- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Isolation of Microglia and Analysis of Protein Expression by Flow Cytometry: Avoiding the Pitfall of Microglia Background Autofluorescence
Published: Vol 11, Iss 14, Jul 20, 2021 DOI: 10.21769/BioProtoc.4091 Views: 7017
Reviewed by: Guillermo A. GomezAlka MehraMeenal SinhaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation and Ex Vivo Testing of CD8+ T-Cell Division and Activation Using Mouse Splenocytes
Melissa Dolan [...] John M.L. Ebos
Aug 20, 2025 3860 Views

Detection of Autophagy in Human Peripheral Blood Mononuclear Cells Using Guava® Autophagy and Flow Cytometry
Melanie Scherer [...] Jörg Bergemann
Sep 20, 2025 1413 Views

Protocol for the Isolation and Analysis of Extracellular Vesicles From Peripheral Blood: Red Cell, Endothelial, and Platelet-Derived Extracellular Vesicles
Bhawani Yasassri Alvitigala [...] Lallindra Viranjan Gooneratne
Nov 5, 2025 1442 Views
Abstract
Microglia are a unique type of tissue-resident innate immune cell found within the brain, spinal cord, and retina. In the healthy nervous system, their main functions are to defend the tissue against infectious microbes, support neuronal networks through synapse remodeling, and clear extracellular debris and dying cells through phagocytosis. Many existing microglia isolation protocols require the use of enzymatic tissue digestion or magnetic bead-based isolation steps, which increase both the time and cost of these procedures and introduce variability to the experiment. Here, we report a protocol to generate single-cell suspensions from freshly harvested murine brains or spinal cords, which efficiently dissociates tissue and removes myelin debris through simple mechanical dissociation and density centrifugation and can be applied to rat and non-human primate tissues. We further describe the importance of including empty channels in downstream flow cytometry analyses of microglia single-cell suspensions to accurately assess the expression of protein targets in this highly autofluorescent cell type. This methodology ensures that observed fluorescence signals are not incorrectly attributed to the protein target of interest by appropriately taking into account the unique autofluorescence of this cell type, a phenomenon already present in young animals and that increases with aging to levels that are comparable to those observed with antibodies against highly abundant antigens.
Keywords: Microglia isolationBackground
Microglia are a type of tissue-resident macrophages residing in the central nervous system (CNS) and account for 10% to 15% of all cells within the CNS. While displaying some canonical macrophage activities, such as the phagocytosis of debris and apoptotic bodies, microglia are also endowed with functions specific to the CNS microenvironment, such as synaptic remodeling, neuronal support, and oligodendrogenesis (Ransohoff and Khoury, 2016; Clayton et al., 2017; Li and Barres, 2018). This wide array of functions implies the existence of a diverse set of microglia phenotypes, states, and subsets well suited to characterization by single-cell analytical approaches. Fluorescence-based flow cytometry, a technique routinely used in immunological studies, allows for high-throughput, multiparametric analysis of single cells in suspension, isolated from blood or from dissociated tissues. However, the dissociation of CNS tissues generates a large amount of debris, principally myelin fragments, which must be removed before flow cytometry analysis. Commonly cited isolation techniques dissociate CNS tissue through enzymatic digestion and utilize magnetic bead-based strategies to remove myelin or isolate microglia. However, commonly used proteases in enzymatic tissue digestion, such as collagenase and trypsin, can lead to the unintended cleavage of surface antigens on microglia (Autengruber et al., 2012) and promote cellular transcriptional changes during the 37°C incubations required for enzymatic activity (O’Flanagan et al., 2019, Mattei et al., 2020). In addition, magnetic bead isolation is costly, limits study throughput, and, in our hands, does not improve the yield or viability compared to this protocol, which in young (~3 month) mice yields approximately 2.5 × 105 microglia for analysis by flow cytometry. In this study, we describe the preparation of CNS single-cell suspensions for flow cytometry by utilizing a simple mechanical tissue dissociation procedure followed by myelin removal via density centrifugation. Cost-efficient and easy to perform, all steps of this protocol are carried out on ice or at 4°C, limiting cellular changes that would otherwise occur during isolation at higher temperatures.
By utilizing two surface markers (CD45 and CD11b), this protocol yields a population of microglia with uniformly high expression of core the homeostatic markers CX3CR1, P2RY12, and TMEM119 (Burns et al., 2020, Figure 1D) and has also been successfully employed to isolate microglia in multiple states, both homeostatic and activated (Burns et al., 2020, Figure 6–figure supplement 1A), from mice as old as 24 months of age. Although this workflow is optimized for cell analysis by flow cytometry, we highly recommend utilizing an alternative 2-phase Percoll protocol (described in Burns et al., 2020) for fluorescence-activated cell sorting (FACS), as sorting instruments are more sensitive to the amount of debris remaining post-isolation, which can negatively impact FACS purity and yield.
While flow cytometry has become a routine method used in the analysis of immune cells, the analysis of microglia (minimally defined as CD45dim, CD11b+) presents a particular challenge, as they exhibit a uniquely intense level of autofluorescence compared to other cell types, including CNS-resident macrophages (CD45bright, CD11b+), which do not emit any detectable autofluorescence. In addition, the distribution of the autofluorescence signal in microglia is bimodal and biologically dynamic, with about two-thirds of the microglia showing a high autofluorescence signal (autofluorescence-positive) and the remaining third showing no or very low levels of autofluorescence (autofluorescence-negative) (Burns et al., 2020). Interestingly, both autofluorescence subsets are differentially impacted by aging and genetic perturbations, which adds further complexity to their analysis (Burns et al., 2020). In this protocol, we provide key considerations and analytical strategies to avoid the issues associated with microglia autofluorescence.
Materials and Reagents
25G x ¾” butterfly needle (EXELINT, catalog number: 26768)
20 ml luer-lock syringe (BD, catalog number: 302830)
7 ml dounce homogenizer (Wheaton, catalog number: 57542)
15 ml polypropylene conical tubes (ThermoFisher, catalog number: 339651)
96 well v-bottom assay plates (Corning, catalog number: 3897)
96 well 40 µm mesh filter plates (MilliporeSigma, catalog number: MANMN4010)
Cluster tubes (Corning, catalog number: 4411)
10 cm plastic Petri dishes (Fisher Scientific, catalog number: FB0875713)
Ultracomp eBeads Plus Compensation Beads (ThermoFisher, catalog number: 01-333-42)
Fetal bovine serum (FBS) (ThermoFisher, catalog number: 26140079)
10% FBS/HBSS
Flow staining buffer (ThermoFisher, catalog number: 00-4222-26)
DAPI (ThermoFisher, catalog number: 62248)
Percoll (GE Healthcare, catalog number: 17-0891-01)
10× Hanks Balanced Salt Solution (ThermoFisher, catalog number: 14185052)
1 M HEPES (ThermoFisher, catalog number: 15630080)
1× HBSS (ThermoFisher, catalog number: 14175095)
0.5 M EDTA (ThermoFisher, catalog number: 15575020)
TruStain FcX (Biolegend, catalog number: 101320)
Anti-CD45 BV785 (Biolegend, catalog number: 103149)
Anti-CD11b BV510 (Biolegend, catalog number: 101263)
33% Isotonic Percoll (see Recipes)
1× Hanks Balanced Salt Solution (HBSS) with 25mM HEPES (see Recipes)
Phosphate buffered solution (PBS) with 3mM EDTA (see Recipes)
Fc receptor blocking solution (see Recipes)
2× Microglia Antibody Panel (see Recipes)
Equipment
Disposable scalpels (Fisher Scientific, catalog: 3120032) or common single-edge razor blades
13 mm extra fine Bonn scissors (Fine Science Tools, catalog number: 14084-08)
Iris forceps (Fine Science Tools, catalog number: 11370-31)
7 ml glass homogenizer (Wheaton, catalog number: 57542)
Refrigerated tabletop centrifuge (ThermoFisher Sorvall Legend XTR, Rotor TX-1000)
5-laser LSR Fortessa X-20 (Becton Dickinson)
Vacuum line for aspirating
Procedure
Isolation of microglia
Keep all solutions ice-cold through the procedure.
Immediately following CO2 euthanasia, open the chest cavity to expose the heart. Insert the 25G butterfly needle into the left ventricle, make a small incision in the right atrium, and slowly perfuse the mouse with 20 ml of PBS with 3 mM EDTA.
Cut and peel back the skin to expose the skull. Using scissors, cut the spine at the base of the skull. Starting from the brain stem, cut rostrally along the sagittal suture. Peel the two halves of the skull away to the side. Using tweezers, scoop out the brain and transfer into a 15 ml conical tube containing 5 ml of cold HBSS with 25 mM HEPES and keep on ice.
Transfer the brain to a fresh Petri dish on ice and mince the tissue with a scalpel or razor blade into pieces approximately 1 mm in size.
Transfer the minced tissue into the 7 ml dounce homogenizer and add 5 ml of HBSS with 25 mM HEPES.
The 7 ml dounce homogenizer is supplied with two pestles of slightly different sizes and labeled by the manufacturer. Using the pestle marked “loose,” gently disrupt the tissue, on ice, for approximately 10 strokes. Repeat with the pestle marked “tight” for another 10 strokes.
Pour the single-cell suspension into a fresh 15 ml conical tube. Rinse the homogenizer with 5 ml of HBSS with 25 mM HEPES and transfer to the same 15 ml conical tube.
Centrifuge the single-cell suspension at 600 × g for 5 min at 4°C.
Aspirate the supernatant and gently resuspend the cell pellet in 1 ml of 100% FBS.
Add 9 ml of 33% isotonic Percoll solution and mix.
Gently add 1 ml of 10% FBS/HBSS over the cell suspension.
Centrifuge the cell suspension at 800 × g for 15 min at 4°C with full acceleration and no brake.
Carefully aspirate the resulting myelin layer located at the interface and down to the cell pellet.

Figure 1. Percoll isolation of microglia. A 30% Percoll cell-suspension overlaid with 1 ml 10% FBS/HBSS solution, A. before centrifugation and B. after centrifugation.Resuspend the cell pellet in 1 ml of HBSS with 25 mM HEPES.
Add 9 ml of HBSS with 25 mM HEPES and centrifuge the single-cell suspension at 600 × g for 5 min at 4°C with full acceleration and brake on.
Aspirate the cell pellet in a final volume of 1 ml of HBSS with 25 mM HEPES. Cells are now ready for antibody staining for flow cytometry.
Fluorochrome-conjugated antibody staining for flow cytometry analysis of cell surface markers on microglia
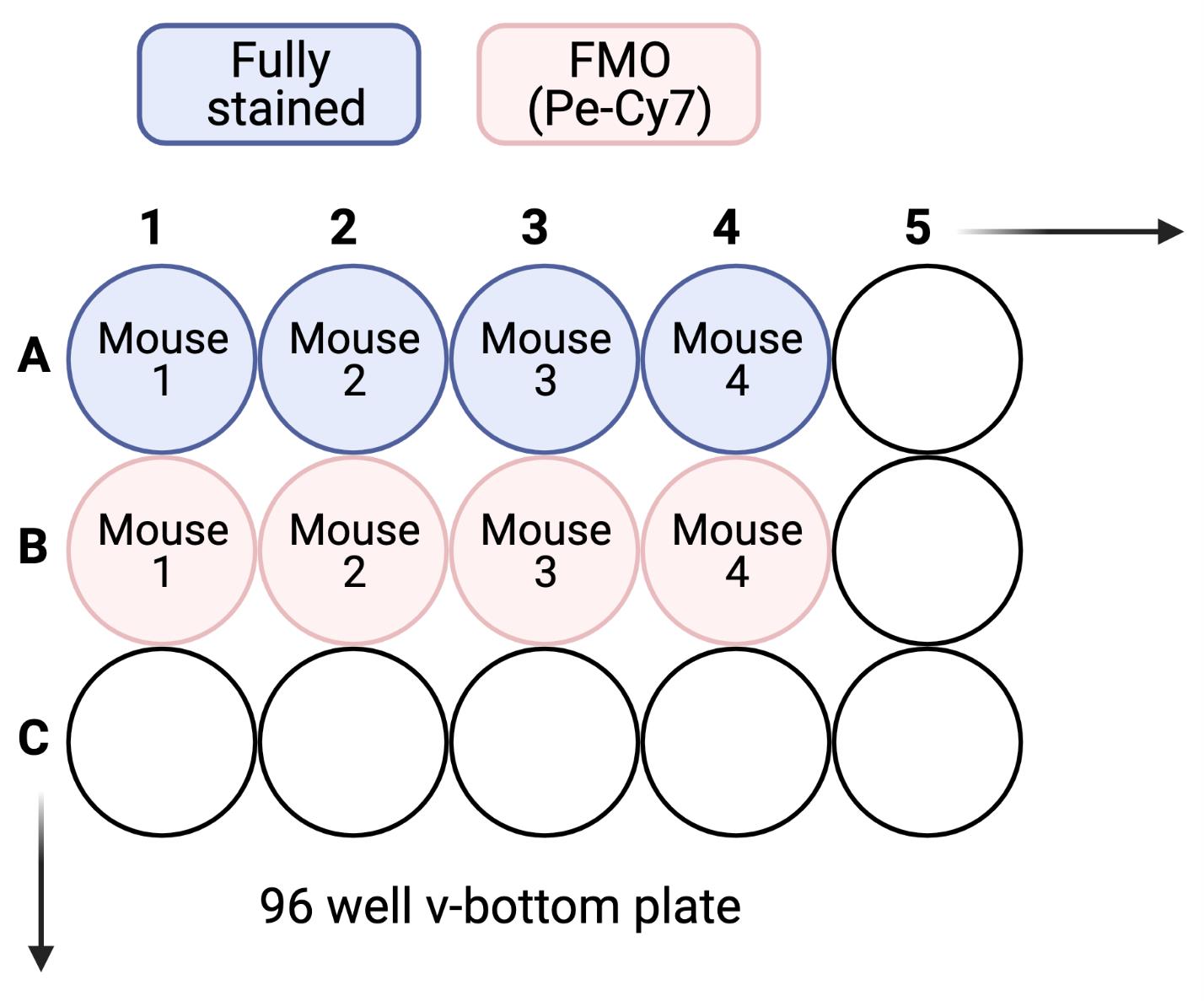
Figure 2. Example plate layout for anti-CD11c PE-Cy7 stained microglia and accompanying FMO controlsTransfer 150 µl (approximately 1/7) of the single-cell suspension (approximately 300,000 live cells) from Step A16 to a well of a 96 well v-bottom plate. Repeat for any additional samples. Make sure to include wells for fluorescence-minus-one (FMO) controls for antigens of interest. For example, if the staining panel includes anti-CD11c PE-Cy7, include a well where cells are stained with all other antibodies EXCEPT for anti-CD11c PE-Cy7. This will account for background fluorescence in that specific channel for each sample.
Centrifuge the plate at 300 × g for 3 min at 4°C.
In one swift motion, decant the plate by flicking into a sink. Do not dab the plate dry. Do not flick the plate twice.
Resuspend the cells in the plate with 25 µl of diluted FcBlock solution. Incubate at 4°C for 10 min.
Add 25 µl of the 2× Microglia Antibody Panel to each well and mix well by pipetting. Incubate at 4°C for 30 min.
Add 150 µl of Flow Staining Buffer to each well. Centrifuge the plate at 300 × g for 3 min at 4°C.
In one swift motion, decant the plate by flicking into a sink. Do not dab the plate dry. Do not flick the plate twice.
Resuspend the cells in 200 µl of Flow Staining Buffer. Centrifuge the plate at 300 × g for 3 min at 4°C.
In one swift motion, decant the plate by flicking into a sink. Do not dab the plate dry. Do not flick the plate twice.
Repeat Steps B8 and B9.
Resuspend the cells in 200 µl of Flow Staining Buffer containing 0.1 µg/ml DAPI.
Transfer the samples to a 40 µm mesh filter plate and centrifuge for 1 min at 100 × g to bring the samples to the lower chamber.
Transfer the samples to cluster tubes and keep on ice, protected from light. Promptly proceed with Section C: single-color compensation controls.
Preparation of single-color compensation controls
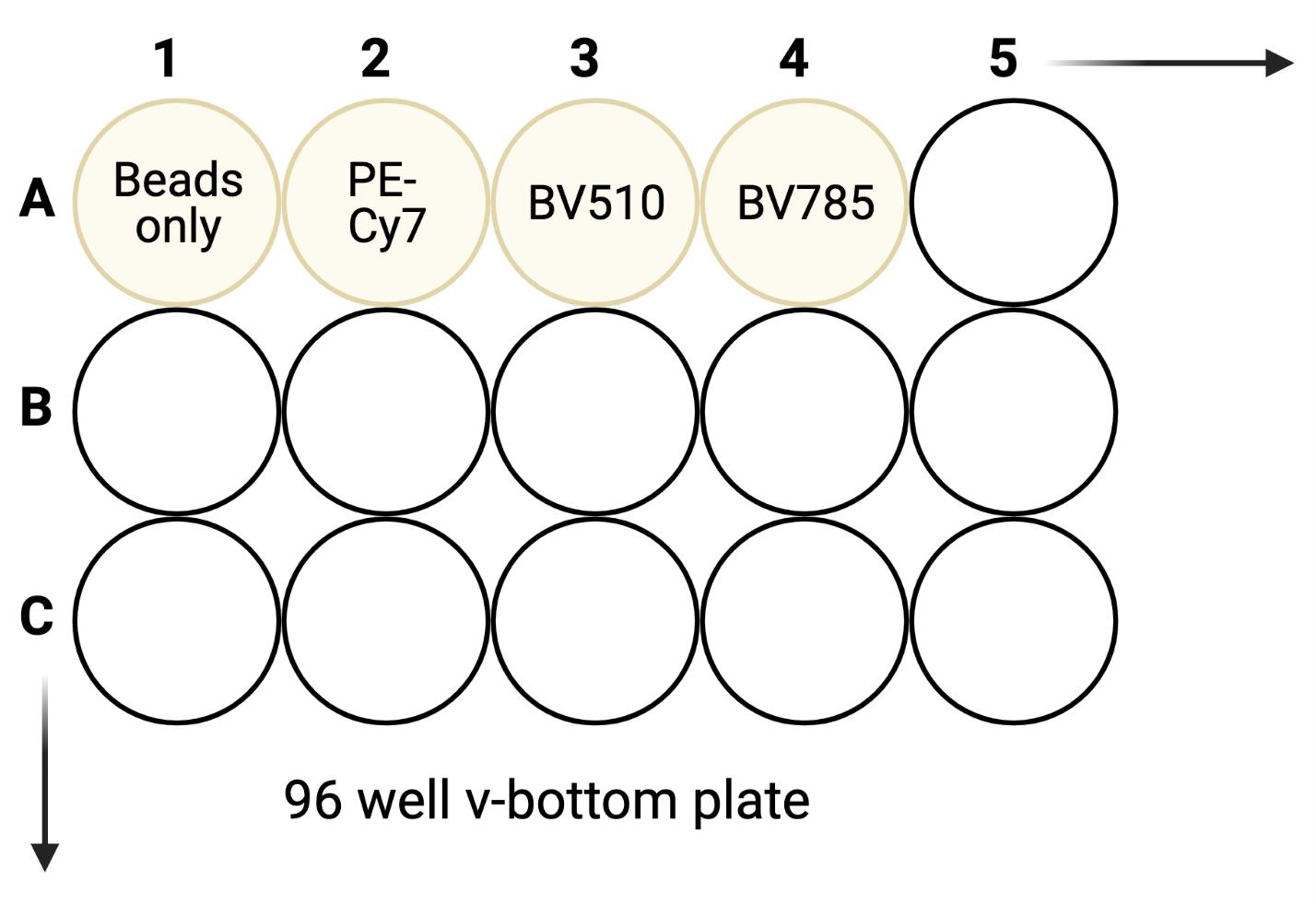
Figure 3. Sample plate layout for compensation control beadsFor the number of fluorochromes used, add one drop of compensation beads to the same number of wells on a separate 96 well v-bottom plate. Include one extra well of beads for an unstained compensation control. Refer to Figure 3 for a sample layout.
Add 150 µl of Flow Staining Buffer to each well.
Add 1 µl of a single fluorochrome-conjugated antibody to a single well. Repeat for the remaining fluorochromes and wells. Do not add any antibody to the well containing the unstained compensation control. Incubate 15 min at room temperature, protected from light.
Centrifuge the plate at 300 × g for 3 min at 4°C.
In one swift motion, decant the plate by flicking into a sink. Do not dab the plate dry. Do not flick the plate twice.
Resuspend the beads in 200 µl of flow staining buffer and transfer them to cluster tubes. Acquire promptly on the flow cytometer along with stained samples from Section B.
Sample acquisition on a 5-laser LSR Fortessa X-20
In Diva, create your experiment and select the detection channels corresponding to the fluorochrome panel used to stain your samples in Section B. For microglia, it is highly recommended to include an additional unoccupied detection channel (e.g., 488 nm laser, 710/50 nm bandpass) to record background autofluorescence.
Create compensation controls for your experiment in Diva and acquire each individual single-color control, including the unstained beads, from Section C.
Calculate the compensation matrix and then run the samples of interest from Section B.
Data analysis
Identification of microglia from a CNS single-cell suspension
After sample acquisition, data analysis is performed in FlowJo V10. If a compensation matrix was not generated on the flow cytometer during sample acquisition, complete the compensation wizard in FlowJo before continuing. By utilizing the hierarchical gating strategy illustrated in Figure 4, singlets are gated first, followed by live cells (DAPI-); lastly, microglia are identified by their relatively low CD45 expression and high CD11b expression.
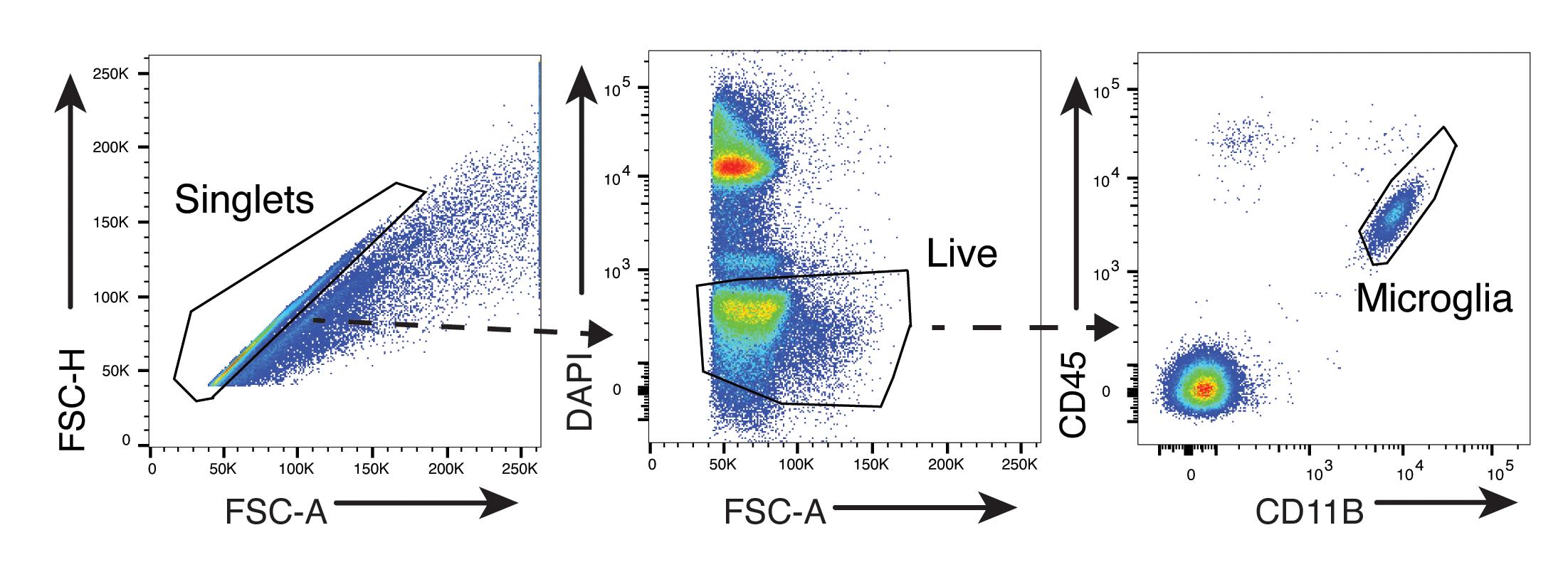
Figure 4. Hierarchical gating strategy to identify microglia from a CNS single-cell suspension. FSC, forward scatter. Reproduced from Burns et al., 2020.Analysis of protein target expression in microglia
Microglia subsets were recently identified based on their high or low/negligible levels of autofluorescence (Burns et al., 2020). Because of the unusually high intensity, broad spectral properties, and bimodality of the autofluorescence signal in the microglia population (Figure 5), flow cytometry analysis of microglia poses specific challenges as experimental observations can be very easily confounded by background autofluorescence. Experiments should include “fluorescence-minus-one” controls (FMO) for antigen-fluorochrome combinations of interest to avoid this issue. In addition, when working with microglia, it is critical to include one or multiple unoccupied cytometer channels (i.e., with no fluorescent antibody/dyes in those channels) during sample acquisition, preferably a channel equivalent to PerCP-Cy5.5 (488 nm blue laser line, 710/50 nm bandpass filter), which is one of the most sensitive channels for microglia autofluorescence (Figure 5).
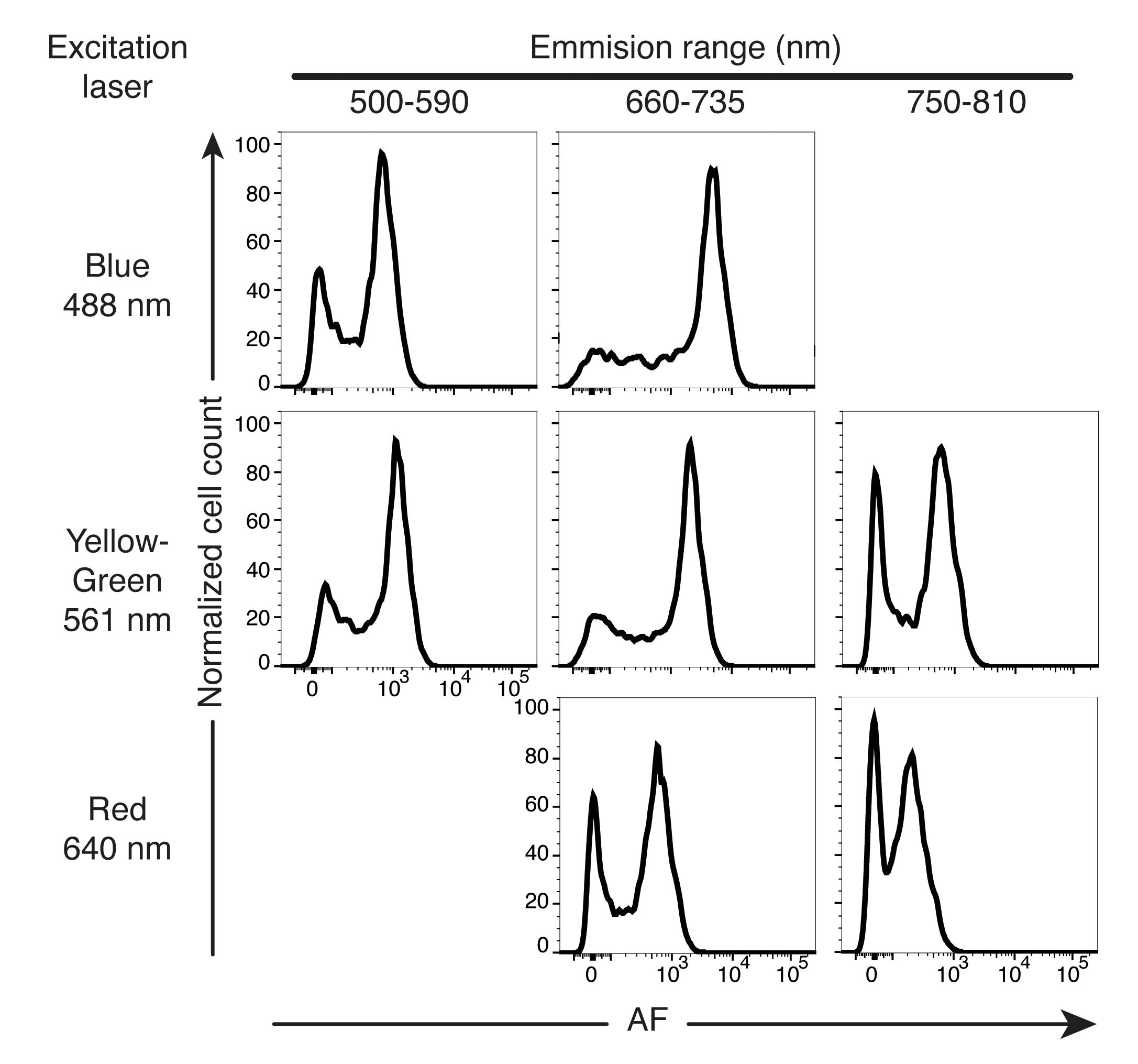
Figure 5. Representative flow cytometry histograms of autofluorescence intensity of the entire microglia population, in a single sample, from multiple combinations of excitation lasers and emission filters. AF, autofluorescence. Reproduced from Burns et al., 2020.To illustrate these challenges, we present two examples where analysis of target protein expression in microglia can be easily confounded by autofluorescence if proper controls are not included to consider the fraction of the fluorescence signal that is attributable to autofluorescence rather than the target protein marker of interest.
In this first example, we selected the surface antigen CD11c, which is an integrin transcriptionally expressed at relatively high levels on a small subset of microglia during development, injury, disease, and aging. In Figure 6A, CD45dimCD11b+ microglia stained with anti-CD11c PE-Cy7 showed a clear bimodal distribution of signal in the PE-Cy7 channel, indicative at face value of positive CD11c staining in 57% of the microglia population. However, the histogram from the FMO control sample revealed a similar, bimodal pattern of fluorescence intensity with 44% of microglia gated as positive, reflective of the cellular autofluorescence in this cytometer channel (Figure 6A). When overlaid with the histogram from the CD11c PE-Cy7 stained sample, it was not possible to delineate an adequate gate to identify CD11c+ microglia given the bimodality of the autofluorescence signal in microglia and the existence of two subsets of microglia, one with high levels of autofluorescence and one with no or very low levels of autofluorescence (Figure 6B). Instead, visualizing the data in an XY dot plot format using an empty cytometer channel to measure autofluorescence (Blue laser, 710/50 nm) on one axis against the cytometer channel for the target antigen of interest on the other axis (CD11c PE-Cy7 in this example) allowed us to distinguish CD11c+ microglia previously confounded by the autofluorescence signal and use distinct thresholds for CD11c positivity for the autofluorescent negative and positive microglia populations (Figure 6C). Using this strategy, CD11c was found to be only expressed in 15% of the overall microglial population (Figure 6C). This result is markedly different from the one that mistakenly identified 57% of microglia as CD11c+ while ignoring the autofluorescence signal (Figure 6A). In imaging studies of young, healthy CD11c-YFP reporter mice, YFP+ cells were present as a minor population in the adult brain (Sato-Hashimoto et al., 2019), and in a recent review (Benmamar-Badel et al., 2020), it was estimated that approximately 2% of microglia are CD11c+ in these animals. Therefore, analyzing microglial CD11c expression by flow cytometry without taking AF into account could lead to misleading conclusions.
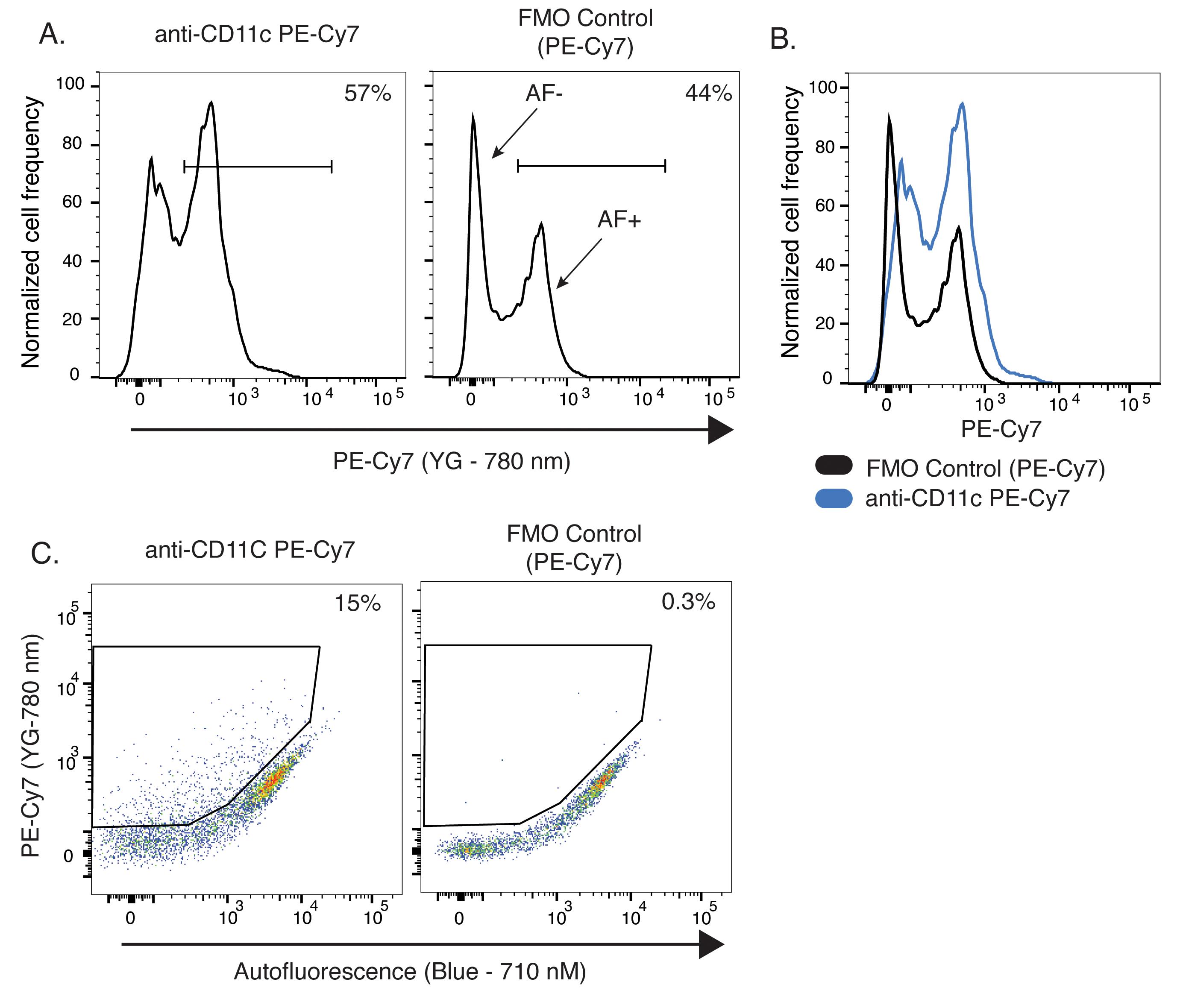
Figure 6. Analysis of microglia from 5-month-old mice by flow cytometry. A. Histograms of fluorescence signal from FMO control and CD11c-stained microglia samples in the PE-Cy7 channel (Yellow-green laser, 780/60 nm). B. Overlay of the two previous histograms. C. Pseudocolor dot plots of signal in the PE-Cy7 fluorescence channel versus an unoccupied fluorescence channel (Blue laser, 710/50 nm). AF+, autofluorescence-positive; AF-, autofluorescence-negative; FMO, fluorescence-minus-one; Blue, 488 nm laser; YG, 561 nm yellow-green laser.As a second example, we analyzed the impact of microglia autofluorescence on the detection of proteins expressed in the entire microglia population (as opposed to a subset) with modest to high levels of expression at the single-cell level. For this, we specifically chose the receptor family for the Fc region of IgG immunoglobulins: CD16, CD32, and CD64. Fc-receptors comprise a large family of transmembrane proteins expressed at the plasma membrane of immune cells that bind the Fc region of antibodies and activate downstream signaling cascades. Microglia are known to express the gamma family of activating Fc receptors, which includes CD16/32 and CD64 (Pellerin et al., 2021). Fcer1g-deficient mice lack the common signaling FCER1G chain required for surface expression of all activating Fc receptors. When stained with FITC conjugated anti-CD16/32 antibody and without considering microglia autofluorescence, wild-type microglia showed a clear positive staining with a bi-modal distribution (Figure 7A). However, microglia from Fcer1g-/- mice retained a large fraction of this signal with a wider bimodal distribution, indicating that a significant fraction of the signal detected was not attributable to CD16/32 expression but to autofluorescence (Figure 7A). Accordingly, when the analytical strategy described in the prior example was used, and anti-CD16/32 stained samples were displayed on dot plots featuring CD16/32 expression on the y-axis against an empty channel on the x-axis, a large fraction of the signal associated with CD16/32 expression in Figure 7A became clearly attributable to the microglia autofluorescence signal in the Fcer1gKO mice (Figure 7B). Inclusion and overlay of each sample’s accompanying FMO control revealed the accurate expression levels of CD16/32 and the expected lack of CD16/32 staining in microglia from Fcer1gKO mice (Figure 7B). Gating autofluorescence-positive (AF+) and autofluorescence-negative (AF-) microglia subsets (Burns et al., 2020) using the autofluorescence channel further allowed the analysis of CD16/32 expression levels on these AF microglia subsets (Figure 7C).
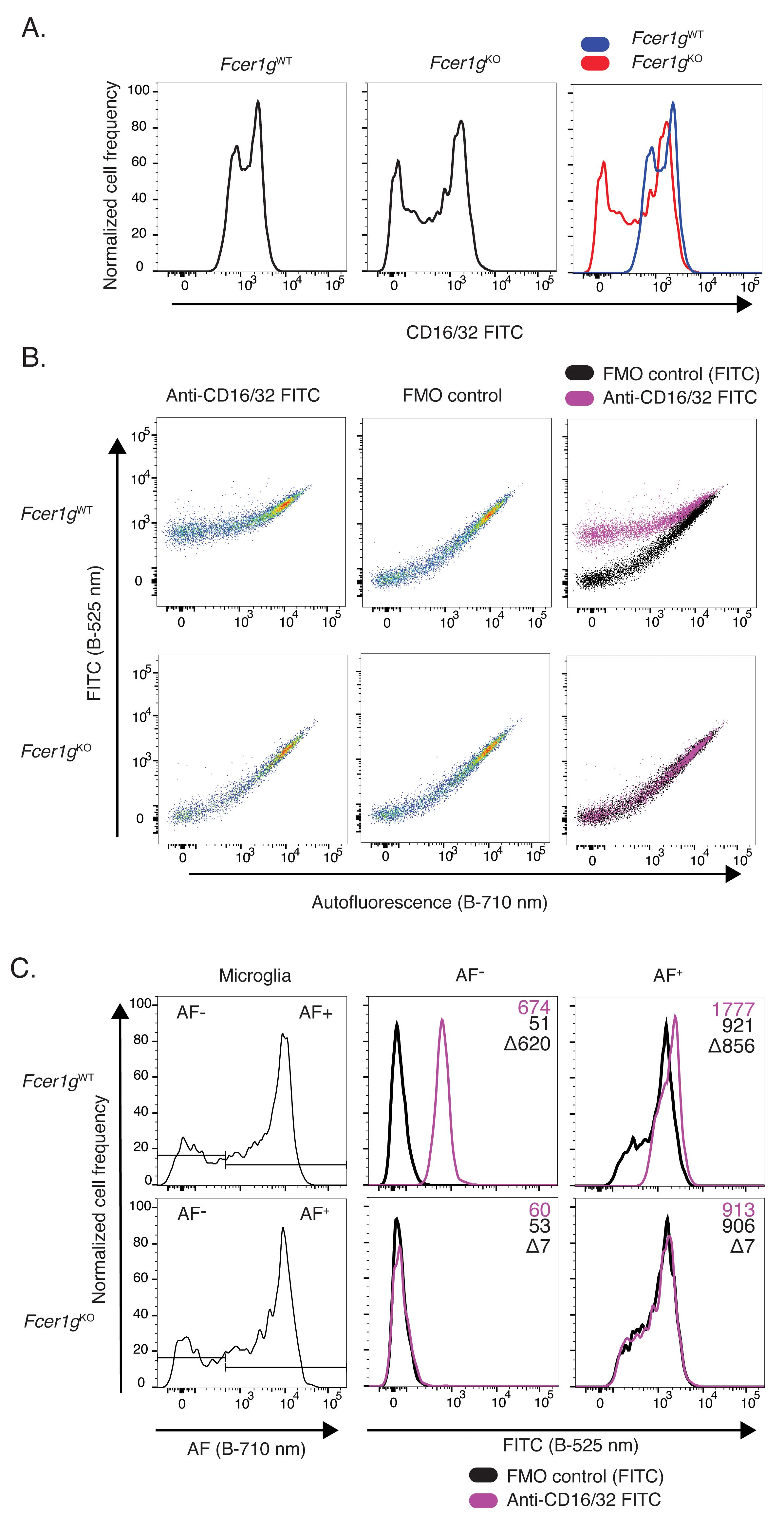
Figure 7. Analysis of Fc-receptor (CD16/32) surface expression levels in 6-month-old Fcer1g wild-type and knockout microglia. A. Individual and overlaid histograms of CD16/32 surface levels on microglia isolated from wild-type and Fcer1gKO animals. B. Individual and superimposed pseudocolor dot-plots of fluorescence signal detected in PerCP-Cy5.5 (B-710/50 nm, x-axis) and FITC (B-525/30 nm, y-axis) channels from anti-CD16/32 FITC stained and FMO control microglia isolated from wild-type or Fcer1gKO mice. C. Gating of microglia AF subsets by AF intensity in the B-710/50 nm channel. Histograms of FITC signal intensity in autofluorescence+ (AF+) and autofluorescence- (AF-) gated microglia subsets. Inset labels indicate geomean fluorescence intensity in the FITC channel for indicated population and the calculated difference between anti-CD16/32 FITC stained and FMO control. AF, autofluorescence; FMO, Fluorescence minus one; B, 488 nm blue laser.In contrast to the modest levels of expression of CD16/32, microglia express high levels of CD64, which exceed the autofluorescence intensity level seen in microglia. As a result, the detection of this highly expressed antigen is much less subject to autofluorescence confounding issues. This is highlighted by the bright, unimodal CD64 signal observed on wild-type microglia, which clearly surpasses the signal observed in Fcer1gKO microglia (Figure 8A). This is confirmed by displaying the data in a pseudocolor dot plot, as the signal observed in anti-CD64 stained samples exceeds that of background autofluorescence (Figure 8B). Gating AF+ and AF- microglia subsets using the autofluorescence channel further allowed the analysis of CD64 expression levels on these AF microglia subsets (Figure 8C). These results highlight that autofluorescence represents a major confounding factor for the detection of protein expression in microglia using fluorescence-based methods, such as flow cytometry, and that care should be taken to account for autofluorescence using empty cytometer channels to achieve accurate detection and quantification of protein expression in this cell type.
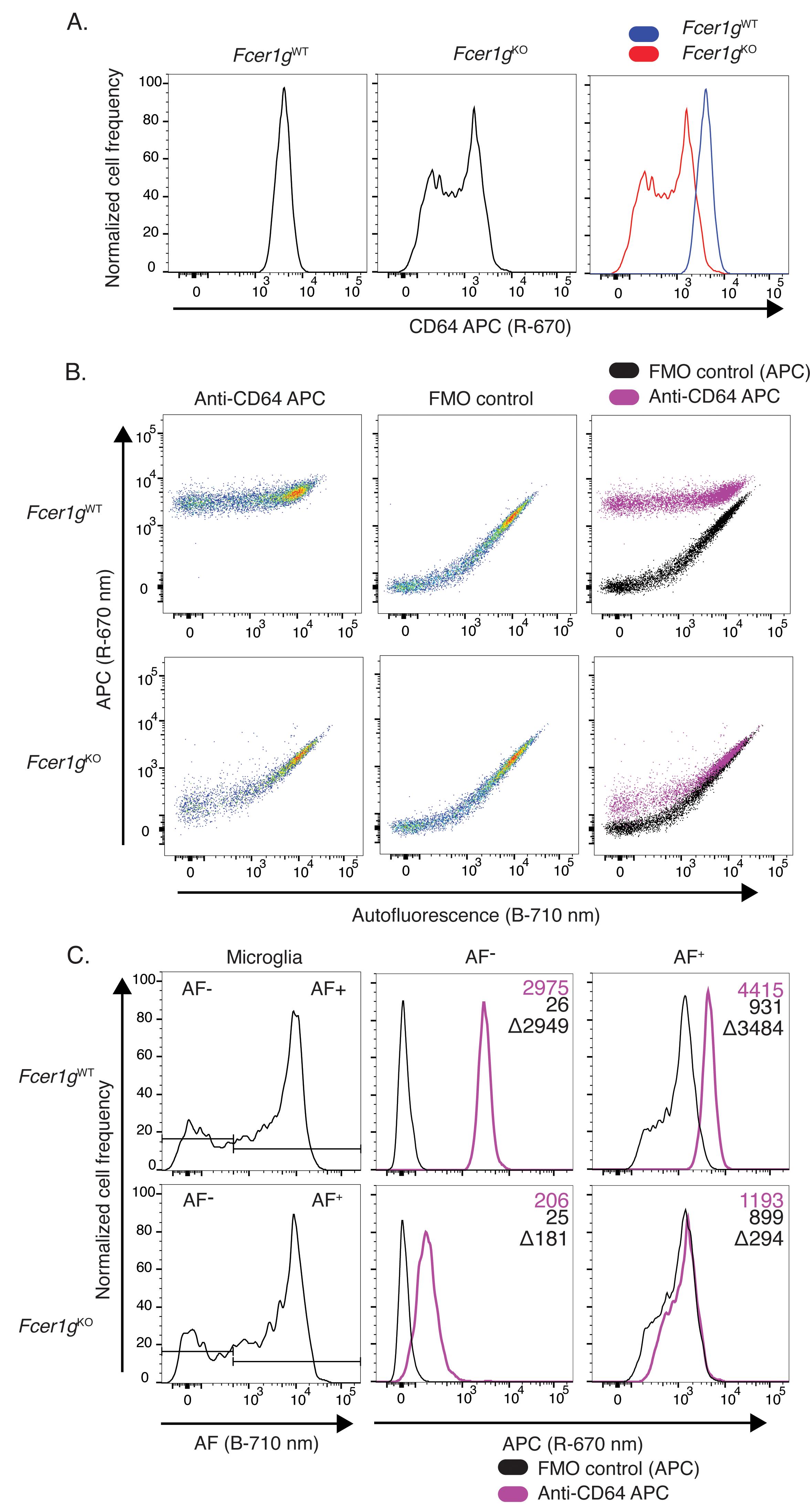
Figure 8. Analysis of Fc receptor (CD64) surface expression levels in 6-month-old Fcerg1 wild-type and knockout microglia. A. Individual and overlaid histograms of CD64 surface levels on microglia isolated from wild-type and Fcer1gKO animals. B. Individual and superimposed pseudocolor dot-plots of fluorescence signal detected in PerCP-Cy5.5 (B-710/50 nm, x-axis) and APC (R-670/30 nm, y-axis) channels from anti-CD64 APC stained and FMO control microglia isolated from wild-type or Fcer1gKO mice. C. Gating of microglia AF subsets by AF intensity in the B-710/50 nm channel. Histograms of APC signal intensity in AF+ and AF- gated microglia subsets. Inset labels indicate geomean fluorescence intensity in the APC channel for indicated population and the calculated difference between anti-CD64 APC stained and FMO control. AF, autofluorescence; FMO, Fluorescence minus one; B, 488 nm blue laser; R, 640 nm red laser.
Notes
During the wash steps of the flow cytometry staining procedure, flicking the plate twice or dabbing it dry after flicking may dislodge the cell pellet and increase cell loss.
Although microglia AF is detectable in all flow cytometer excitation/emission combinations we have tested, we have found that fluorescence detection channels for red-laser (640 nm) excited fluorochromes exhibit relatively lower levels of AF signal, which may minimize the confounding effects of cell autofluorescence on protein marker detection. In contrast, we found that blue-laser (488 nm) fluorescence detection channels were the more sensitive to detect microglia autofluorescence, and PerCP-Cy5.5 (710/50 nm bandpass) provided the highest resolution between AF microglia subsets.
If the microglial expression of an antigen of interest is expected to be low, it is advisable to test with brighter fluorochromes (e.g., AlexaFluor 647); however, relative fluorochrome intensity will vary depending on the cytometer instrument ultimately used.
When generating single-color compensation controls, microglia should not be used, as the brightness level and bimodal nature of autofluorescence will result in an aberrant compensation matrix. Ideally, compensation beads should be used, but if unavailable, a surrogate non-autofluorescent cell population, such as splenocytes, may be used instead.
On the cytometer, PMT voltage levels should not be decreased to artificially minimize microglial autofluorescence, as this will also decrease the signal from antigens of interest.
Although we do not explicitly cover the methods to probe intracellular antigens, such as LAMP1 and Ki-67, we have successfully used the eBioscience FoxP3 Transcription Factor (ThermoFisher, catalog number: 00-5523-00) staining kit, per manufacturer protocol. However, because this strong fixation/permeabilization protocol alters the detection of several key markers to identify microglia, we recommend using the nuclear antigen PU.1 to accurately identify microglia within these fixed and permeabilized single-cell suspensions.
For murine samples, additional antibodies against microglia-specific cell surface markers have been successfully used (Burns et al., 2020, Figure 1), including anti-P2RY12 (Biolegend, catalog number: 848004), anti-TMEM119 (AbCam, catalog number: 225494), and anti-CX3CR1 (Biolegend, catalog number: 149023)
Although this protocol has been applied primarily to mouse brain tissue, we have successfully used it for mouse spinal cord and brain tissue from rats and cynomolgus monkeys. When isolating CNS microglia from species other than mice, the tissue weight being processed should be kept under < 450 mg.
Aging critically impacts levels of autofluorescence observed in microglia. Although autofluorescence can be detected with a bimodal distribution in microglia as early as postnatal day 30, autofluorescence levels dramatically increase with aging and become more and more confounding when mice get into mature adulthood and older (Burns et al., 2020).
Recipes
33% isotonic Percoll
9 ml of Percoll
10× Hanks Balanced Salt Solution
20 ml of 1× HBSS with 25 mM HEPES
HBSS with 25 mM HEPES
12.5 ml 1 M HEPES
487.5 ml 1× HBSS
PBS with 3 mM EDTA
6 ml 0.5 M EDTA
494 ml 1× HBSS
Fc receptor blocking solution (for 10 samples)
245 µl Flow Staining buffer
5 µl TruStain FcX
2× microglia antibody panel (for 10 samples)
2.5 µl of 0.2 mg/ml anti-CD45 BV785
2.5 µl of 0.2 mg/ml anti-CD11b BV510
245 µl Flow Staining buffer
DAPI working solution
1 µl of 1 mg/ml DAPI solution
10 ml of Flow Staining buffer
Acknowledgments
We thank the authors of Burns et al. (2020), from which this protocol was originally derived from.
Competing interests
JCB and MM are full-time employees of Biogen and Biogen shareholders. RMR is a full-time employee of Third Rock Ventures. No authors were provided compensation or free products by any vendors utilized in this protocol.
Ethics
This study was performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Research animals at Biogen were housed in an AAALAC accredited facility and handled according to an approved institutional animal care and use committee (IACUC) protocol (#756).
References
- Autengruber, A., Gereke, M., Hansen, G., Hennig, C. and Bruder, D. (2012). Impact of enzymatic tissue disintegration on the level of surface molecule expression and immune cell function. Eur J Microbiol Immunol (Bp) 2(2): 112-120.
- Benmamar-Badel, A., Owens, T. and Wlodarczyk, A. (2020). Protective Microglial Subset in Development, Aging, and Disease: Lessons From Transcriptomic Studies. Front Immunol 11: 430.
- Burns, J. C., Cotleur, B., Walther, D. M., Bajrami, B., Rubino, S. J., Wei, R., Franchimont, N., Cotman, S. L., Ransohoff, R. M. and Mingueneau, M. (2020). Differential accumulation of storage bodies with aging defines discrete subsets of microglia in the healthy brain. Elife 9: e57495.
- Clayton, K. A., Van Enoo, A. A. and Ikezu, T. (2017). Alzheimer's Disease: The Role of Microglia in Brain Homeostasis and Proteopathy. Front Neurosci 11: 680.
- Li, Q. and Barres, B. A. (2018). Microglia and macrophages in brain homeostasis and disease. Nat Rev Immunol 18(4): 225-242.
- Mattei, D., Ivanov, A., van Oostrum, M., Pantelyushin, S., Richetto, J., Mueller, F., Beffinger, M., Schellhammer, L., Vom Berg, J., Wollscheid, B., Beule, D., Paolicelli, R. C. and Meyer, U. (2020). Enzymatic Dissociation Induces Transcriptional and Proteotype Bias in Brain Cell Populations. Int J Mol Sci 21(21).
- O'Flanagan, C. H., Campbell, K. R., Zhang, A. W., Kabeer, F., Lim, J. L. P., Biele, J., Eirew, P., Lai, D., McPherson, A., Kong, E., Bates, C., Borkowski, K., Wiens, M., Hewitson, B., Hopkins, J., Pham, J., Ceglia, N., Moore, R., Mungall, A. J., McAlpine, J. N., Team, C. I. G. C., Shah, S. P. and Aparicio, S. (2019). Dissociation of solid tumor tissues with cold active protease for single-cell RNA-seq minimizes conserved collagenase-associated stress responses. Genome Biol 20(1): 210.
- Pellerin, K., Rubino, S. J., Burns, J. C., Smith, B. A., McCarl, C. A., Zhu, J., Jandreski, L., Cullen, P., Carlile, T. M., Li, A., Rebollar, J. V., Sybulski, J., Reynolds, T. L., Zhang, B., Basile, R., Tang, H., Harp, C. P., Pellerin, A., Silbereis, J., Franchimont, N., Cahir-McFarland, E., Ransohoff, R. M., Cameron, T. O. and Mingueneau, M. (2021). MOG autoantibodies trigger a tightly-controlled FcR and BTK-driven microglia proliferative response. Brain. doi: 10.1093/brain/awab231.
- Ransohoff, R. M. and El Khoury, J. J. (2016). Microglia in health and disease. Cold Spring Harb. Perspect. Biol 8(1): a020560.
- Sato-Hashimoto, M., Nozu, T., Toriba, R., Horikoshi, A., Akaike, M., Kawamoto, K., Hirose, A., Hayashi, Y., Nagai, H., Shimizu, W., Saiki, A., Ishikawa, T., Elhanbly, R., Kotani, T., Murata, Y., Saito, Y., Naruse, M., Shibasaki, K., Oldenborg, P. A., Jung, S., Matozaki, T., Fukazawa, Y. and Ohnishi, H. (2019). Microglial SIRPalpha regulates the emergence of CD11c(+) microglia and demyelination damage in white matter. Elife 8: e42025.
Article Information
Copyright
Burns et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Burns, J. C., Ransohoff, R. M. and Mingueneau, M. (2021). Isolation of Microglia and Analysis of Protein Expression by Flow Cytometry: Avoiding the Pitfall of Microglia Background Autofluorescence. Bio-protocol 11(14): e4091. DOI: 10.21769/BioProtoc.4091.
- Burns, J. C., Cotleur, B., Walther, D. M., Bajrami, B., Rubino, S. J., Wei, R., Franchimont, N., Cotman, S. L., Ransohoff, R. M. and Mingueneau, M. (2020). Differential accumulation of storage bodies with aging defines discrete subsets of microglia in the healthy brain. Elife 9: e57495.
Category
Immunology > Immune cell isolation > Myeloid cell
Neuroscience > Cellular mechanisms > Microglia
Cell Biology > Cell-based analysis > Flow cytometry
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









