- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Analysis of Soluble Sugar Content in Minute Quantities of Rice Tissues by GC-MS
Published: Vol 11, Iss 13, Jul 5, 2021 DOI: 10.21769/BioProtoc.4077 Views: 4842
Reviewed by: Runlai HangWeiyan JiaYu Liu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Maize Seedlings Colonization with Serendipita indica and Its Colonization Efficiency Analysis
Om Prakash Narayan [...] Atul Kumar Johri
Oct 20, 2023 2266 Views

Botrytis cinerea in vivo Inoculation Assays for Early-, Middle- and Late-stage Strawberries
Piao Yang [...] Ye Xia
Oct 20, 2023 2761 Views
Abstract
Soluble sugars play key roles in plant growth, development, and adaption to the environment. Characterizing sugar content profiling of plant tissues promotes our understanding of the mechanisms underlying these plant processes. Several technologies have been developed to quantitate soluble sugar content in plant tissues; however, it is difficult with only minute quantities of plant tissues available. Here, we provide a detailed protocol for gas chromatography mass spectrometry (GC-MS)-based soluble sugar profiling of rice tissues that offers a good balance of sensitivity and reliability, and is considerably more sensitive and accurate than other reported methods. We summarize all the steps from sample collection and soluble sugar extraction to derivatization procedures of the soluble extracted sugars, instrumentation settings, and data analysis.
Keywords: GC-MSBackground
Sucrose, as the major product of photosynthesis in plants, is transported from sources to sinks by three steps, phloem loading, long-distance transport in phloem, and phloem unloading, which are mediated by both plasmodesmata and sugar transporters (Zhu et al., 2013; Milne et al., 2018). Sucrose is then transformed into other sugars such as glucose and fructose. These soluble sugars are involved in energy and material metabolism and signaling, plant growth and development, and plant responses to biotic and abiotic stresses (Bezrutczyk et al., 2018; Yamada and Osakabe, 2018; Wei et al., 2020). On the other hand, pests regulate plant sugar partitioning to facilitate their infection of, and damage to, plants (Hofmann et al., 2009; Ibraheem et al., 2014; Sosso et al., 2019). It is highly beneficial for researchers to understand the mechanisms of plant-pest interactions in order to investigate changes in soluble sugar content of plant tissues during pest infection or damage.
Several methods have been developed to quantitate soluble sugars, such as the anthrone-sulfuric acid colorimetric method, high-performance liquid chromatography (HPLC), liquid chromatography-mass spectrometry (LC-MS), and gas chromatography-mass spectrometry (GC-MS) (Montero et al., 2004; Jan et al., 2006; Magwaza and Opara, 2015; Ye et al., 2019). Among these methods, LC-MS and GC-MS are widely used for sugar quantitation. In comparison with LC, GC shows higher resolution at the high-performance level; although, a dervatization step is needed to convert sugars into volatile compounds (Romani et al., 1994; Cataldi et al., 2000). It has been reported that approximately 50 mg fresh weight of leaves and roots from two-year-old olive plants can be used to quantitate the soluble sugar content of these tissues by GC-MS (Cataldi et al., 2000), which, as we learned, is the highest sensitivity of GC-MS used in characterizing soluble sugar content of plant tissues. Here, we describe in detail a new GC-MS protocol, with some important modifications, that is able to quantitate the three soluble sugars, sucrose, glucose, and fructose, in plant tissues with approximately 15 mg fresh weight. This method was modified by us and has been successfully applied in our current work.
Materials and Reagents
1.5-ml centrifuge tubes (Axygen, catalog number: AXYMCT150C)
2.0-ml centrifuge tubes (Axygen, catalog number: AXYMCT200C)
Rice (Oryza sativa cv Nipponbare)
Liquid nitrogen
Methoxyamine hydrochloride (98%, Sigma-Aldrich, catalog number: 226904-1G)
Pyridine (Macklin, catalog number: P816290)
N-methyl-N-(trimethylsilyl)trifluoroacetamide (Santa Cruz Biotechnology, catalog number: 24589-78-4)
Fructose (Shanghai Yuanye Bio-Technology Co., Ltd, catalog number: B21896)
Glucose (Sigma-Aldrich, catalog number: G7528)
Sucrose (DR EHRENSTRORFER, catalog number: CDCT-C16901100)
Ribitol (99%, Sigma-Aldrich, catalog number: A5502-5G)
Helium gas (v/v ≥ 99.999%, Gassoon)
Methoxyamine hydrochloride solution (see Recipes)
Internal standard solution (see Recipes)
Equipment
Gas chromatograph (SHIMADZU, model: GAS CHROMATOGRAPH GC-2010 Plus)
Glas-Col evaporator (Guangzhou Pengxin Technology, model: FD5-series).
SH-Rxi-5Sil MS Cap (SHIMADZU, catalog number: R221-75954-30)
Auto sampler vials with caps (Agilgent, catalog number: 5182-0715)
Glass inserts (BioReags, catalog number: 571200)
Centrifuge (Eppendorf, model: Centrifuge 5424 R)
Vortex (Haimen Kylin-Bell, model: QL-901)
Vacuum freeze drier (Gold Sim, model: FD5-series freeze dryer)
1-ml transfer pipettes (Axygen, catalog number: T-1000-B)
200-μl transfer pipettes (Crystalgen, catalog number: 23-2346)
Software
GC-MSsolution software (Shimadzu GC-MS QP-2010)
NIST17 mass spectra search program (version 2d, build 2017) (National Institute of Standards and Technology, Gaithersburg, MD)
Microsoft Excel
Procedure
Sample handling
Sample fresh rice root tissue from 15-day-old rice seedlings (approximately 15 mg fresh weight per sample) and transfer to a 2-ml tube. Grow the rice seedlings under a 12 h/12 h light/dark cycle with 75% relative humidity, a light intensity of 110 μmol·m-2·s-1, and a temperature of 28°C.
Immediately freeze root samples in liquid nitrogen and grind thoroughly using a pestle and mortar.
Add 1.5 ml 75% methanol to each tube and vortex briefly.
Place the samples in an incubator at 70°C for 15 min and then centrifuge at 10,000 × g for 10 min at room temperature.
For each sample, transfer 1.2 ml supernatant to a 1.5-ml tube.
Add 100 μl ribitol (50 μg/ml, as an internal standard) to each tube and vortex briefly, then centrifuge at 10,000 × g for 30 s at room temperature.
Dry samples in a vacuum freeze drier.
Note: Samples need to be thoroughly dried before the following derivatization.
Add 50.0 μl methoxyamine hydrochloride to each tube to dissolve the samples, pipette up and down several times, and centrifuge at 10,000 × g for 2 min at room temperature.
Transfer the solutions from the tubes to an auto sampler vial with a glass insert.
Place the samples in an incubator at 65°C for 2 h.
Add 50.0 μl N-methyl-N-(trimethylsilyl) trifluoroacetamide to each auto sampler vial and place in an incubator at 65°C for another 2 h.
Note: Perform the derivatization at 65°C (Steps A10 and A11) instead of a lower temperature; this is necessary for quantitating sugar content in minute quantities of rice tissues by GC-MS and significantly increases the sensitivity and accuracy of this method.
Analyze the samples by GC-MS.
Instrument Parameters
Set the GC parameters as follows: Carrier gas, helium (61.3 kPa); gas flow rate, 30 ml/min; GC interface temperature, 230°C.
GC oven temperature gradient program: Begin at 70°C; hold at 70°C for 2 min; ramp to 200°C at a rate of 5°C/min; hold at 200°C for 3 min; ramp to 300°C at a rate of 10°C/min; hold at 300°C for 3 min. Total run time: 44 min.
MS parameters: Source temperature, 250°C; mass-to-charge ratio (m/z) range, 60-600; scanning speed, 0.4 scans/s; ionization energy, 70 eV.
Note: The above instrumental parameters are based on the Shimadzu GC-MS QP-2010, which may be different and require modification when using a different GC-MS instrument.
Data analysis
The four standard compounds, fructose, glucose, sucrose, and ribitol, are analyzed by GC-MS, where both the GC-MS solution software and the NIST17 mass spectra search program are used to confirm the peak assignments and obtain the retention times (see Figure 1). Results are reported as ‘‘Positive’’ when the final concentrations of the target sugars extracted from rice tissues range from 2.5 μg/ml to 500.0 μg/ml.
Note: For peak quantitation, the chromatographic integral (CI) of a target sugar was normalized to the CI of ribitol of the same sample (CItarget sugar/CIribitol); The normalized CI of the target sugar is used in the comparison of the target sugar content among different samples; The correlation coefficients of calibration curves developed should be higher than 0.97, approximately 15 mg fresh weight of rice tissues should yield reliable data. The final sugar concentrations are reliable in the range of 2.5-500.0 μg/ml.
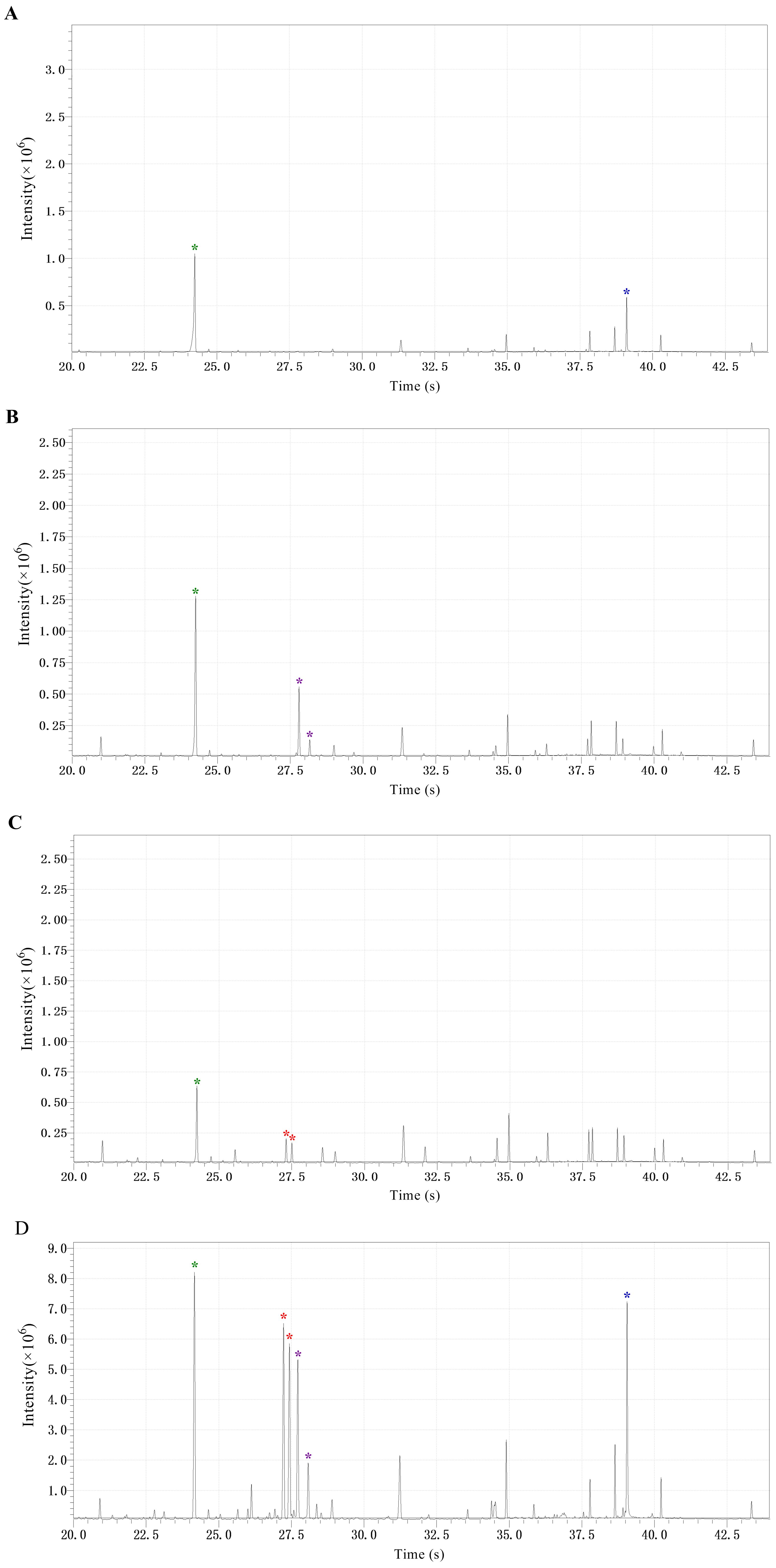
Figure 1. Chromatograms of ribitol and the three soluble sugars in the tested samples. (A) Chromatograms of ribitol and sucrose, indicated by the green and blue asterisk, respectively. (B) Chromatograms of ribitol and glucose, indicated by the green and voilet asterisk, respectively. (C) Chromatograms of ribitol and fructose, indicated by the green and red asterisk, respectively. (D) Chromatograms of ribitol and the three sugars (sucrose, glucose, and fructose) extracted from rice roots, marked with the colored asterisks as described above.
Recipes
Methoxyamine hydrochloride solution
Dissolve 15 mg methoxyamine hydrochloride in 1.0 ml pyridine. It must be freshly prepared and used at room temperature.
Internal standard solution
Prepartion of the internal standard (50 μg/ml ribitol): Dissolve 1.0 mg ribitol in 20 ml deionized water and vortex briefly. Store at -80°C until use.
Acknowledgments
We acknowledge financial support from the National Nature Science Foundations of China (31501613) and the National Key Research and Development Program of China (2018YFD0201201-4 & 2018YFD0200500). This work was adapted and modified from Xu et al., 2021.
Competing interests
The authors declare no conflicts of interest.
References
- Bezrutczyk, M., Yang, J., Eom, J. S., Prior, M., Sosso, D., Hartwig, T., Szurek, B., Oliva, R., Vera-Cruz, C., White, F. F., Yang, B. and Frommer, W. B. (2018). Sugar flux and signaling in plant-microbe interactions. Plant J 93(4): 675-685.
- Cataldi, T. R., Margiotta, G., Iasi, L., Di Chio, B., Xiloyannis, C. and Bufo, S. A. (2000). Determination of sugar compounds in olive plant extracts by anion-exchange chromatography with pulsed amperometric detection. Anal Chem 72(16): 3902-3907.
- Hofmann, J., Kolev, P., Kolev, N., Daxbock-Horvath, S., Grundler and F. M. W. (2009). The Arabidopsis thaliana sucrose transporter gene AtSUC4 is expressed in Meloidogyne incognita-induced root galls. J Phytopathol 157(4): 256-261.
- Ibraheem, O., Botha, C.E.J., Bradley, G., Dealtry, G. and Roux, S. (2014). Rice sucrose transporter1 (OsSUT1) up-regulation in xylem parenchyma is caused by aphid feeding on rice leaf blade vascular bundles. Plant Biol 16(4): 783-791.
- Jan, L., Nicolas, S., Joachim, K. (2006). Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1(1): 387-396.
- Magwaza, L.S. and Opara, U.L. (2015). Analytical methods for determination of sugars and sweetness of horticultural products-A review. Sci Hortic 184: 179-192.
- Milne, R.J., Grof, C.P.L. and Patrick, J.W. (2018). Mechanisms of phloem unloading: shaped by cellular pathways, their conductances and sink function. Curr Opin Plant Biol 43: 8-15.
- Montero, C.M., Dodero, M.C.R., Sanchez, D.A.G. and Barroso, C.G. (2004). Analysis of low molecular weight carbohydrates in food and beverages: A review.Chromatographia 59(1-2): 15-30.
- Romani, A., Baldi, A., Tattini, M. and Vincieri, F.F. (1994). Extraction, purification procedures and HPLC-RI analysis of carbohydrates in Olive (Olea-Europaea L) plants. Chromatographia 39(1-2): 35-39.
- Sosso, D., van der Linde, K., Bezrutczyk, M., Schuler, D., Schneider, K., Kamper, J. and Walbot, V. (2019). Sugar partitioning between Ustilago maydis and its host Zea mays L during infection. Plant Physiol 179(4): 1373-1385.
- Wei, X.Y., Nguyen, S.T.T, Collings, D.A. and McCurdy, D.W. (2020). Sucrose regulates wall ingrowth deposition in phloem parenchyma transfer cells in Arabidopsis via affecting phloem loading activity. J Exp Bot 71(16): 4690-4702.
- Xu, L.H., Xiao, L.Y., Peng, D.L., Xiao, X.Q., Huang, W.K., Gheysen, G. and Wang, G.F. (2021). Plasmodesmata play pivotal role in sucrose supply to Meloidogyne graminicola-caused giant cells in rice. Mol Plant Pathol 22(5): 539-550.
- Yamada, K. and Osakabe, Y. (2018). Sugar compartmentation as an environmental stress adaptation strategy in plants. Semin Cell Dev Biol 83: 106-114.
- Ye, H., Ma, Y.Q., Wang, X., Gao, Y.X., Zhang, K., and Xue, Y. (2019). Determination of soluble sugar from sweet corn cob by microtiter method of the anthrone-sulfuric acid colorimetric assay. Food Sci Technol 44(11): 327-333.
- Zhu, X.G., Wang, Y. and Long, S.P. (2013). e-photosynthesis: a comprehensive dynamic mechanistic model of C3 photosynthesis: from light capture to sucrose synthesis. Plant Cell Environ 36(9): 1711-1727.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Xu, L., Xiao, L., Xiao, X., Xiao, Y. and Wang, G. (2021). Analysis of Soluble Sugar Content in Minute Quantities of Rice Tissues by GC-MS. Bio-protocol 11(13): e4077. DOI: 10.21769/BioProtoc.4077.
Category
Plant Science > Plant physiology > Biotic stress
Biochemistry > Carbohydrate > Disaccharide
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










