- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Ethanol-induced Sedative Behavior: An Assay to Investigate Increased Dopamine Signaling in Caenorhabditis elegans
Published: Vol 11, Iss 13, Jul 5, 2021 DOI: 10.21769/BioProtoc.4083 Views: 3426
Reviewed by: Oneil Girish BhalalaShaarika SarasijaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

An ex vivo Model of Paired Cultured Hippocampal Neurons for Bi-directionally Studying Synaptic Transmission and Plasticity
Ruslan Stanika and Gerald J. Obermair
Jul 20, 2023 2200 Views

Local Iontophoretic Application for Pharmacological Induction of Long-Term Synaptic Depression
Borys Olifirov [...] Pavel Belan
Jun 5, 2025 1899 Views

Labeling Postsynaptic Densities for Super-Resolution Microscopy With Minimal Signal-Loss and Offset
Sheng-Yang Ho [...] Johannes W. Hell
Nov 5, 2025 1577 Views
Abstract
Dopamine (DA) signaling affects locomotion, feeding, learning, and memory in C. elegans. Various assays have been developed to study the proteins involved in these behaviors; however, these assays show behavioral output only when there is a drastic change in DA levels. We designed an assay capable of observing behavioral output even with only slight alterations in DA levels. To achieve this, we designed a behavioral paradigm where we combined C. elegans movement with ethanol (EtOH) administration. The behavioral response to alcohol/EtOH and susceptibility to alcohol-use disorders (AUDs) have been linked to DA. Our assay correlates an increase in DA levels due to EtOH and movement obstruction due to a dry surface to a circular sedative behavior, which we designated as EtOH-induced sedative (EIS) behavior. We successfully utilized this assay to assign physiological and behavioral functions to a DA autoreceptor, DOP-2.
Keywords: EtOH (Ethanol)Background
Alcohol is a widely abused drug with a plethora of associated diseases that can impact societal functioning. Multiple studies have focused on unravelling the mode of action and effect of this drug; however, the neuronal mechanisms underlying alcohol susceptibility and disinhibition are unclear. Studies across various species have demonstrated that alcohol intake increases the release of the neurotransmitter DA that induces the reward pathway (Imperato and Di Chiara, 1986; Weiss et al., 1996; Baik, 2013). Although C. elegans does not mimic all the complexities of the mammalian system, it has been successfully modeled for studying alcohol-dependent neuronal behaviors. Studies show that C. elegans display diverse aspects of alcohol responses (Davies et al., 2003 and 2004). Previously, investigations in C. elegans have revealed that there is a dose-dependent decline in the locomotor activity upon acute and chronic alcohol exposure at a concentration of 400-500 mM (Davies et al., 2003; Lee et al., 2009). The DA system in C. elegans is involved in feeding, movement, learning, and memory; and similar to that of mammals, signals through two receptor subfamilies D1-like and D2-like receptors. Mutants of the D2-like receptor, dop-2, exhibited no obvious phenotype when analyzed for DA-dependent behaviors, despite being expressed in all dopaminergic neurons and predicted to be an autoreceptor (Chase et al., 2004). We devised an assay utilizing the EIS behavior observed in mutants of dop-2 to investigate the neuronal circuitry involved in regulating locomotory behavior under the influence of EtOH (Pandey et al., 2021).
Materials and Reagents
60-mm Petri dishes (Tarsons, catalog number: 460061)
Spreader (Tarsons, catalog number: 920081)
99.99% platinum wire (Sigma-Aldrich, catalog number: 267201)
C. elegans: N2 (wild-type (WT) and dop-2 (vs105) adult animals with 3-4 eggs (University of Minnesota, Caenorhabditis Genetic Center)
Escherichia coli OP50 (University of Minnesota, Caenorhabditis Genetic Center)
Ethanol (EtOH) (Fisher chemical, catalog number: UN1170)
Cholesterol (SRL Sisco Research Laboratories, catalog number: 54181)
Calcium chloride dihydrate (CaCl2·2H2O) (Sigma-Aldrich, catalog number: C3306)
Magnesium sulfate (MgSO4) (Sigma-Aldrich, catalog number: M7506)
Potassium phosphate, monobasic (KH2PO4) (Sigma-Aldrich, catalog number: P5379)
Potassium phosphate, dibasic (K2HPO4) (Sigma-Aldrich, catalog number: P8281)
Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S7653)
Bacto-agar (HiMedia Laboratories, catalog number: GRM026)
Bacto-peptone (BD, catalog number: 211677)
400 mM EtOH (see Recipes)
5 mg/ml cholesterol (see Recipes)
1 M CaCl2 stock solution (see Recipes)
1 M MgSO4 stock solution (see Recipes)
1 M KPO4, pH 6.0 stock solution (see Recipes)
Nematode growth medium (NGM) agar plates (see Recipes)
Equipment
Pipettes (Eppendorf, model: Research® plus, catalog number: 2231000224)
2-L glass conical flask (DWK Life Sciences, DURAN, catalog number: 2121763)
Autoclave (Equitron-7431 SLEFA)
Microscope (ZEISS, model: Stemi 2000 C)
Software
GraphPad Prism v6 (GraphPad Software)
ImageJ (developed by the National Institutes of Health)
Procedure
Prepare 60-mm NGM plates for the maintenance of C. elegans. Seed the plates with E. coli OP50 (serves as food for C. elegans) under sterile conditions in a bacterial hood and allow to grow overnight at 37°C.
Synchronize the C. elegans strains to be analyzed by bleaching. Collect the animals from the NGM plates using 1 ml M9 solution in 2-ml microcentrifuge tubes (MCT). Add 400 µl freshly prepared bleach solution (1:1 ratio of 5 N sodium hydroxide and sodium hypochlorite) to the tubes and vortex at medium speed for 5 min. Spin the MCT at 6,097 × g (4000 rpm) for 60 s and discard the supernatant. Wash the pellet again with fresh M9 and plate on 60-mm NGM plates.
To perform the assay, prepare fresh 60-mm (NGM) plates containing 8 ml media one day before the assay and store at 4°C until use.
On the day of the assay, dry the plates in the laminar airflow for 3-4 h with the lids open.
Spread 400 mM (196 µl) 100% EtOH on the dry 60-mm plates with 8 ml media using a glass spreader. Dry plates without EtOH (– EtOH) are used as control plates.
Seal the EtOH assay plates with parafilm to avoid any loss of ethanol and place at 20°C for 2 h to allow the EtOH to equilibrate across the plates.
The EtOH assay plates are now ready and can be used for the assay.
Transfer 10 animals of the genotype being studied from a seeded plate (+ food) to an unseeded plate (– food) for 15-20 s (to get rid of the food). Next, transfer the animals to the assay plate or control plate and leave undisturbed for 2 h at 20°C.
Observe the C. elegans after a 2-h time interval. Count the number of animals on the plates and make 1-min videos for each animal to observe and analyze the locomotory behavior by counting the number (anterior and posterior) and amplitude of body bends to characterize the behavior.
The number of body bends are scored manually using ImageJ while maintaining identical parameters when analyzing anterior and posterior body bends (Figure 1A).
Calculate the amplitude of the body bends manually using the NIH ImageJ software, quantitating the anterior and posterior body bends separately. To quantitate the amplitude of the body bends, measure the distance between the deepest angle of the body bend. Draw a tangential line from the tip of the head to the midsection of the body and measure the vertical distance from the line to the body as the amplitude of the anterior body bend (Figure 1B). Correspondingly, from the midsection to the tail of the animal, measurements are obtained for the posterior body bend amplitude (Figure 1B). Normalize the measurements to the length of the animal to derive the measured values in micrometers.
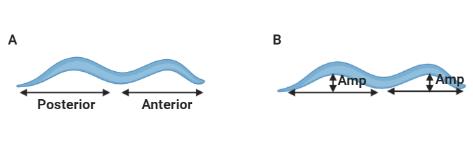
Figure 1. Representative image used for the analysis of the amplitude of body bends. A. Image represents the evaluation of anterior and posterior body bends separately. B. Image indicates the amplitude of body bends; a double-sided arrow depicts the amplitude (Amp) in the anterior and posterior regions.Perform timeline-based analysis of behavior. Starting from EtOH exposure, different time points (30, 60, 120, 300 min), including overnight exposure (16 h), are analyzed (Figure 2A).
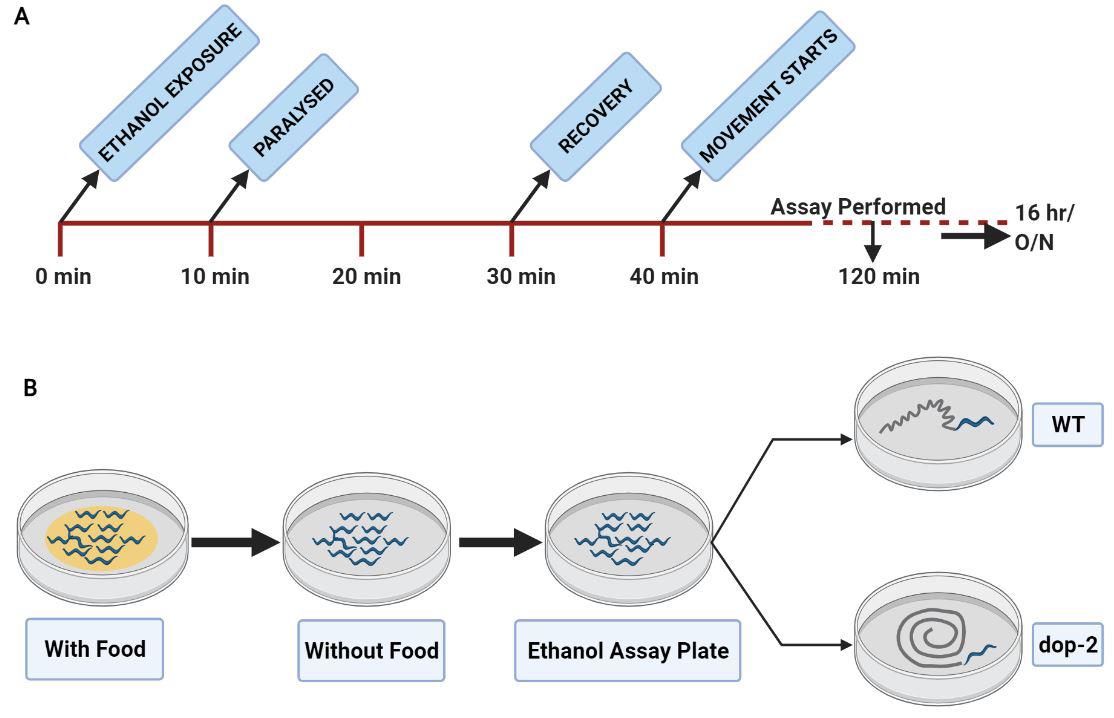
Figure 2. Illustration of the timeline for EtOH treatment of C. elegans to study EIS behavior. A. The timepoints denote different phases of WT animals upon EtOH exposure, as indicated in the timeline. Experiments to test the number and amplitude of body bends are performed at ~2 h. B. Illustration of EIS behavior, where WT animals recover from EtOH-induced paralysis and show normal sinusoidal movement, while dop-2 mutant animals show EtOH-induced sedative behavior (Pandey et al., 2021).WT animals show normal behavior 2 h after EtOH paralysis (Video 1). While dopaminergic autoreceptor, dop-2, mutant animals show EtOH-induced sedative (EIS) behavior at the same time point (Figure 2B and Video 2).
Video 1. A representative video of WT animals showing a single C. elegansthat has recovered from EtOH-induced paralysis. The animal can be seen moving in normal sinusoidal wave-like patterns, with no observable defects in locomotion (Pandey et al., 2021).Video 2. A representative video of the EIS behavior shown by dop-2 mutant C. elegans upon EtOH exposure at the 2-h time point. The animal shows defects in locomotion and moves by dragging its posterior region (Pandey et al., 2021).Recovery from the EIS behavior can be observed upon transferring the mutant animals to normal NGM plates with food after a 2-h exposure to EtOH. Mutants of dop-2 recover from the EIS phenotype 1 h after being transferred.
Data analysis
Calculate the number of anterior and posterior body bends separately in a 1-min video at the 2-h time point.
Software: GraphPad Prism v6.
Transfer 10 animals to each assay plate and perform the experiment in triplicate for each strain.
Statistical analysis: Use one way-ANOVA to determine statistical P-values.
Notes
Drying the NGM plates perfectly is important, and the surface of the plates should be free from any dust or other surface aberrations for proper visualization of the tracks.
Seal the plates with parafilm to avoid loss of ethanol and appropriate equilibration.
Recipes
5 mg/ml cholesterol
Add 500 mg cholesterol to 100 ml 95% ethanol and mix by rotating at room temperature for a few hours to dissolve.
Store at 4°C.
1 M CaCl2 stock solution
Dissolve 14.7 g CaCl2·2H2O in 100 ml ddH2O and autoclave for 15 min at 121°C.
Store at 4°C.
1 M MgSO4 stock solution
Dissolve 12.04 g MgSO4 in 100 ml ddH2O and autoclave for 15 min at 121°C.
Store at 4°C.
1 M KPO4, pH 6.0 stock solution
Mix 108.3 g KH2PO4 and 35.6 g K2HPO4 in 500 ml ddH2O.
Adjust the pH to 6.0 by adding NaOH; finally, make up the volume to 1 L.
Aliquot the solution and autoclave for 15 min at 121°C.
Store at 4°C.
Nematode growth medium (NGM) agar plate
Add 3 g NaCl, 16 g Bacto-agar, and 2.5 g Bacto-peptone to 975 ml ddH2O in a 2-L flask.
Autoclave for 50 min at 121°C.
Allow the NGM agar to cool to 55-60°C. Add 1 ml 5 mg/ml cholesterol, 1 ml 1 M CaCl2, 1 ml 1 M MgSO4, and 25 ml 1 M KPO4, pH 6.0.
400 mM ethanol
Spread 196 ml ethanol on 8 ml media in a 60-mm plate.
Seal the plate with parafilm and allow equilibration at 20°C.
Acknowledgments
This protocol was adapted from Pandey et al. (2021). AS thanks the Council of Scientific and Industrial Research (CSIR) – University Grants Commission (UGC) for a graduate fellowship. PP acknowledges support from a Department of Science and Technology (DST) – Women of Science (WOS-A) grant as well as past funding from the Department of Biotechnology (DBT) Bio-CARe, the Indian Institute of Science Education and Research (IISER) Mohali, and an IA grant awarded to KB. KB was an Intermediate Fellow of the India Alliance (IA) and is currently an IA Senior Fellow. KB thanks the Alliance for funding support. KB also thanks DBT, DST – Science and Engineering Research Board (SERB), and the Ministry of Human Resources Development (MHRD) – Scheme for Transformational and Advanced Research in Sciences (STARS) for funding support.
Funding
This work was supported by the DBT/Wellcome Trust India Alliance fellowships [grant numbers IA/S/19/2/504649 and IA/I/12/1/500516] awarded to KB and partially supported by DBT, MHRD–STARS, and DST–SERB grants [BT/PR24038/BRB/10/1693/2018, STARS/APR2019/BS/454/FS and SERB/F/7047] as well as a DBT-IISc partnership grant to KB. PP is supported by a DST WOS-A grant [SR/WOS-A/LS-285/2018] and was earlier supported by a DBT Bio-CARe grant [BioCARe/01/10167].
Competing interests
The authors declare no conflicts of interest or competing interests.
References
- Baik, J. H. (2013). Dopamine signaling in reward-related behaviors. Front Neural Circuits 7: 152.
- Chase, D. L., Pepper, J. S. and Koelle, M. R. (2004). Mechanism of extrasynaptic dopamine signaling in Caenorhabditis elegans. Nat Neurosci 7(10): 1096-1103.
- Davies, A. G., Pierce-Shimomura, J. T., Kim, H., VanHoven, M. K., Thiele, T. R., Bonci, A., Bargmann, C. I. and McIntire, S. L. (2003). A central role of the BK potassium channel in behavioral responses to ethanol in C. elegans. Cell 115(6): 655-666.
- Davies, A. G. and McIntire, S. L. (2004). Using C. elegans to screen for targets of ethanol and behavior-altering drugs. Biol Proced Online 6: 113-119.
- Imperato, A. and Di Chiara, G. (1986). Preferential stimulation of dopamine release in the nucleus accumbens of freely moving rats by ethanol. J Pharmacol Exp Ther 239(1): 219-228.
- Lee, J., Jee, C. and McIntire, S. L. (2009). Ethanol preference in C. elegans. Genes Brain Behav 8(6): 578-585.
- Pandey, P., Singh, A., Kaur, H., Ghosh-Roy, A. and Babu, K. (2021). Increased dopaminergic neurotransmission results in ethanol dependent sedative behaviors in Caenorhabditis elegans. PLoS Genet 17(2): e1009346.
- Weiss, F., Parsons, L. H., Schulteis, G., Hyytia, P., Lorang, M. T., Bloom, F. E. and Koob, G. F. (1996). Ethanol self-administration restores withdrawal-associated deficiencies in accumbal dopamine and 5-hydroxytryptamine release in dependent rats. J Neurosci 16(10): 3474-3485.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Singh, A., Babu, K. and Pandey, P. (2021). Ethanol-induced Sedative Behavior: An Assay to Investigate Increased Dopamine Signaling in Caenorhabditis elegans. Bio-protocol 11(13): e4083. DOI: 10.21769/BioProtoc.4083.
Category
Cell Biology > Cell signaling > Synaptic transmision
Neuroscience > Behavioral neuroscience
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link