- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Antisense Oligo Pulldown of Circular RNA for Downstream Analysis
(*contributed equally to this work) Published: Vol 11, Iss 14, Jul 20, 2021 DOI: 10.21769/BioProtoc.4088 Views: 5187
Reviewed by: Florian KarrethGopal PandiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Efficient Large DNA Fragment Knock-in by Long dsDNA with 3′-Overhangs Mediated CRISPR Knock-in (LOCK) in Mammalian Cells
Wenjie Han [...] Jianqiang Bao
Oct 20, 2023 2800 Views

Leveraging Circular Polymerization and Extension Cloning (CPEC) Method for Construction of CRISPR Screening Libraries
Bengisu Dayanc [...] Serif Senturk
Feb 20, 2025 2832 Views

Selective Isolation of TOP3B•mRNA Covalent Intermediates Using Denaturing Oligo-dT Pulldown
Julia E. Warrick and Michael G. Kearse
Mar 5, 2026 55 Views
Abstract
Circular RNAs (circRNAs) are a large family of noncoding RNA molecules that have emerged as novel regulators of gene expression by sequestering microRNAs (miRNAs) and RNA-binding proteins (RBPs). Several computational tools have been developed to predict circRNA interaction with target miRNAs and RBPs with a view to studying their potential effect on downstream target genes and cellular physiology. Biochemical assays, including reporter assays, AGO2 pulldown, ribonucleoprotein pulldown, and biotin-labeled RNA pulldown, are used to capture the association of miRNAs and RBPs with circRNAs. Only a few studies have used circRNA pulldown assays to capture the associated miRNAs and RBPs under physiological conditions. In this detailed protocol, the circRNA of interest (e.g., circHipk2) was captured using a biotin-labeled antisense oligo (ASO) targeting the circHipk2 backsplice junction sequence followed by pulldown with streptavidin-conjugated magnetic beads. The specific enrichment of circRNA was analyzed using reverse transcription quantitative PCR (RT-qPCR). Furthermore, the ASO pulldown assay can be coupled to miRNA RT-qPCR and western blotting analysis to confirm the association of miRNAs and RBPs predicted to interact with the target circRNA. In summary, the specific pulldown of circRNA using this quick and easy method makes it a useful tool for identifying and validating circRNA interaction with specific miRNAs and RBPs.
Keywords: CircRNAsBackground
The RNA family can be broadly classified into coding (mRNAs) and noncoding RNAs. Interestingly, the vast majority of cellular RNAs are noncoding RNAs, including rRNA, lincRNA, miRNA, snRNA, snoRNA, tRNA, piRNA, and poorly characterized circular (circ) RNAs (Palazzo and Lee, 2015). The use of next-generation RNA sequencing and bioinformatics tools has uncovered more than a hundred thousand circRNAs in humans (Vromman et al., 2020). CircRNAs are generated from pre-mRNAs by head-to-tail splicing of specific exons by a process called backsplicing, which is regulated by transcription speed, RBPs, and inverted repeat sequences. Their length can vary from less than 100 nucleotides to a few thousand nucleotides. Depending on the sequence of circRNA origin from the parent gene, circRNAs have been classified into exonic circular RNA (circRNA), lariat-derived circular intronic RNA (ci-RNA), stable intronic sequence RNA (sisRNA), and exonic-intronic circular RNA (EIcircRNA) (Guria et al., 2019). Although a vast number of circRNAs have been identified in different cellular systems and disease models, only a fraction of circRNAs has been functionally characterized (Vromman et al., 2020). Recent evidence suggests that circRNAs play a critical role in regulating cellular events by interacting with miRNAs and RBPs (Guria et al., 2019). The majority of studies use computational tools to predict the association of circRNAs with cellular miRNAs and proteins that could regulate the expression of downstream target genes (Dudekula et al., 2016). Several biochemical techniques are currently used to analyze the circRNA-miRNA interaction, including luciferase reporter assays, AGO2 pulldown, and fluorescence in situ hybridization (FISH). Similarly, ribonucleoprotein pulldown assays using an antibody against the predicted RBP and biotin-labeled RNA pulldown assays to capture the target RBPs are used to validate the interaction of circRNA with specific RBPs. However, some of these methods are indirect assays to conclude whether the circRNA is associated with target miRNAs or RBPs. Our previous studies have successfully used circRNA pulldown assays with biotin-labeled antisense oligos targeting the circRNA backsplice junction sequence. Furthermore, circRNA-pulldown followed by RT-qPCR and western blotting analysis identified the miRNA or RBP associated with the circRNA of interest (Abdelmohsen et al., 2017; Panda et al., 2017; Pandey et al., 2020). Here, we adapted the published method to successfully pull down circHipk2 in βTC6 cells. The detailed protocol for the pulldown of circRNA to analyze the associated miRNA or RBP is described here.
Materials and Reagents
Nuclease-free 1.5-ml microcentrifuge tubes (Tarson, catalog number: 500010)
Nuclease-free 2-ml microcentrifuge tubes (Tarson, catalog number: 500020)
10 µl, 20 µl, 200 µl, and 1 ml tips (Tarson, catalog numbers: 524053, 528101, 528104, 528106)
96-well PCR plates (Thermo Fisher Scientific, catalog number: 4483485)
Nuclease-free water (Thermo Fisher Scientific, catalog number: 10977023)
0.25% Trypsin (Thermo Fisher Scientific, catalog number: 25200056)
Phosphate-buffered saline (PBS) (Sigma, catalog number: P4417-50TAB)
1 M Tris-HCl, pH 7.5 (Thermo Fisher Scientific, catalog number: 15567027)
1 M Tris-HCl, pH 8.0 (Thermo Fisher Scientific, catalog number: 15568025)
2 M Potassium chloride (Thermo Fisher Scientific, catalog number: AM9640G)
1 M Magnesium chloride (Thermo Fisher Scientific, catalog number: AM9530G)
5 M Sodium chloride (Thermo Fisher Scientific, catalog number: AM9759)
Nonidet P-40 (Amresco, catalog number: M158-100ML)
0.5 M EDTA, pH 8.0 (Amresco, catalog number: E177-100ML)
Triton X-100 (SRL, catalog number: 64518)
Murine RNase inhibitor (NEB, catalog number: M0314S, 40 U/µl)
20× Protease inhibitor (Sigma, catalog number: S8830)
Streptavidin Dynabeads (NEB, catalog number: S1420S)
TRIzol (Thermo Fisher Scientific, catalog number: 15596018)
Ethanol (HiMedia, catalog number: MB228)
Chloroform (SRL, catalog number: 96764)
Isopropanol (SRL, catalog number: 38445)
GlycoBlueTM Coprecipitant (15 mg/ml) (Thermo Fisher Scientific, catalog number: AM9515)
Maxima reverse transcriptase (Thermo Fisher Scientific, catalog number: EP0743)
dNTP set, 100 mM solutions (Thermo Fisher Scientific, catalog number: R0181)
Random primers (Thermo Fisher Scientific, catalog number: 48190011)
PowerUp SYBR Green master mix (Thermo Fisher Scientific, catalog number: A25778)
MicroAmp optical adhesive film (Thermo Fisher Scientific, catalog number: 4311971)
Divergent DNA oligo primers synthesized by SIGMA for PCR amplification of circHipk2 (circHipk2-F: 5’-TCCAGACAACCGTACCGAGT-3’ and circHipk2-R: 5’-GGCACTTGATTGAAGGGTGT-3’)
Custom antisense oligos labeled with biotin-TEG synthesized by SIGMA for control (Ctrl-ASO: 5’-TGCGTAACGAACGACGAATCGTCGCAGATC-3’[BtnTg]) and circHipk2 (circHipk2-ASO: 5’-CATGTGAGGCCATACCGGTAGTATCTGGAT-3’[BtnTg])
3× SDS loading dye (NEB, catalog number: B7703S)
Polysome extraction buffer (PEB) (see Recipes)
2× Tris, EDTA, NaCl, Triton (TENT) buffer (see Recipes)
1× TENT (see Recipes)
Equipment
Manual pipette set, 2 µl, 20 µl, 200 µl, and 1 ml (various manufacturers)
Vortex mixer (Tarson, catalog number: 3001)
Magnetic stand (Tarson, catalog number: S1509S)
Tube rotator (Tarson, catalog number: 3071)
Refrigerated centrifuge (Eppendorf, model: 5810R)
Benchtop microfuge (Tarson, catalog number: 1010)
Thermomixer (Eppendorf, catalog number: 5384000012)
PCR machine (various manufacturers)
QuantStudio 3 real-time PCR system (Thermo Fisher Scientific, catalog number: A28567)
Software
UCSC genome browser (https://genome.ucsc.edu/)
Primer3 webtool (https://bioinfo.ut.ee/primer3/)
GeneRunner (http://www.generunner.net/)
QuantStudio 3 and 5 system software
Circinteractome website (https://circinteractome.nia.nih.gov/)
Procedure
Oligo design
Obtain the mature sequence of the circRNA of interest from the RNA sequencing data or the UCSC genome browser using the genome coordinates of backsplice sites. Here, we retrieved the sequence of mouse circHipk2 (chr6|38818229|38819313|-) from the mouse mm10 UCSC genome browser (Figure 1).
Note: The mature splice sequence of multiexonic circRNA can be obtained by combining all the exon sequences between the genomic coordinates of the backsplice site.
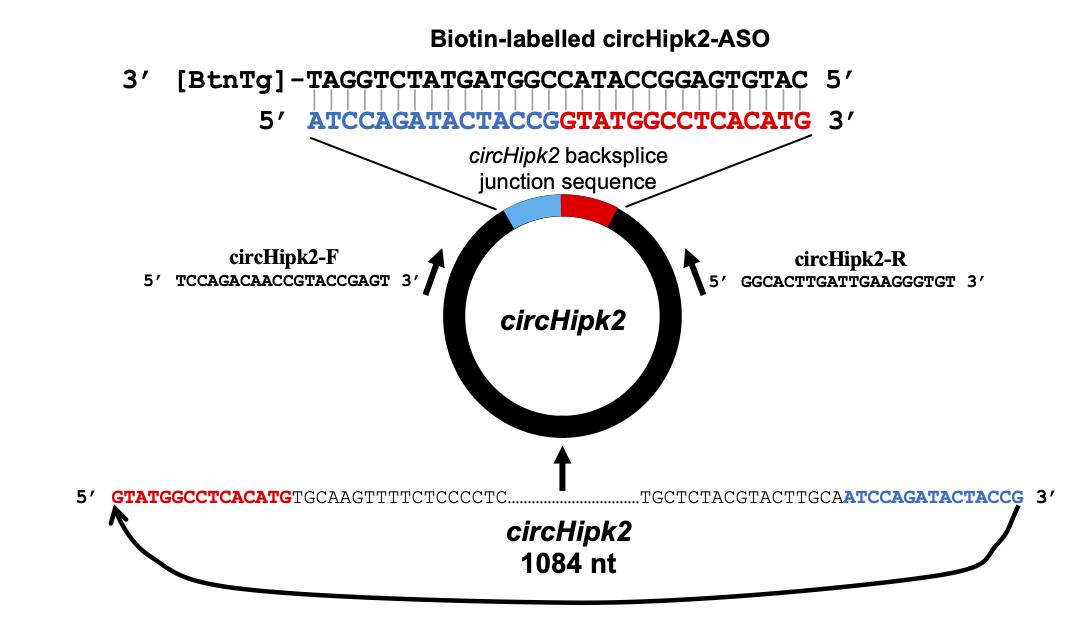
Figure 1. Design of the divergent primers and biotin-labeled antisense oligo targeting circHipk2As shown in Figure 1, join the last 15 nucleotides of the circRNA sequence to the upstream of the first 15 nucleotides to obtain the 30-nucleotide sequence spanning the circRNA backsplice junction.
To design ASO, generate the reverse complement sequence of the 30-nucleotide junction sequence using GeneRunner. Have this sequence synthesized with biotin-TEG added to the 3’ end by the preferred vendor.
Design primers for the target housekeeping gene mRNAs or rRNAs using the Primer 3 web tool.
Design the divergent primer for the target circRNA as described previously (Panda and Gorospe, 2018) or using the circinteractome website for human circRNAs (Dudekula et al., 2016).
Cell lysis
Take one 100-mm dish of ~70% confluent βTC6 cells and discard the culture media.
Note: A minimum of 5 million cells should be used for the pulldown assay. A higher amount of cells may help to obtain better pulldown of rare or low copy number circRNAs.Wash the cells three times with 5-10 ml ice-cold 1× PBS.
Harvest the cells by trypsinization or scraping with a cell scraper.
Pellet the cells by centrifuging at 1,000 × g for 2 min at 4°C and discard the supernatant.
Resuspend the cell pellet in 1 ml ice-cold polysome extraction buffer (PEB).
Note: Lysis with PEB releases the cytoplasmic fraction, not the nuclear fraction.Immediately add 5 µl RNase inhibitor and 50 µl 20× protease inhibitor.
Mix well by pipetting ten times and keep on ice for 15 min, pipetting or vortexing for a few seconds every 4-5 min until the cells are lysed.
Centrifuge the lysate at 12,000 × g for 10 min at 4°C.
Collect 900 µl supernatant and proceed to the hybridization step (Figure 2).
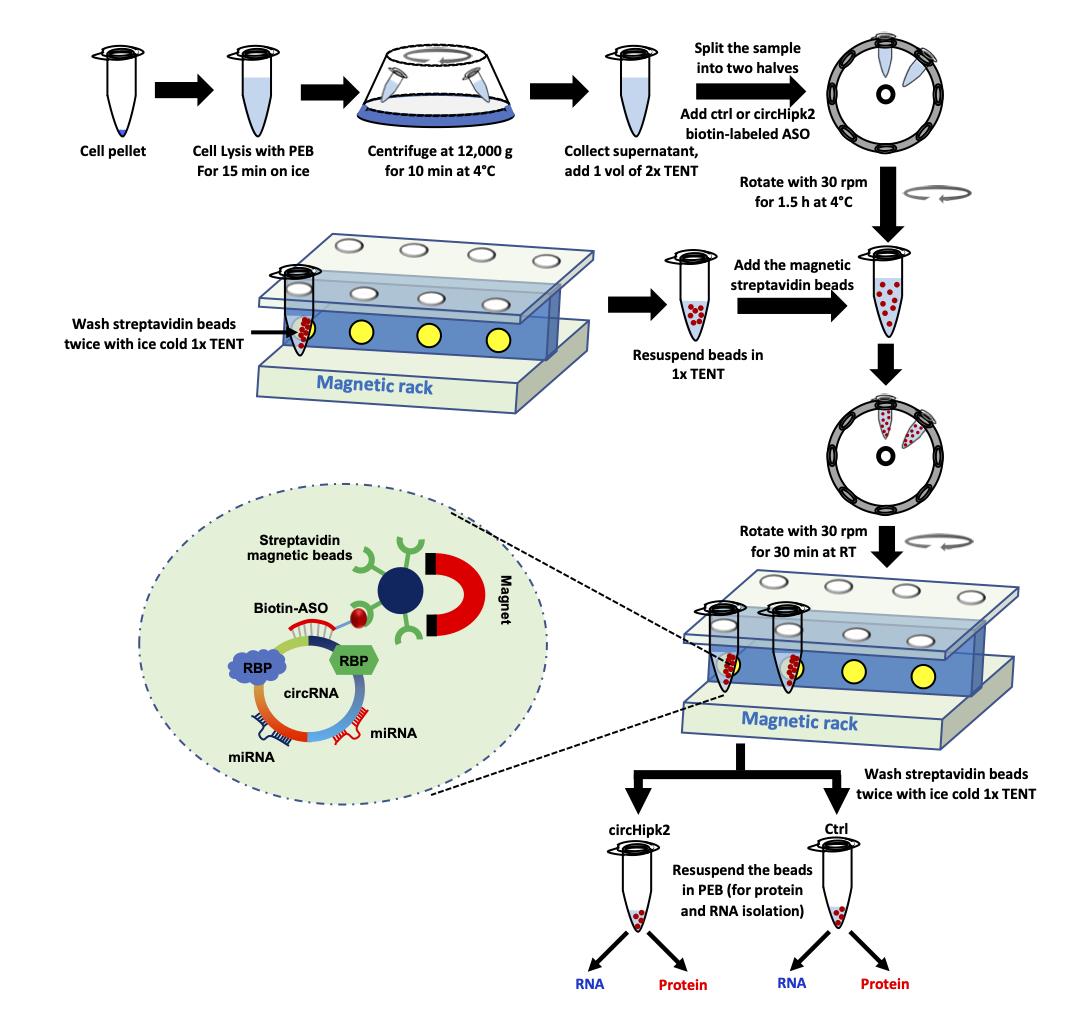
Figure 2. Schematic of the pulldown of circRNA using biotinylated-ASO targeting circHipk2
Antisense oligo hybridization
Add an equal volume (900 µl) of ice-cold 2× Tris, EDTA, NaCl, Triton (TENT) buffer to the supernatant collected in the above step in a 2-ml microcentrifuge tube.
Divide the mixture into two tubes (one for the control-ASO pulldown and the other for the circRNA-ASO pulldown).
Add 1 µl 100 µM (100 pmol) ctrl-ASO and circRNA-ASO to the control and circRNA pulldown tube, respectively.
Note: The manufacturer's datasheet gives the streptavidin bead binding capacity. The ASO amount should be less than the streptavidin bead binding capacity for the pulldown in the next step. A higher amount of ASO may reduce the pulldown efficiency.Incubate the reactions on a rotor at 30 rpm for 90 min at 4°C.
Note: The ASO hybridization reaction can be performed at room temperature or at 37°C to facilitate ASO binding to target circRNA; however, this may affect RNA integrity in samples with high levels of endogenous RNase.
Streptavidin bead preparation
Start the streptavidin bead preparation 15 min before the ASO hybridization step is completed.
Mix well and place 100 µl magnetic streptavidin beads (50 µl for each reaction) into a new tube.
Place the magnetic beads on a magnetic stand for 30 s and discard the supernatant.
Resuspend the magnetic beads in 500 µl ice-cold 1× TENT buffer and place the tube on the magnetic stand for 30 s.
Rotate the tubes 180º on the magnetic rack twice to wash the beads (Video 1).
Video 1. Magnetic beads washingDiscard the supernatant.
Repeat the washing steps (Steps D4-D6) twice.
Resuspend the magnetic streptavidin beads in 100 µl ice-cold 1× TENT buffer.
Circular RNA pulldown
Add 50 µl washed magnetic streptavidin beads into each hybridization reaction tube (mentioned in Step C4) after completing the 90-minute hybridization step.
Add 1 µl RNase inhibitor and 5 µl 20× protease inhibitor to the tubes and mix well by pipetting.
Rotate both the control and circRNA ASO tubes at room temperature for 30 min on a tube rotator at 30 rpm.
Centrifuge the tubes briefly to bring all the liquid samples to the bottom of the tube without pelleting the beads.
Note: High-speed centrifugation may pellet the beads, which may affect the washing steps.
Place the tubes on a magnetic rack for 30 s and discard the supernatant.
Resuspend the magnetic beads in 500 µl ice-cold 1× TENT buffer and place the tube on the magnetic stand for 30 s.
Rotate the tube twice on the magnetic stand to wash the beads (Video 1).
Allow the beads to settle toward the magnet and discard the buffer.
Repeat the washing steps (steps 6-8) twice.
Note: The number of washes may be decided depending on the fold enrichment of the target circRNA. We found that two to three washes are suitable to enrich circHipk2 in our pulldown assays.After the last wash, centrifuge the tube for a few seconds to settle the beads at the bottom.
Place the tube on the magnetic stand for 30 s and discard the remaining supernatant.
Resuspend the beads in 30 µl ice-cold PEB.
Take 15 µl beads to a fresh tube for RNA isolation and RT-qPCR to analyze the pulldown efficiency of circRNA using the ASO and to detect the circRNA-associated miRNAs.
Add 7 µl 3× SDS loading dye to the remaining 15 µl beads and mix by pipetting, then heat at 95°C for 5 min. This sample can be immediately used for western blotting to identify interacting RBPs or stored at -20°C.
RNA and cDNA preparation from the pulldown sample
Add 250 µl TRIzol reagent to the 15 µl beads for RNA isolation and mix well by pipetting.
Note: Any similar RNA isolation reagent such as TriSure, RNAzol, TRI reagent, or TriPure can be used to prepare RNA. Column-based RNA isolation kits should be avoided since miRNAs are not purified well using the standard RNA isolation kits. Otherwise, use a kit that can isolate both long RNAs and miRNAs together.
Add 50 µl chloroform and vortex for 15 s.
Centrifuge at 12,000 × g for 15 min at 4°C, then collect 100 µl aqueous layer into a new tube.
Add 100 µl isopropanol and 0.5 µl glycoblue as a co-precipitant.
Mix well and keep at room temperature for 10 min, then centrifuge at 12,000 × g for 10 min at 4°C.
Discard the supernatant, add 500 µl 75% ethanol to the RNA pellet, and vortex the tube for a few seconds.
Centrifuge at 12,000 × g for 5 min at room temperature.
Discard the supernatant and air-dry the RNA pellet for 3-5 min with the lid open.
Note: Excessive drying of the RNA pellet or residual ethanol may inhibit the solubility of the pellet.
Dissolve the pellet in 20 µl nuclease-free water. The RNA can be stored at -20°C for future use or used immediately for cDNA synthesis.
Prepare a 20-μl cDNA synthesis reaction containing 13 μl prepared RNA, 0.5 μl Maxima reverse transcriptase, 0.5 μl RNase inhibitor, 1 μl 10 mM dNTP mix, 1 μl random primers, and 4 μl 5× RT buffer.
Note: The cDNA can be prepared using any standard reverse transcription kit and random primers.
Mix the reaction gently and incubate for 10 min at room temperature followed by 1 h at 50°C.
Incubate the reaction at 85°C for 5 min to inactivate the reverse transcriptase.
Dilute the cDNA with 250 µl nuclease-free water. This can be stored at -20°C or used immediately for PCR analysis.
circRNA enrichment analysis by RT-qPCR
Mix 10 μl 100 μM forward and reverse primer stock with 980 μl nuclease-free water to obtain a final concentration of 1 μM primer mix.
Prepare 20-μl reactions in a 96-well plate containing 5 μl cDNA, 5 μl primer mix, and 10 μl 2× SYBR Green mix. Three technical replicates should be prepared.
Seal the plate with an optical adhesive cover and vortex for a few seconds.
Centrifuge the plate for a few seconds to bring the reactions to the bottom of the wells.
Perform quantitative PCR on a QuantStudio 3 Real-Time PCR System with the following reaction setup: initial cycle for 2 min at 95°C followed by 40 cycles of 5 s at 95°C and 20 s at 60°C.
Obtain the average Ct value of the technical replicates for each target in the control and circRNA pulldown samples.
Calculate the percentage (%) enrichment of the target circRNA in the pulldown sample as compared with the control pulldown sample using the delta-CT method (Figure 3) (Livak and Schmittgen, 2001).
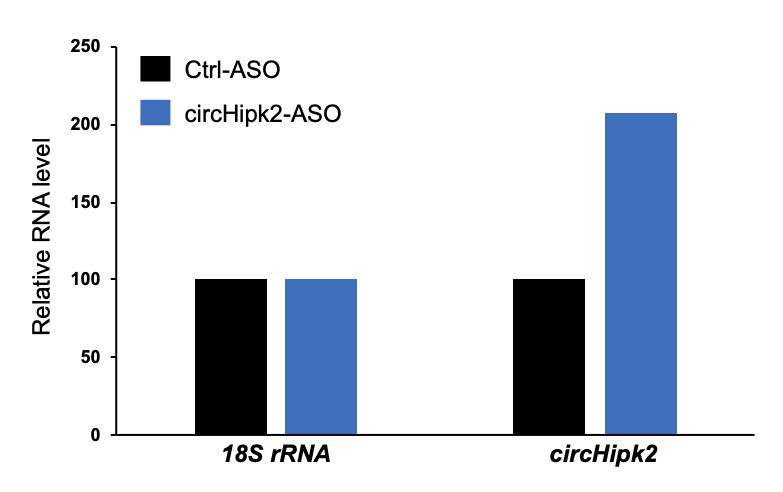
Figure 3. Example data showing the percentage enrichment of circHipk2 in the circRNA-ASO pulldown sample relative to the control ASO pulldown sample
Detection of interacting miRNAs and RBPs
After confirming the enrichment of target circRNA in the circRNA ASO pulldown sample, associated miRNAs and RBPs can be analyzed.
The miRNAs associated with the target circRNA can be analyzed by RT-qPCR. The RNA prepared in the above step can be used to produce miRNA cDNA followed by qPCR analysis to study their specific enrichment in the circRNA pulldown samples.
Perform standard western blotting to detect circRNA-associated RBPs. Briefly, subject the input, control ASO, and circRNA ASO pulldown samples mixed with SDS dye to SDS-PAGE, and transfer the proteins to nitrocellulose membrane. Incubate the membrane with primary antibody against the predicted RBP followed by the appropriate secondary antibody and detect the chemiluminescence signals.
Data analysis
This method describes circular RNA pulldown using an antisense oligo specifically targeting the backsplice junction sequence of the target circRNA. However, the success of this pulldown assay depends on the availability of the circRNA junction sequence, which may be inaccessible due to secondary structures or junction-interacting RBPs, preventing the ASO from binding. Moreover, the PEB used for cell lysis is good for releasing the cytoplasmic fraction, while lysis of the nucleus for the pulldown of nuclear-localized circRNAs remains to be standardized. Since most circRNAs are cytoplasmic in nature (Jeck et al., 2013), we used PEB for the cell lysis and pulldown assay. Use the delta-Ct method to analyze the enrichment of target circRNA in the circRNA ASO pulldown sample compared with the control pulldown using a loading control such as 18S rRNA or Gapdh mRNA (Livak and Schmittgen, 2001). As shown in Figure 3, the circHipk2 levels are more than 2-fold higher in the circHipk2 ASO pulldown than in the control ASO pulldown. Alternatively, the efficiency of circRNA pulldown can be measured by comparing the pulldown samples with the flow through or input. After checking the enrichment of circRNA in the pulldown samples, the remaining RNA can be subjected to miRNA RT-qPCR analysis to check for the specific enrichment of miRNAs predicted to interact with the circRNA of interest. Furthermore, the other half of the pulldown sample may be used for western blotting analysis to evaluate the RBPs associated with the target circRNA. This is a promising method to pulldown the circular RNA of interest and study the circRNA-associated miRNAs and RBPs, which are critical factors for circRNA-mediated gene regulation.
Recipes
Polysome extraction buffer (PEB)
Reagents Stock Vol required for 250 ml 20 mM Tris-HCl, pH 7.5 1 M 5 ml 100 mM KCl 2 M 12.5 ml 5 mM MgCl2 1 M 1.25 ml 0.5% Nonidet P-40 10% 12.5 ml Adjust the volume to 250 ml with nuclease-free water 2× Tris, EDTA, NaCl, Triton (TENT) buffer
Reagents Stock Vol required for 100 ml 20 mM Tris-HCl pH 8.0 1 M 2 ml 2 mM EDTA pH 8.0 0.5 M 0.4 ml 500 mM NaCl 5 M 10 ml 1% v/v Triton X-100 20% 5 ml Adjust the volume to 100 ml with nuclease-free water 1× TENT
Mix equal volumes of PEB/water and 2× TENT
Acknowledgments
This research was supported by intramural funding from the Institute of Life Sciences and the Wellcome Trust/DBT India Alliance Intermediate Fellowship (IA/I/18/2/504017) provided to ACP. DD and AD were supported by Junior Research Fellowships from the University Grant Commission, India. This protocol was adapted from previously published papers (Abdelmohsen et al., 2017 and Panda et al., 2017). The authors thank our colleagues at the Institute of Life Sciences, Bhubaneswar, for proofreading the article.
Competing interests
The authors declare no conflicts of interest.
References
- Abdelmohsen, K., Panda, A. C., Munk, R., Grammatikakis, I., Dudekula, D. B., De, S., Kim, J., Noh, J. H., Kim, K. M., Martindale, J. L. and Gorospe, M. (2017). Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol 14(3): 361-369.
- Dudekula, D. B., Panda, A. C., Grammatikakis, I., De, S., Abdelmohsen, K. and Gorospe, M. (2016). CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol 13(1): 34-42.
- Guria, A., Sharma, P., Natesan, S. and Pandi, G. (2019). Circular RNAs-The Road Less Traveled. Front Mol Biosci 6: 146.
- Jeck, W. R., Sorrentino, J. A., Wang, K., Slevin, M. K., Burd, C. E., Liu, J., Marzluff, W. F. and Sharpless, N. E. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19(2): 141-157.
- Livak, K. J. and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25(4): 402-408.
- Palazzo, A. F. and Lee, E. S. (2015). Non-coding RNA: what is functional and what is junk? Front Genet 6: 2.
- Panda, A. C. and Gorospe, M. (2018). Detection and Analysis of Circular RNAs by RT-PCR. Bio-protocol 8(6): e3241.
- Panda, A. C., Grammatikakis, I., Kim, K. M., De, S., Martindale, J. L., Munk, R., Yang, X., Abdelmohsen, K. and Gorospe, M. (2017). Identification of senescence-associated circular RNAs (SAC-RNAs) reveals senescence suppressor CircPVT1. Nucleic Acids Res 45(7): 4021-4035.
- Pandey, P. R., Yang, J. H., Tsitsipatis, D., Panda, A. C., Noh, J. H., Kim, K. M., Munk, R., Nicholson, T., Hanniford, D., Argibay, D., Yang, X., Martindale, J. L., Chang, M. W., Jones, S. W., Hernando, E., Sen, P., De, S., Abdelmohsen, K. and Gorospe, M. (2020). circSamd4 represses myogenic transcriptional activity of PUR proteins. Nucleic Acids Res 48(7): 3789-3805.
- Vromman, M., Vandesompele, J. and Volders, P. J. (2021). Closing the circle: current state and perspectives of circular RNA databases. Brief Bioinform 22(1): 288-297.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Das, D., Das, A. and Panda, A. C. (2021). Antisense Oligo Pulldown of Circular RNA for Downstream Analysis. Bio-protocol 11(14): e4088. DOI: 10.21769/BioProtoc.4088.
Category
Molecular Biology > RNA > RNA-protein interaction
Cancer Biology > General technique > Molecular biology technique
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











