only for indication and not as an established performance, it has been observed in some applications that:
Blood sample can be venous or capillary.
Blood can be anticoagulated with EDTA or heparin.
Thanks to the gentle lysis reagent and added fixative, blood can be processed up to 3-4 days after sampling when stored at RT or 2-8°C. Moreover, samples can be analyzed on the cytometer up to 3 days after processing when stored at 2-8°C.
The lysis solution can include 0.05% fixative solution.
The lysis buffer (lysis solution containing fixative solution) can be stored for up to one month at RT.
The targeted marker can be any membrane marker.
Antibodies can be liquid or dried depending on product availability.
WARNING:
If a liquid cocktail is used, anantibody volume higher than 100 µl could impact lysis efficiency.
If a commercial DURAClone format is used, the lysis volume should be increased to 1 ml (for 2-50 µl blood) or 2 ml (for 50-100 µl blood as per IFU).
Antibodies can be premixed with the lysis buffer up to a few days prior.
Recipes
Note: To be adapted depending on each application; performances are not established.
Easy batch preparation of lysis buffer
One bottle Versalyse solution (100 ml)
500 µl fixative solution (10×)
Mix and store at RT
Batch preparation of the staining and lysis buffer for n tests
Pipet (n+1) × 500 µl lysis buffer
Add (n+1) × 10 µl myeloid activation cocktail (or any other panel at its optimal dose)
Mix well and distribute 510 µl per reaction tube, store in the dark
Tube is now ready to receive the blood sample
Acknowledgments
IAB is the recipient of a CIFRE Ph.D. grant (N°2018/1212) from the ANRT (National Agency for Research and Technology). This work was supported by Beckman Coulter through donation of the research reagents used in the flow cytometry experiments and participation of the four employees: IAB, PB, JMB, and FM. The method described was modified from ‘VersaLyse and the mechanism of ammonium chloride lysis’ (van Agthoven, 2007) and resulted in recent publications (Bourgoin et al., 2019a, 2019b, 2020a, 2020b, 2020c, and 2021; Bedin et al., 2020; Michel et al., 2020).
Competing interests
There are no conflicts of interest or competing interests.
Ethics
All enrolled patients provided informed consent, and the procedures followed were in accordance with the Helsinki Declaration. Routine care of the subjects was not modified; analyses were performed on anonymized left-over blood, and all data collected in the study were part of routine clinical practice and retrieved from subject records. Results of this study had no influence on subject management.
References
- Bedin, A.S., Makinson, A., Picot, M.C., Mennechet, F., Malergue, F., Pisoni, A., Nyiramigisha, E., Montagnier, L., Bollore, K. and Debiesse, S. J. (2020). Monocyte CD169 expression as a biomarker in the early diagnosis of COVID-19. J Infect Dis 223(4): 562-567.
- Bourgoin, P., Biechele, G., Ait Belkacem, I., Morange, P. E. and Malergue, F. (2020a). Role of the interferons in CD64 and CD169 expressions in whole blood: Relevance in the balance between viral- or bacterial-oriented immune responses. Immun Inflamm Dis 8(1): 106-123.
- Bourgoin, P., Hayman, J., Rimmele, T., Venet, F., Malergue, F. and Monneret, G. (2019a). A novel one-step extracellular staining for flow cytometry: Proof-of-concept on sepsis-related biomarkers. J Immunol Methods 470: 59-63.
- Bourgoin, P., Lediagon, G., Arnoux, I., Bernot, D., Morange, P. E., Michelet, P., Malergue, F. and Markarian, T. (2020b). Flow cytometry evaluation of infection-related biomarkers in febrile subjects in the emergency department. Future Microbiol 15: 189-201.
- Bourgoin, P., Soliveres, T., Ahriz, D., Arnoux, I., Meisel, C., Unterwalder, N., Morange, P. E., Michelet, P., Malergue, F. and Markarian, T. (2019b). Clinical research assessment by flow cytometry of biomarkers for infectious stratification in an Emergency Department. Biomark Med 13(16): 1373-1386.
- Bourgoin, P., Soliveres, T., Barbaresi, A., Loundou, A., Belkacem, I. A., Arnoux, I., Bernot, D., Loosveld, M., Morange, P. E., Michelet, P., Malergue, F. and Markarian, T. (2021). CD169 and CD64 could help differentiate bacterial from CoVID-19 or other viral infections in the Emergency Department. Cytometry A 99(5): 435-445.
- Bourgoin, P., Taspinar, R., Gossez, M., Venet, F., Delwarde, B., Rimmele, T., Morange, P. E., Malergue, F. and Monneret, G. (2020c). Toward Monocyte HLA-DR Bedside Monitoring: A Proof of Concept Study. Shock 55(6): 782-789.
- Brown, M. and Wittwer, C. (2000). Flow cytometry: principles and clinical applications in hematology. Clin Chem 46(8 Pt 2): 1221-1229.
- Michel, M., Malergue, F., Belkacem, I. A., Bourgoin, P., Morange, P.-E., Arnoux, I., Miloud, T., Million, M., Tissot-Dupont, H., Mege, J. L., Busnel, J. M. and Vitte, J. (2020). An ultra-sensitive, ultra-fast whole blood monocyte CD169 assay for COVID-19 screening. medRxiv.
- van Agthoven, A. (2007). VersaLyse and the mechanism of ammonium chloride lysis. Int J Lab Hematol 29: 65-66.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Belkacem, I. A., Bourgoin, P., Busnel, J. M., Galland, F. and Malergue, F. (2021). One-step White Blood Cell Extracellular Staining Method for Flow Cytometry. Bio-protocol 11(16): e4135. DOI: 10.21769/BioProtoc.4135.
Category
Immunology > Immune cell staining > Flow cytometry
Biochemistry > Protein > Expression
Cell Biology > Cell-based analysis > Flow cytometry
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link