- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Method for Rapid Enzymatic Cleaning for Reuse of Patch Clamp Pipettes: Increasing Throughput by Eliminating Manual Pipette Replacement between Patch Clamp Attempts
Published: Vol 11, Iss 14, Jul 20, 2021 DOI: 10.21769/BioProtoc.4085 Views: 4443
Reviewed by: Alexandros C KokotosAkira KarasawaCarmelo Bellardita

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Capacitance Measurements of Exocytosis From AII Amacrine Cells in Retinal Slices
Espen Hartveit and Margaret L. Veruki
Jan 5, 2025 2323 Views

Visualization of Gap Junction–Mediated Astrocyte Coupling in Acute Mouse Brain Slices
Nine F. Kompier [...] Fritz G. Rathjen
Feb 20, 2025 2262 Views

Ultrafast Isolation of Synaptic Terminals From Rat Brain for Cryo-Electron Tomography Analysis
Rong Sun and Qiangjun Zhou
Sep 5, 2025 3559 Views
Abstract
The whole-cell patch-clamp method is a gold standard for single-cell analysis of electrical activity, cellular morphology, and gene expression. Prior to our discovery that patch-clamp pipettes could be cleaned and reused, experimental throughput and automation were limited by the need to replace pipettes manually after each experiment. This article presents an optimized protocol for pipette cleaning, which enables it to be performed quickly (< 30 s), resulting in a high yield of whole-cell recording success rate (> 90%) for over 100 reuses of a single pipette. For most patch-clamp experiments (< 30 whole-cell recordings per day), this method enables a single pipette to be used for an entire day of experiments. In addition, we describe easily implementable hardware and software as well as troubleshooting tips to help other labs implement this method in their own experiments. Pipette cleaning enables patch-clamp experiments to be performed with higher throughput, whether manually or in an automated fashion, by eliminating the tedious and skillful task of replacing pipettes. From our experience with numerous electrophysiology laboratories, pipette cleaning can be integrated into existing patch-clamp setups in approximately one day using the hardware and software described in this article.
Graphic abstract:
Rapid enzymatic cleaning for reuse of patch-clamp pipettes
Background
Whole-cell patch-clamp recordings allow unprecedented access to electrical activity, neuronal morphology, and gene expression at the single-cell level (Jiang et al., 2015; Gouwens et al., 2019). However, because this method requires a great amount of skill and care to perform correctly, it remains one of the most difficult in neuroscience. A crucial step in this method is the formation of a tight, high resistance (e.g., >1 GΩ) connection between the cell membrane and the glass pipette known as a gigaseal. Gigaseal formation requires a clean pipette surface, and even small contaminants (e.g., cell debris or dust) can disrupt this process (Hamill et al., 1981). For this reason, patch-clamp experimenters need to replace glass pipettes after each recording attempt (Figure 1A), requiring additional time and attention (e.g., removal, fabrication, filling, and installation of pipettes). We have previously found that contrary to decades of this ubiquitous practice in the field, patch-clamp pipettes can be cleaned and reused, enabling many patch clamp recordings with a single pipette (Kolb et al., 2016).
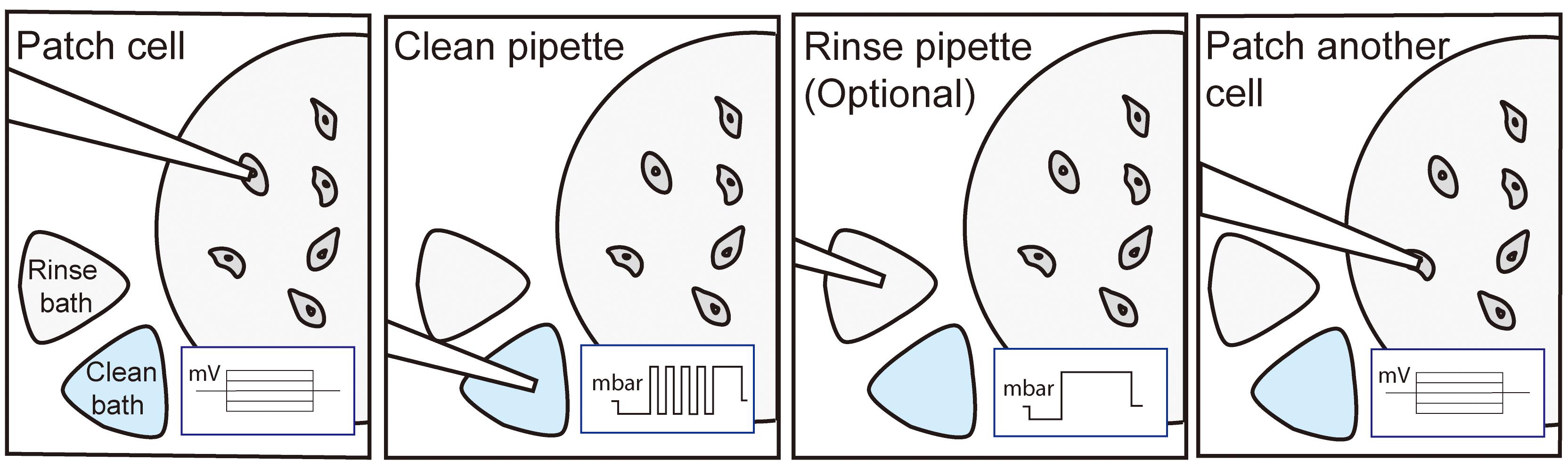
Pipette cleaning is a robust, simple process involving the following steps: (1) attempt whole-cell patch-clamp recording; (2) retract patch-clamp pipette and move towards bath containing cleaning solution; (3) with tip submerged in cleaning solution, cycle positive and negative pressures to remove cell debris from pipette tip; (4) position pipette over new target cell for second patch-clamp attempt; and (5) repeat steps 1-4 until the experiment is completed or the pipette fails (e.g., tip breakage, clog, evaporation of cleaning solution, or user error). In this protocol, we will describe the use of simple hardware and software to automate pipette cleaning, which will assist other labs in implementing pipette cleaning in either fully automated or “push-to-clean” methods (Figure 1B).

Figure 1. Pipette cleaning methods. A. Process flow chart for traditional manual patch clamping without pipette cleaning. Removing, filling, and installing fresh pipettes takes between 60-120 s. B. Process flow chart for manual patching with automated cleaning (“push-to-clean”). Automated cleaning can be run in as little as 30 s. C. Close up images of pipette being moved from the experimental chamber (left) to the cleaning bath (middle) and to the rinse bath (right) before returning to the experimental bath to patch another cell. Scale bar is 25 mm. D. Custom experimental chamber for pipette cleaning featuring fluid inlet and outlet, inset for ground wire, and external baths for cleaning and rinsing solutions. Scale bar is 1 cm.
We published our initial discovery of this method in 2016 and used it to develop the first fully autonomous patch-clamp robot capable of recording dozens of cells with no human supervision in 2019 (Kolb et al., 2016 and 2019). This pipette cleaning method has been used by us, our collaborators, and other groups to make large-scale patch-clamp studies (i.e., single-cell electrophysiology and connectomics in rodents and humans and high throughput screening) more efficient (Peng et al., 2019; Koos et al., 2020) and to make complex experiments (i.e., in vivo patch-clamp) simpler (Suk et al., 2017; Stoy et al., 2020). In addition, since our initial report, we have discovered that 2% w/v Tergazyme is a superior cleaning solution (Figure 2A) and that the rinsing step from the original method is unnecessary (Figure 5). These improvements to the method have increased the whole-cell recording yield ~20%, increased the number of total cleans with a single pipette by a factor of 10 in HEK 293 cells (Figure 4), and decreased the time needed for each round of cleaning to <30 s, faster than manual pipette replacement (~1-2 min). Building on these gains in efficiency, we improved whole-cell yield to ~90% in HEK 293 cells by optimizing the position of the pipette tip relative to the cell membrane (Figure 2A-C) (Stoy et al., 2020). Briefly, by varying the distance the pipette was advanced into the cell, we found a strong relationship between distance and gigaseal probability, which reached ~100% at a range of 1-2 µm below the cell surface (defined as the z-axis point where pipette resistance increased 0.1 MΩ from initial resistance) (Stoy et al., 2020). When the patcherBot was programmed to attempt gigasealing at this position, the whole-cell recording yield increased significantly, as shown in the “Optimized” trace of Figure 2A (P = 0.044, Kolmorogov-Smirnov test). This method is used by the patcherBot in Figure 3 and Figure 4. Overall, this improved pipette cleaning method enables automated patch-clamp experiments to operate completely unattended for 3-4 h at yields of up to 90% and throughputs of up to 15 whole-cell recordings per hour, surpassing the output of highly skilled human experimenters (Figure 2, Figure 3). We were able to achieve up to 100 whole-cell recordings in ~13 h with a single pipette (Figure 4). We have also developed a simple hardware and software package for “push-to-clean” semi-automated patch-clamp experiments, enabling electrophysiologists to easily integrate this method into their hardware setup (Figure 1B-D, Figure 6, and Supplemental Information).
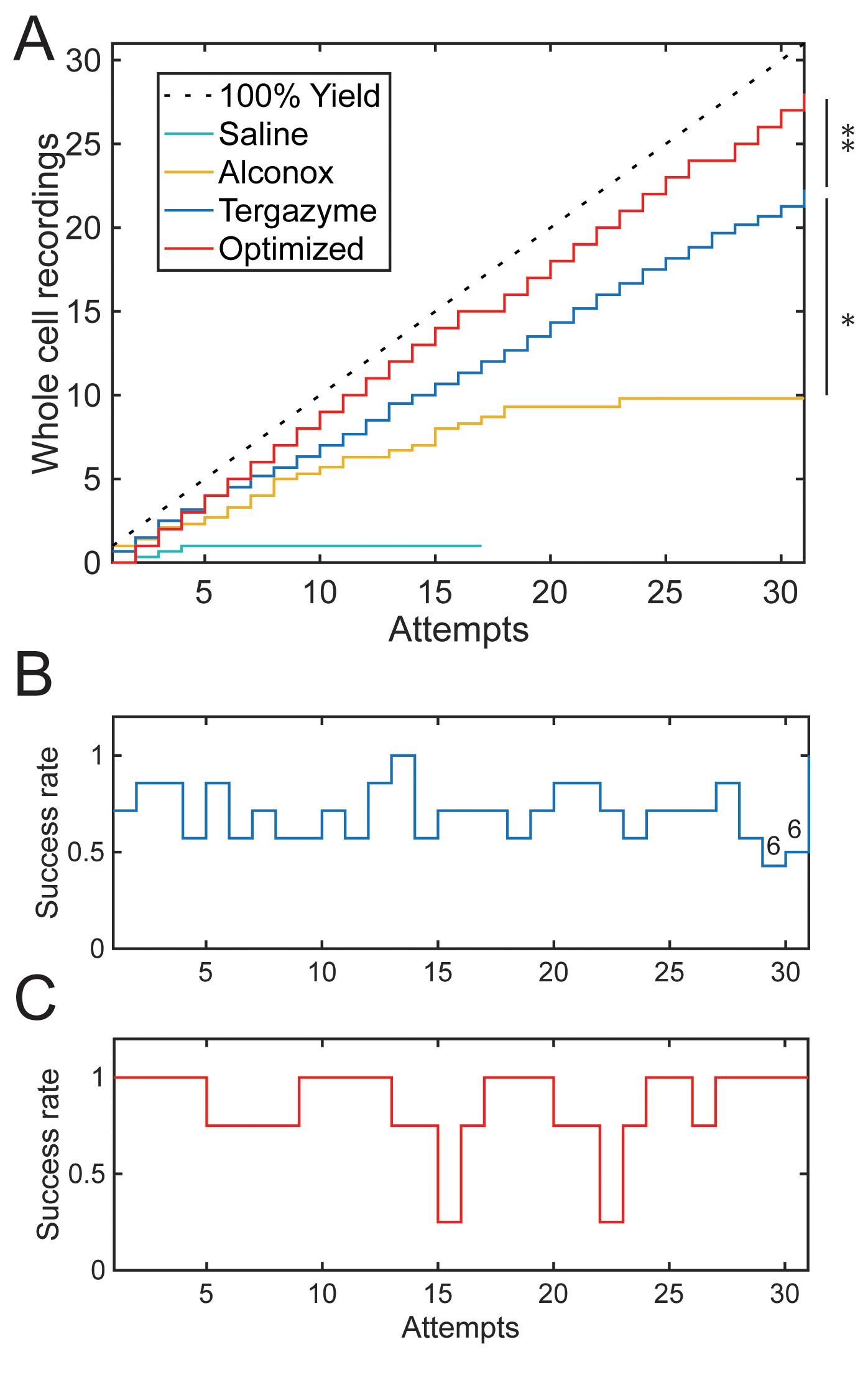
Figure 2. Improvements to pipette cleaning. A. Each trace represents the number of whole-cell recordings in HEK 293 cells as a function of the number of recording attempts with a single pipette. Each trace is the average of at least three pipettes. The “Saline” trace is a negative control (i.e., cleaning solution replaced with extracellular solution), and a 100% theoretical maximum is included for reference. The “Alconox” trace shows the performance of 2% w/v Alconox cleaning, which decreases as a function of the number of attempts. The “Tergazyme” trace shows no decrease in yield for 30 attempts with 2% w/v Tergazyme. The “Optimized” trace represents 2% w/v Tergazyme cleaning with optimized pipette positioning relative to the cell surface for gigasealing. The “Tergazyme” performance is superior to that of Alconox (*, P = 1.375E-5, Kolmogorov-Smirnov test). The “Optimized” performance is superior to that of “Tergazyme” (**, P = 0.04368, Kolmogorov-Smirnov test). B. Success rate of whole-cell patch-clamp as a function of the number of cleans using 2% w/v Tergazyme shows no significant decrease in likelihood of subsequent whole-cell recording (Odds ratio (OR) = 1.0067, CI: 0.97-1.04, P = 0.69, n = 215 attempts, each attempt is for n = 7 pipettes, except attempts 29 and 30, which are for n = 6). C. Optimized indentation with Tergazyme cleaning shows no significant decrease in likelihood of subsequent whole-cell recording (OR = 1.00, CI: 0.94-1.06, P = 0.95, n = 124 attempts, each attempt is for n = 4 pipettes).
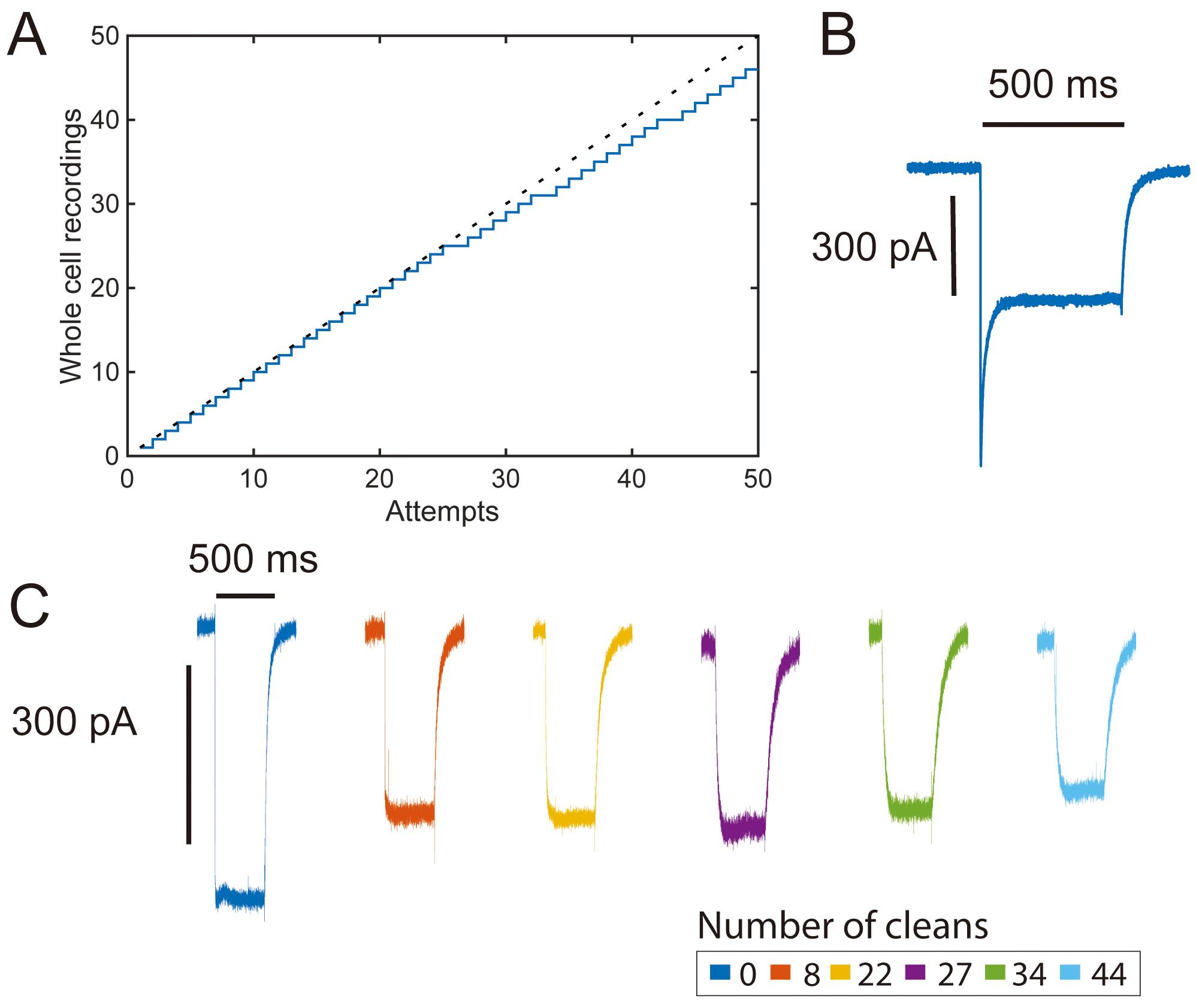
Figure 3. High-throughput opsin screening with pipette cleaning. A. Yield curve for a single pipette channelrhodopsin-2 (ChR-2) characterization experiment (46/51 attempts, 90% yield). B. Representative photocurrent trace (voltage clamp) in response to an initial pulse of 500 ms 480 nm LED pulse recorded from transiently transfected HEK 293 cell showing a large peak photocurrent response. C. Photocurrent traces (voltage clamp) from cells recorded in the middle of a series of 500 ms light pulses showing steady-state photocurrents over many pipette cleans. Large initial photocurrent responses are typical of ChR-2 (B) and are reduced in subsequent stimulation pulses (C) (Lin et al., 2009).
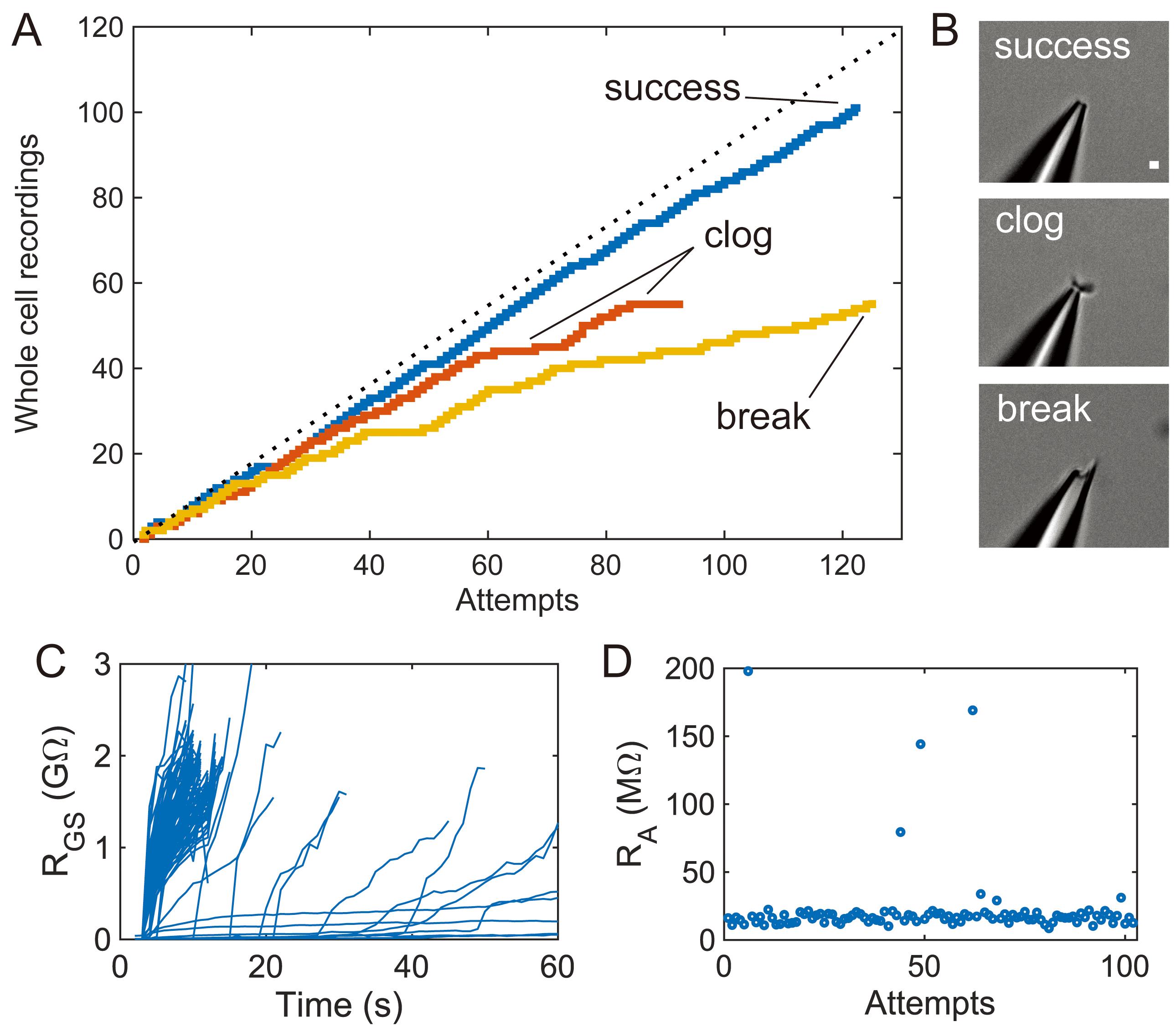
Figure 4. Upper limits of pipette cleaning with 2% w/v Tergazyme. A. Yield curves for individual pipettes showing cleaning for over 90 recording attempts with associated failure modes. The “success” trace shows effective pipette cleaning, the “clog” trace shows reversible pipette tip clogs that cause low yield over time, and the “break” trace shows experiments terminated by broken pipette tips. Theoretical maximum (100% yield) included for reference. B. Representative pipette images taken at 40× magnification for each failure mode in (A). Scale bar is 1 µm. C. Individual gigaseal resistance traces from the “success” trace (n = 122 gigaseal attempts). D. Access resistance of cells recorded in the “success” trace (n = 101 whole-cell recordings).
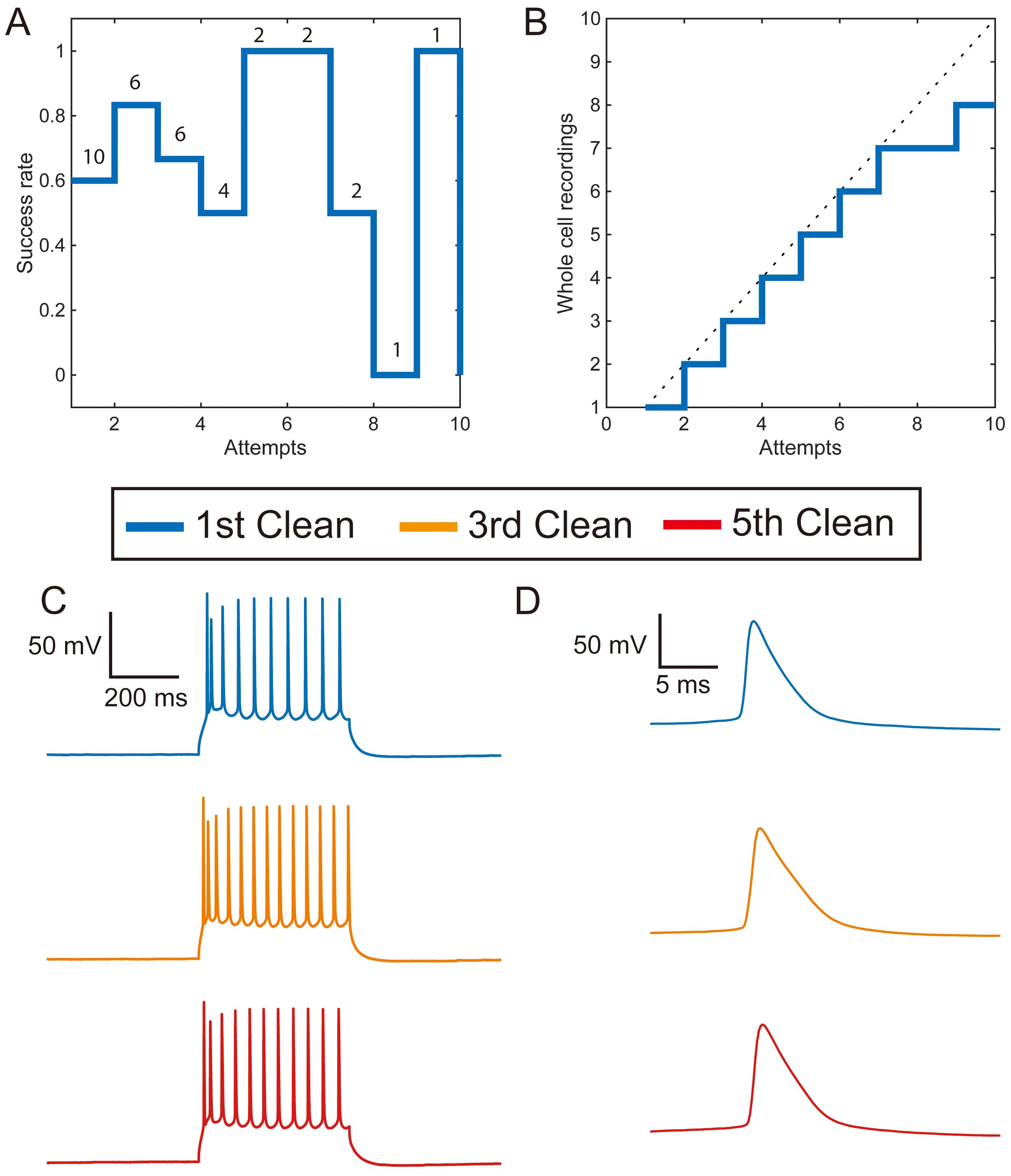
Figure 5. Tergazyme cleaning is effective without a rinsing step in acute mouse brain slices. A. The success rate of patching without a rinse step does not decrease significantly with the number of cleans (OR = 1.14, CI: 0.87-1.41, P = 0.34, n = 36 attempts). The number of pipettes used for each experiment is noted above each number of reuses. B. Yield plot for a single pipette using 2% w/v Tergazyme without rinsing. C. Representative recordings of evoked action potential firing in current clamp from three neurons recorded with a single pipette cleaned in 2% w/v Tergazyme without rinsing. D. Enlarged single evoked action potentials from the neurons in (C).
A robust and easy-to-implement method for automated pipette cleaning is of interest to all laboratories using the patch-clamp method. Although no negative side effects have been measured in cleaning experiments to date (Kolb et al., 2016 and 2019; Peng et al., 2019), for experiments where the possibility of exposing sensitive cells to cleaning residues is of particular concern (e.g., pharmacology or single-channel recordings), the method can be easily adapted to further minimize this risk (see Notes). We believe that this improved pipette cleaning method will be especially useful to labs working in the areas of multi-pipette patch clamping and high-throughput screening (see Notes, Figure 3).
The pipette cleaning method described in this protocol enables pipette reuse for patch clamping. By eliminating the need to fabricate, fill, install, and remove pipettes throughout experiments, experimenters can save valuable time and attention from these labor-intensive tasks. We show that 2% w/v Tergazyme enables up to and over 100 cells to be recorded with a single pipette (5-8 MΩ, Warner Instruments), eliminating the need for experimenters to replace pipettes over the course of an experimental day.
Materials and reagents
Borosilicate pipette glass with filament (Warner Instruments, catalog number: 64-0793)
Syringe, 5 ml (VWR, catalog number: BD309646)
Syringe filter, 0.2 µm (VWR, catalog number: 10218-486)
23G needle (VWR, catalog number: 89134-098)
Tergazyme (Alconox, catalog number: 1304-1)
Equipment
The equipment listed is in addition to standard patch-clamp electrophysiology equipment (e.g., amplifier, digitizer, headstage, micromanipulator, microscope, and pipette puller). Specific details of the patch-clamp rig used in this paper are described in detail elsewhere (Kolb et al., 2016 and 2019).
Cleaning dish (3D print or mill according to CAD files in SI, with appropriate changes for microscope stage)
Pressure control box (detailed plans and parts list on autopatcher.org, direct order from Neuromatic Devices, neuromaticdevices.com)
Software
Depending on the level of automation desired, download either (1) and (2) for full automation or only (2) to enable “push-to-clean” for manual patch clamping with cleaning.
Autopatcher software (downloadable at autopatcher.org). This software enables full automation of the cell detection, gigasealing, and break-in functions.
Push-to-clean software (downloadable at Github, https://github.com/mightenyip/Pipette-Cleaning-Software). The terminology “push-to-clean” is defined as an otherwise manual electrophysiology rig that includes a button-actuated pipette cleaning function. The button initiates a series of pipette position and pressure changes to clean the pipette for reuse.
Procedure
Before the first experiment
Manufacture cleaning dish according to plans and microscope stage dimensions (Figure 1C, CAD files in Supplementary Information Appendix B) in-house, using an on-demand production service (e.g., Protolabs, protolabs.com), or purchase from a commercial supplier (e.g., Neuromatic Devices).
Install pressure control box on existing patch-clamp electrophysiology rig
The pressure control box can be built from scratch according to plans presented by Kodandaramaiah et al. (2016). Schematics, instructions, and parts lists are also available for download at autopatcher.org.
A cleaning-compatible pressure control box can be purchased directly from Neuromatic Devices (neuromaticdevices.com).
Download and install software from autopatcher.org. Perform initial software setup. Detailed instructions are provided in Supplementary Information Appendix A.
Register the manipulator according to the manufacturer and COM port (see Supplementary Information Figures S1-S2).
Register the pressure control box to the specified COM port (see Supplementary Information Figures S1-S2).
Before each experiment
Prepare biological samples for patch-clamp recording. Methods are referenced for experimental preparations in which pipette cleaning has been validated by us or in other published reports.
For in vitro HEK 293 cells, follow Kolb et al. (2016). Pipette cleaning works well with wild-type cells, stably transfected lines, and transient transfections (Kolb et al., 2016 and 2019)
For rodent neuron culture recording, follow Kaech and Banker (2006) and Kolb et al. (2016). Cleaning for this preparation is verified in Kolb et al. (2016 and 2019).
For acute brain tissue slices recording, follow Jiang et al. (2015). Cleaning for this preparation is verified in the following reports Kolb et al. (2016 and 2019).
For in vivo mouse recording in anesthetized preparations, follow Bagal et al. (2013). Cleaning for this preparation is verified in Kolb et al. (2016 and 2019) and Stoy et al. (2020).
For acute human brain tissue slices, follow Ting et al. (2018) and Peng et al. (2019). Cleaning for this preparation is verified in Peng et al. (2019).
For human cerebral organoids, follow Mariani et al. (2015) and Qian et al. (2016). We have verified the cleaning in this preparation in unpublished experiments.
Prepare electrophysiology rig for the patch-clamp experiment and prepare pipettes as appropriate for experiment. Detailed guides for patch-clamp rig setup, denoising, and troubleshooting are provided elsewhere (Perin and Markram, 2013; Wang et al., 2015; Kodandaramaiah et al., 2016).
Load software for a push-to-clean patch-clamp experiment (Supplementary Information Appendix C).
Make 2% w/v Tergazyme cleaning solution.
Prepare 2% w/v Tergazyme solution in room temperature deionized water.
Mix solution until all Tergazyme powder is dissolved.
Note: Because Tergazyme is an enzymatic detergent, the enzymatic component degrades over time. The manufacturer recommends making fresh solutions and using them within 8 h for maximum efficacy.
Fill cleaning and rinsing bath reservoirs with filtered solutions
Using a syringe with a 0.2 µm filter and 23G needle, fill the appropriate bath reservoir with freshly made 2% w/v Tergazyme (or extracellular solution for rinsing).
Be careful not to overfill the cleaning bath reservoir, as this can cause Tergazyme solution to flow into the experimental chamber, potentially damaging the cells.
Note: To ensure there is no fluid exchange between the cleaning bath and the experimental bath, insert the tip of the pipette into the cleaning bath and monitor the square wave pulse in the voltage clamp. If there is no electrical contact between the ungrounded cleaning bath and the grounded experimental bath, you will see capacitive transients, similar to when the tip of the pipette is in the air. If there is electrical contact, you will see a square wave pulse, similar to when the tip is submerged in the experimental bath. To resolve this, use a task wipe to remove a small amount of fluid from the cleaning bath until electrical contact is eliminated.
In the software interface, calibrate manipulators in reference to cells and cleaning baths (Figure 6).
Select and save “exp bath location” position above target cell.
Select and save “location above baths” position directly above the cleaning bath reservoir.
Select and save “cleaning bath location” position with tip safely submerged in cleaning solution.
Select and save “wash bath” position with tip safely submerged in rinsing solution (i.e., extracellular solution) if desired.
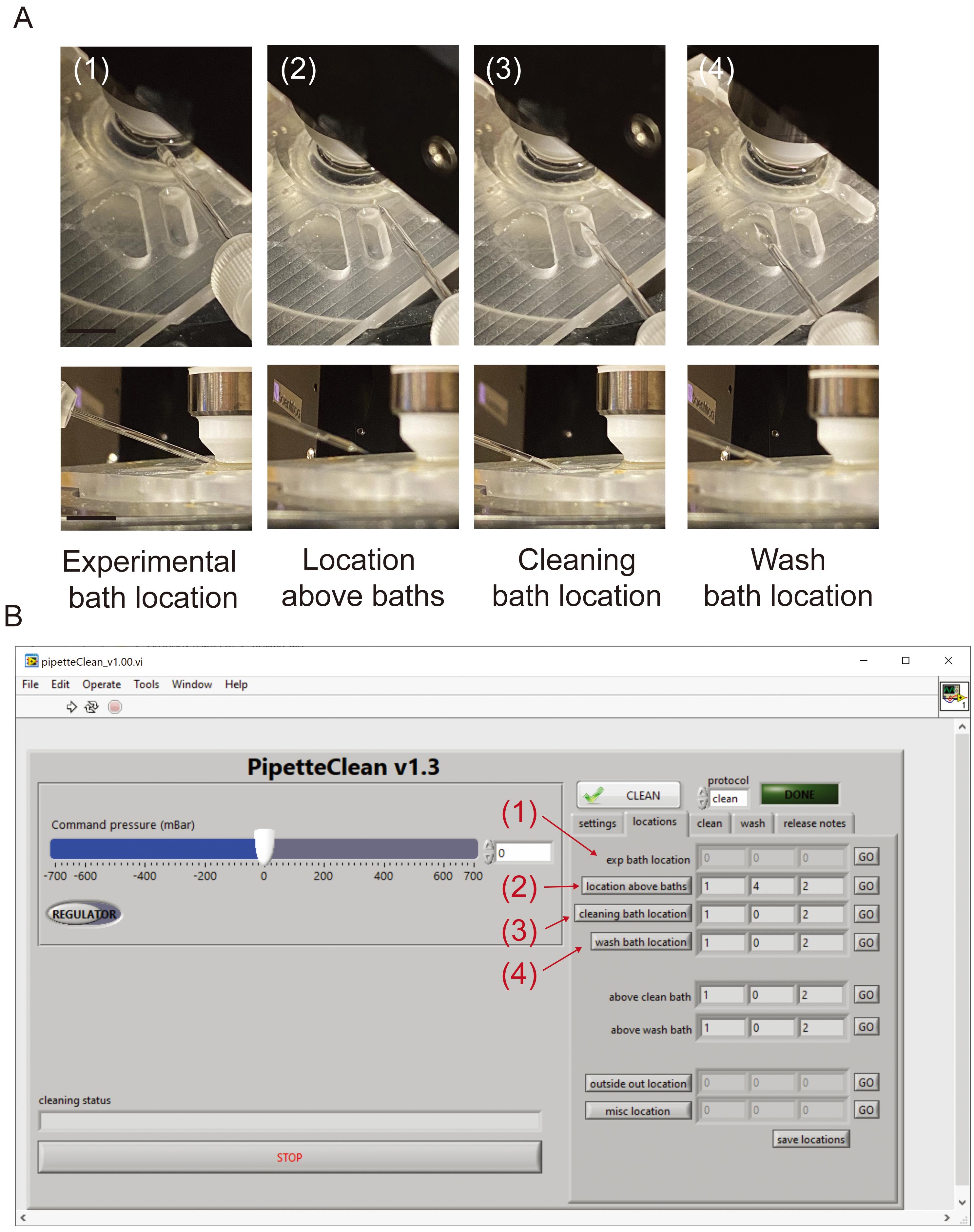
Figure 6. Calibration of pipettes for pipette cleaning. Positions used by the push-to-clean software for each cleaning attempt. Images in (A) refer to saved position values in (B). Briefly, (1) refers to position directly above target cells or tissue, (2) refers to a z-location above the clean and rinse baths, (3) refers to the position where the tip is submerged in cleaning solution, and (4) refers to the position where the pipette tip is submerged in the rinsing solution.
Patch-clamp experiment
Attempt patch-clamp recording on target cell
Initiate pipette cleaning using the software interface by clicking the “clean” button. The functions performed by the software are as follows:
Pipette retracts from cell to “location above baths” position under light positive pressure (+50 mbar).
Pipette moves from “location above baths” position to “cleaning bath location” position until contact is made between the pipette tip and the cleaning solution.
Note: Touching the pipette to the surface of the cleaning solution can be detected as a change in the capacitance of a square wave pulse at the pipette tip. This can often be observed prior to the pipette visibly touching the surface. Visible confirmation of pipette-fluid contact is also sufficient to begin cleaning.
With pipette tip in cleaning bath, suction is applied (-345 mbar) for 5 s.
Five rounds of alternating pulses of suction (-345 mbar for 1 s) and positive pressure (+700 mbar for 1 s) are applied.
Positive pressure is applied (+700 mbar) for 5 s to expel any residual cleaning solution from pipette tip.
Pipette is retracted from cleaning bath to “location above baths” position and then to either “exp bath location” position or “wash bath” position (optional).
Rinse the pipette (optional)
Pipette is moved from “cleaning bath location” position to “location above baths” position under positive pressure (+ 200 mbar).
Pipette is moved down from “location above baths” position toward “wash bath” position until contact is made between the pipette tip and the cleaning solution.
With pipette tip in rinse bath, apply 3 s of suction (-345 mbar) followed by 10 s of positive pressure (+700 mbar).
Retract pipette from “wash bath” position to “location above baths” position under positive pressure (+200 mbar).
Attempt patch-clamp recording on next target cell
Repeat steps A-D until the end of the experiment or failure of the pipette (e.g., tip breakage, clog, evaporation of cleaning solution, or user error).
Note: If the pipette appears to be clogged (i.e., visible internal clog observed in pipette tip or increase in resistance) or broken (i.e., visible broken tip or decrease in resistance), then replace it and repeat calibration.
Data analysis
Data from patch-clamp experiments using pipette cleaning can be processed in the same way as traditional patch-clamp experiments using software tools like pClamp (Molecular Devices) or Matlab (Mathworks). A useful analysis to characterize the efficacy of pipette cleaning is a yield curve, with the number of attempts on the x-axis and the gigaseal or whole-cell yield on the y-axis (Figure 2A, Figure 3A, Figure 4A, and Figure 5B). By comparing yield curves to ideal yields in which 100% of cells one attempts to patch result in a whole-cell patch-clamp configuration, it is possible to diagnose problems with cleaning yield, clogs, breaks, or other failure modes (Figure 4A). Methods and experiments can be compared from yield curve data using the Kolmogorov-Smirnov test. In addition, it is important to verify that cleaning does not cause a decrease in patching yield. Success rate plots that show the probability of obtaining a whole-cell for a defined number of cleaning attempts are also useful (Figure 1B-C, Figure 5A). Data from these plots can be modeled using linear regression (e.g., the mnrfit function in MATLAB). Odds ratios, 95% confidence intervals, and p-values test for deviations from initial performance.
Notes
How can pipette cleaning be used for multi-pipette experiments? Peng et al. (2019) recently demonstrated that multi-pipette connectivity studies in both rodent and human brain slices can be greatly accelerated by using pipette cleaning to both increase yield and extend the number of connections tested per tissue [see Figures 3 and 5 of Peng et al. (2019)]. These systems rely on routinely achieving simultaneous whole-cell recordings with all available pipettes to efficiently test for inter-neuronal connections, but obtaining simultaneous whole-cell recordings on all available pipettes is difficult and dependent on experimenter skill (n.b., a study by Perin and Markram (2013) found that with a 12 pipette rig, novice users achieved an average of 4.8 ± 1.7 out of 12 possible whole-cell recordings per attempt, whereas experts achieved 9.6 ± 1.4 out of 12 possible whole-cell recordings per attempt). Implementing a single round of pipette cleaning with 2% w/v Alconox and no rinsing resulted in significant improvements in the success rate (i.e., the ratio of actual to possible number of simultaneous whole-cell recordings) relative to no cleaning for both 8- and 10-manipulator patch-clamp recording setups, from 85 ± 13% to 97 ± 5% and 79 ± 11% to 92 ± 6%, respectively (Peng et al., 2019). Furthermore, once the first round of simultaneous whole-cell recordings was obtained, pipette cleaning allowed for additional surrounding neurons to be patched, increasing the total number of connections that can be tested in a single sample from 140 ± 24% to 244 ± 52% with an 8 pipette rig (Peng et al., 2019). Using this method could enable faster, more efficient collection of large datasets required for understanding neuronal connectivity (Goriounova et al., 2018; Gouwens et al., 2019).
How can pipette cleaning be used for high throughput screening? Large-scale efforts are also underway to discover and characterize drug candidates (Dunlop et al., 2008; Bagal et al., 2013), engineer improved genetic tools for neuroscience (Piatkevich et al., 2018; Yang et al., 2019), and understand human mutations in ion channel proteins (Swanger et al., 2016; Ogden et al., 2017). However, many of these projects are limited by the low throughput of traditional patch-clamp experiments (Park et al., 2013; Cho et al., 2019). For most screening experiments, manual patch clamping without cleaning has a throughput of 8-10 whole-cell recordings per experimenter per day (Milligan et al., 2009). Using our automated patch clamp system with 2% w/v Tergazyme, we have demonstrated routine throughputs of 10 whole-cell recordings per pipette per hour and daily throughputs of up to 100 whole-cell recordings (Figure 5) (Kolb et al., 2019). To further show the utility of pipette cleaning for functional screening, we performed a pilot experiment using HEK 293 cells transiently transfected with channelrhodopsin-2 (ChR-2). In this experiment, 46 whole-cell recordings were obtained from 51 patch-clamp attempts with a single pipette cleaned using 2% w/v Tergazyme (Figure 3).
What are expected yields using this method? Our yield and recording quality of patch-clamp recordings were comparable to manual pipette replacement. For experiments with HEK 293 cells, whole-cell recording yield was consistently 70-90% using the patcherBot. The ability to perform high-yield, high-throughput experiments in a fully automated Tergazyme cleaning system has also enabled us to iteratively improve our automated method in HEK 293 cells. For example, we compared 2% w/v Tergazyme solution to 2% w/v Alconox and saline in a randomized experiment where the operator was blinded to the identity of the cleaning solution (Figure 2). Following that experiment, we randomly varied the depth of pipette indentation into the cell membrane and found an optimal range that increased whole-cell recording yield to ~90% (Figure 2).
How long can cleaned pipettes be used? We have also found that Tergazyme cleaning is effective on single pipettes reused over 100 times, using the patcherBot (Figure 5) (Kolb et al., 2019). Because typical throughput for patch-clamp electrophysiology experiments is in the range of 10-30 recordings per day, it is likely that pipettes only need to be replaced once per day, except in cases where pipettes are broken or clogged.
How does the cleaning method fail? Using a single pipette cleaned with our improved 2% w/v Tergazyme cleaning solution, we achieved 102 whole-cell recordings in 122 patch-clamp attempts over a 13 h automated experiment. In our attempts to find the failure point of 2% w/v Tergazyme cleaning, pipette breakage or internal clogs were more likely to cause failure than an inability to clean the pipette. Internal pipette clogs are thought to form from environmental dust of particulates in the pipette solution. Clogs tended to form as a function of the duration of positive pressure applied and were more likely to occur over long experiments. Clogs can be diagnosed from flat portions in the yield curve that are unlikely to result from chance. Some clogs are reversible (see a representative trace in Figure 4). Pipettes can also fail after a tip breakage, which typically occurs if a target cell is missed. To determine an approximate failure point of cleaning, consider each patch-clamp attempt as an independent event with a probability equal to the gigaseal recording failure rate and determine the number of cleaning attempts until the probability is less than or equal to 0.01. For example, with a gigaseal failure rate of 30% (i.e., gigaseal success rate = 70%), the likelihood of a sequence of four failures to gigaseal has a probability of <1%.
Does cleaning transfer residual enzyme or detergent to the cells? One concern of patch-clamp experimenters interested in implementing cleaning is the possibility of contamination from residual cleaning solution in the pipette after cleaning. Our initial study addressed this concern with two types of experiments (Kolb et al., 2016). First, we performed electrospray ionization mass spectrometry (ESI-MS) on fresh pipettes and pipettes cleaned with 2% w/v Alconox and found that no Alconox residues were detectable. For this experiment, we used 2% w/v Alconox, and the limit of detection was 147 ng/ml. Secondly, we performed patch-clamp experiments on HEK 293 cells expressing the γ-aminobutyric acid type A Receptor (GABAAR), which is known to be sensitive to extracellular application of detergents. In these experiments, we did not find any statistically significant differences in GABAAR electrophysiology between cleaned and fresh pipettes.
Is the rinsing step (Procedure Step C. Rinse the pipette) required? Interestingly, recent experiments by Peng et al. (2019) (using human brain slices) and by our lab (using mouse brain slices, Figure 5) have provided evidence that the rinse step (Procedure, Step C. Rinse the pipette (optional) ) is unnecessary. Thus, in this method description, we label it as “optional.” However, if the possibility of contamination is still a concern in a particular experiment, we suggest rinsing as described in Procedure Step C. Rinse the pipette (optional) with the following modifications as needed to minimize risk:
Add time and cycles to the rinsing step to remove residual Tergazyme by dilution.
Reduce the concentration of Tergazyme in the cleaning solution. A 2% w/v Tergazyme solution is effective at cleaning pipettes up to 100 times with no measurable degradation in yield (Figure 4). This suggests that lower concentrations of Tergazyme will still be effective for pipette cleaning, with a potential trade-off in the maximum number of cleans per pipette.
Increase the perfusion rate of the external solution so that any residual Tergazyme is removed from the experimental chamber quickly.
Acknowledgments
This work was supported by the National Institutes of Health (NIH) grants R01NS102727, R01DA029639, U01MH106027, and R01 EY023173. We also acknowledge the original research papers from which this protocol is derived, Kolb et al. (2016 and 2019).
Competing interests
MCY and IK have consulting agreements with Neuromatic Devices, which manufactures pipette pressure control systems. IK, WAS, and CRF are inventors on U.S. Patent 10,830,758 related to pipette cleaning technology and licensed to Sensapex.
Ethics
For the representative work of cleaning using acute rodent brain slices, all animal procedures were in accordance with the US National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee at the Georgia Institute of Technology (A100359, Exp: Dec, 2022). When using cleaning methods for experiments requiring ethics committee approval, please follow the appropriate legal and institutional guidance.
References
- Bagal, S. K., Brown, A. D., Cox, P. J., Omoto, K., Owen, R. M., Pryde, D. C., Slidders, B., Skerratt, S. E., Stevens, E. B., Storer, R. I. and Swain, N. A. (2013). Ion channels as therapeutic targets: A drug discovery perspective. J Med Chem 56 (3): 593-624.
- Cho, Y. K., Park, D., Yang, A., Chen, F., Chuong, A. S., Klapoetke, N. C. and Boyden, E. S. (2019). Multidimensional screening yields channelrhodopsin variants having improved photocurrent and order-of-magnitude reductions in calcium and proton currents. J Biol Chem 294 (11): 3806-3821.
- Dunlop, J., Bowlby, M., Peri, R., Vasilyev, D. and Arias, R. (2008). High-throughput electrophysiology: an emerging paradigm for ion-channel screening and physiology. Nat Rev Drug Discov 7(4): 358-368.
- Goriounova, N. A., Heyer, D. B., Wilbers, R., Verhoog, M. B., Giugliano, M., Verbist, C., Obermayer, J., Kerkhofs, A., Smeding, H., Verberne, M., Idema, S., Baayen, J. C., Pieneman, A. W., de Kock, C. P., Klein, M. and Mansvelder, H. D. (2018). Large and fast human pyramidal neurons associate with intelligence. Elife 7: e41714.
- Gouwens, N. W., Sorensen, S. A., Berg, J., Lee, C., Jarsky, T., Ting, J., Sunkin, S. M., Feng, D., Anastassiou, C. A., Barkan, E., et al. (2019). Classification of electrophysiological and morphological neuron types in the mouse visual cortex. Nat Neurosci 22(7): 1182-1195.
- Hamill, O.P., Marty, A., Neher, E., Sakmann, B. and Sigworth, F.J. (1981). Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflugers Arch Eur J Physiol 391: 85-100.
- Jiang, X., Shen, S., Cadwell, C. R., Berens, P., Sinz, F., Ecker, A.S., Patel, S.and Tolias, A. S. (2015). Principles of connectivity among morphologically defined cell types in adult neocortex. Science 350 (6264): aac9462-aac9462.
- Kaech, S. and Banker, G. (2006). Culturing hippocampal neurons. Nat Protoc 1(5): 2406-2415.
- Kodandaramaiah, S. B., Holst, G. L., Wickersham, I. R., Singer, A. C., Franzesi, G. T., McKinnon, M. L., Forest, C. R. and Boyden, E. S. (2016). Assembly and operation of the autopatcher for automated intracellular neural recording in vivo. Nat Protoc 11(4): 634-654.
- Kolb, I., Landry, C. R., Yip, M. C., Lewallen, C. F., Stoy, W. A., Lee, J., Felouzis, A., Yang, B., Boyden, E. S., Rozell, C. J. and Forest, C. R. (2019). PatcherBot: a single-cell electrophysiology robot for adherent cells and brain slices. J Neural Eng 16(4): 046003.
- Kolb, I., Stoy, W. A., Rousseau, E. B., Moody, O. A., Jenkins, A. and Forest, C. R. (2016). Cleaning patch-clamp pipettes for immediate reuse. Sci Rep 6: 35001.
- Koos, K., Oláh, G., Balassa, T., Mihut, N., Rózsa, M., Qzsvar, A., Tasnadi, E., Barzó, P., Faragó, N., Puskás, L., Molnár, G., Molnár, J., Tamás, G. and Horvath, P. (2020). Automatic deep learning driven label-free image guided patch clamp system for human and rodent in vitro slice physiology. bioRxiv 2020.05.05.078162.
- Lin, J. Y., Lin, M. Z., Steinbach, P. and Tsien, R. Y. (2009). Characterization of engineered channelrhodopsin variants with improved properties and kinetics. Biophys J 96: 1803-1814.
- Mariani, J., Coppola, G., Zhang, P., Abyzov, A., Provini, L., Tomasini, L., Amenduni, M., Szekely, A., Palejev, D., Wilson, M., Gerstein, M., Grigorenko, E. L., Chawarska, K., Pelphrey, K. A., Howe, J. R. and Vaccarino, F. M. (2015). FOXG1-Dependent dysregulation of GABA/Glutamate neuron differentiation in autism spectrum disorders. Cell 162(2): 375-390.
- Milligan, C. J., Li, J., Sukumar, P., Majeed, Y., Dallas, M. L., English, A., Emery, P., Porter, K. E., Smith, A. M., McFadzean, I., Beccano-Kelly, D., Bahnasi, Y., Cheong, A., Naylor, J., Zeng, F., Liu, X., Gamper, N., Jiang, L. H., Pearson, H. A., Peers, C., Robertson, B. and Beech, D. J. (2009). Robotic multiwell planar patch-clamp for native and primary mammalian cells. Nat Protoc 4(2): 244-255.
- Ogden, K. K., Chen, W., Swanger, S. A., McDaniel, M. J., Fan, L. Z., Hu, C., Tankovic, A., Kusumoto, H., Kosobucki, G. J., Schulien, A. J., Su, Z., Pecha, J., Bhattacharya, S., Petrovski, S., Cohen, A. E., Aizenman, E., Traynelis, S. F. and Yuan, H. (2017). Molecular Mechanism of Disease-Associated Mutations in the Pre-M1 Helix of NMDA Receptors and Potential Rescue Pharmacology. PLoS Genet 13(1): e1006536.
- Park, J., Werley, C. A., Venkatachalam, V., Kralj, J. M., Dib-Hajj, S. D., Waxman, S. G. and Cohen, A. E. (2013). Screening fluorescent voltage indicators with spontaneously spiking HEK cells. PLoS One 8(12): e85221.
- Peng, Y., Mittermaier, F. X., Planert, H., Schneider, U. C., Alle, H. and Geiger, J. R. P. (2019). High-throughput microcircuit analysis of individual human brains through next-generation multineuron patch-clamp. Elife 8: e48178.
- Perin, R. and Markram, H. (2013). A computer-assisted multi-electrode patch-clamp system. J Vis Exp 80: e50630.
- Piatkevich, K. D., Jung, E. E., Straub, C., Linghu, C., Park, D., Suk, H. J., Hochbaum, D. R., Goodwin, D., Pnevmatikakis, E., Pak, N., Kawashima, T., Yang, C. T., Rhoades, J. L., Shemesh, O., Asano, S., Yoon, Y. G., Freifeld, L., Saulnier, J. L., Riegler, C., Engert, F., Hughes, T., Drobizhev, M., Szabo, B., Ahrens, M. B., Flavell, S. W., Sabatini, B. L. and Boyden, E. S. (2018). A robotic multidimensional directed evolution approach applied to fluorescent voltage reporters. Nat Chem Biol 14(4): 352-360.
- Qian, X., Nguyen, H. N., Song, M. M., Hadiono, C., Ogden, S. C., Hammack, C., Yao, B., Hamersky, G. R., Jacob, F., Zhong, C., et al.(2016). Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell 165 (5): 1238-1254.
- Stoy, W. M., Yang, B., Kight, A., Wright, N. C., Borden, P. Y., Stanley, G. B. and Forest, C. R. (2020). Compensation of physiological motion enables high-yield whole-cell recording in vivo. J Neurosci Methods 348: 109008.
- Suk, H. J., van Welie, I., Kodandaramaiah, S. B., Allen, B., Forest, C. R. and Boyden, E. S. (2017). Closed-Loop real-time imaging enables fully automated cell-targeted patch-clamp neural recording In vivo. Neuron 95(5): 1037-1047 e1011.
- Swanger, S. A., Chen, W., Well, G., Burger, P., Tankovic, A., Bhattacharya, S., Strong, K . L., Hu, C., Kusumoto, H., Zhang, J., Adams, D. R., Millichap, J. J., Petrovski, S., Traynelis, S. F. and Yuan, H. (2016). Mechanistic Insight into NMDA Receptor Dysregulation by Rare Variants in the GluN2A and GluN2B Agonist Binding Domains. Am J Hum Genet 99 (6): 1261-1280.
- Ting, J. T., Kalmbach, B., Chong, P., de Frates, R., Keene, C. D., Gwinn, R. P., Cobbs, C., Ko, A. L., Ojemann, J. G., Ellenbogen, R. G., Koch, C. and Lein, E. (2018). A robust ex vivo experimental platform for molecular-genetic dissection of adult human neocortical cell types and circuits. Sci Rep 8(1): 8407.
- Wang, G., Wyskiel, D. R., Yang, W., Wang, Y., Milbern, L. C., Lalanne, T., Jiang, X., Shen, Y., Sun, Q. Q. and Zhu, J. J. (2015). An optogenetics- and imaging-assisted simultaneous multiple patch-clamp recording system for decoding complex neural circuits. Nat Protoc 10(3): 397-412.
- Yang, K. K., Wu, Z. and Arnold, F. H. (2019). Machine-learning-guided directed evolution for protein engineering. Nat Methods 16(8): 687-694.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Landry, C. R., Yip, M. C., Kolb, I., Stoy, W. M., Gonzalez, M. M. and Forest, C. R. (2021). Method for Rapid Enzymatic Cleaning for Reuse of Patch Clamp Pipettes: Increasing Throughput by Eliminating Manual Pipette Replacement between Patch Clamp Attempts. Bio-protocol 11(14): e4085. DOI: 10.21769/BioProtoc.4085.
Category
Neuroscience > Basic technology > Acute slice preparation
Biophysics > Electrophysiology > Patch-clamp technique
Biological Sciences > Biological techniques
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








