- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Visualization of Host Cell Kinase Activation by Viral Proteins Using GFP Fluorescence Complementation and Immunofluorescence Microscopy
(*contributed equally to this work) Published: Vol 11, Iss 13, Jul 5, 2021 DOI: 10.21769/BioProtoc.4068 Views: 5041
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

General Maintenance and Reactivation of iSLK Cell Lines
Ariana C. Calderón-Zavala [...] Ekaterina E. Heldwein
Jun 5, 2025 1876 Views

Inducible HIV-1 Reservoir Reduction Assay (HIVRRA), a Fast and Sensitive Assay to Test Cytotoxicity and Potency of Cure Strategies to Reduce the Replication-Competent HIV-1 Reservoir in Ex Vivo PBMCs
Jade Jansen [...] Neeltje A. Kootstra
Jul 20, 2025 2457 Views

Assembly and Mutagenesis of Human Coronavirus OC43 Genomes in Yeast via Transformation-Associated Recombination
Brett A. Duguay and Craig McCormick
Aug 20, 2025 3033 Views
Abstract
Non-receptor protein-tyrosine kinases regulate cellular responses to many external signals and are important drug discovery targets for cancer and infectious diseases. While many assays exist for the assessment of kinase activity in vitro, methods that report changes in tyrosine kinase activity in single cells have the potential to provide information about kinase responses at the cell population level. In this protocol, we combined bimolecular fluorescence complementation (BiFC), an established method for the assessment of protein-protein interactions, and immunofluorescence staining with phosphospecific antibodies to characterize changes in host cell tyrosine kinase activity in the presence of an HIV-1 virulence factor, Nef. Specifically, two Tec family kinases (Itk and Btk) as well as Nef were fused to complementary, non-fluorescent fragments of the Venus variant of YFP. Each kinase was expressed in 293T cells in the presence or absence of Nef and immunostained for protein expression and activity with anti-phosphotyrosine (pTyr) antibodies. Multi-color confocal microscopy revealed the interaction of Nef with each kinase (BiFC), kinase activity, and kinase protein expression. Strong BiFC signals were observed when Nef was co-expressed with both Itk and Btk, indicative of interaction, and a strong anti-pTyr immunoreactivity was also seen. The BiFC, pTyr, and kinase expression signals co-localized to the plasma membrane, consistent with Nef-mediated kinase activation in this subcellular compartment. Image analysis allowed calculation of pTyr-to-kinase protein ratios, which showed a range of responses in individual cells across the population that shifted upward in the presence of Nef and back down in the presence of a kinase inhibitor. This method has the potential to reveal changes in steady-state non-receptor tyrosine kinase activity and subcellular localization in a cell population in response to other protein-kinase interactions, information that is not attainable from immunoblotting or other in vitro methods.
Keywords: Protein-tyrosine kinaseBackground
Non-receptor protein-tyrosine kinases, exemplified by members of the Src and Tec kinase families, regulate many aspects of cell biology including growth, differentiation, and motility in response to diverse stimuli (Amatya et al., 2019). Methods that assess the spatial and temporal aspects of tyrosine kinase signaling at the single cell and population levels are essential to better understanding their function. In this protocol, we provide details of a cell-based method to evaluate protein-protein interaction, kinase activity, and subcellular localization of Tec family kinases in response to the interaction with the HIV-1 accessory protein, Nef. This approach is potentially applicable to many other kinase systems in which protein-protein interactions impact kinase activity.
Nef is a small (27-34 kDa, depending on the subtype) membrane-associated protein unique to the primate lentiviruses HIV-1, HIV-2, and SIV (Foster and Garcia, 2008). HIV-1 Nef enhances viral infectivity, supports high-titer replication in vivo, and promotes immune escape of HIV-infected cells (Basmaciogullari and Pizzato, 2014; Pawlak and Dikeakos, 2015). Rhesus macaques infected with nef-defective SIV exhibit very low viral loads and do not progress to simian AIDS (Kestler et al., 1991), illustrating an essential role for Nef in viral pathogenesis. Along the same lines, individuals infected with nef-defective HIV-1 can remain AIDS-free in the absence of antiretroviral therapy for many years (Deacon et al., 1995; Kirchhoff et al., 1995).
Nef lacks intrinsic biochemical activities, functioning instead through interactions with host cell proteins related primarily to endocytic trafficking and kinase signaling pathways (Staudt et al., 2020).Nef hijacks non-receptor tyrosine kinases of the Src and Tec families normally linked to immune receptor activation to enhance HIV-1 replication. Nef directly activates the Src family members Hck and Lyn by binding to their SH3 domains (Briggs et al., 1997; Trible et al., 2006). Selective inhibition of Nef-mediated Src family kinase activation blocks Nef-dependent enhancement of HIV-1 infectivity and replication (Emert-Sedlak et al., 2009 and 2013). Tec family kinases play essential roles in B- and T-cell receptor signaling (Andreotti et al., 2010), with the interleukin-2 inducible T-cell kinase (Itk) and Bruton's tyrosine kinase (Btk) expressed in primary HIV-1 target cells (CD4+ T cells and macrophages, respectively). Readinger et al. provided the first evidence linking Itk to HIV-1 entry, viral transcription, assembly, and release (Readinger et al., 2008). A subsequent study showed that Nef provides a link between HIV-1 infection and Tec family kinase signaling by demonstrating direct interaction between Nef and both Itk and Btk at the cell membrane (Tarafdar et al., 2014). Treatment of HIV-infected T cells with a selective Itk inhibitor blocked Nef-dependent enhancement of viral infectivity and replication. Importantly, activation of both Src and Tec family kinases is highly conserved across all M group HIV-1 subtypes, consistent with an important function in the HIV-1 life cycle and viral pathogenesis (Narute and Smithgall, 2012; Emert-Sedlak et al., 2013; Tarafdar et al., 2014). For an in-depth review of Nef interactions with host cell tyrosine kinases, see Staudt et al. (2020).
In a recent study, we explored the molecular mechanisms of Tec family kinase activation by the Nef proteins of HIV-1 and SIV (Li et al., 2020). By combining cell-based bimolecular fluorescence complementation (BiFC) (Romei and Boxer, 2019) and anti-phosphotyrosine immunofluorescence microscopy, we found that HIV-1 Nef interacts with Itk and Btk at the cell membrane and results in constitutive kinase activity. For the BiFC assay, Itk or Btk and Nef were fused to complementary, non-fluorescent fragments of the Venus variant (Nagai et al., 2002) of YFP. The kinases were then expressed in 293T cells either alone or together with Nef, followed by immunostaining for Nef and kinase protein expression as well as protein-tyrosine phosphorylation using anti-phosphotyrosine (pTyr) antibodies. Multi-color confocal microscopy enabled simultaneous assessment of Nef-kinase complex formation (BiFC), kinase activity (anti-pTyr immunofluorescence), and kinase protein expression (anti-kinase immunofluorescence). When expressed alone, both Itk and Btk showed a diffuse subcellular staining pattern with the anti-kinase antibody and weak reactivity with the anti-pTyr antibody. In contrast, co-expression with Nef induced a strong BiFC signal with both Itk and Btk, indicative of interaction, and a strong anti-pTyr immunoreactivity was seen. The BiFC, pTyr, and kinase expression signals co-localized to the cell membrane, consistent with Nef-mediated kinase activation in this subcellular compartment. As a control, cells were treated with Tec family kinase inhibitors (Lin et al., 2004; Roskoski, 2016) that suppressed the pTyr signal but did not affect BiFC, demonstrating that interaction of Nef with Tec family kinases at the membrane does not require kinase activity.
Results were quantitated at the single-cell level using the NIH ImageJ image analysis software (Schneider et al., 2012). Kinase expression and tyrosine phosphorylation immunofluorescence signal intensities for at least 100 cells for each condition were expressed as the mean pTyr-to-kinase protein fluorescence intensity ratios. This analysis enabled statistical comparisons of cell populations and showed that cells co-expressing Itk or Btk and Nef had significantly higher fluorescence ratios as compared with those expressing the kinase alone or the inhibitor-treated cells expressing the Nef-kinase complex.
Using the same approach, we also investigated Nef-stimulated Itk and Btk autophosphorylation on their respective activation loop tyrosine residues (pTyr511 and pTyr551, respectively), a step required for Tec family kinase activation (Joseph et al., 2013). Cells were transfected with the Nef and Itk/Btk Venus fusion constructs for BiFC as before, but in this case the cells were stained with phosphospecific antibodies for the activation loop phosphotyrosines in place of the general anti-pTyr antibody. Co-expression with Nef led to significant increases in Itk and Btk activation loop autophosphorylation, which also localized almost exclusively to the cell membrane. SIV Nef (mac239 allele) was also found to strongly induce membrane-associated autophosphorylation of both kinases, demonstrating that Tec family kinase activation is conserved across Nef proteins from diverse primate lentiviruses. A representative confocal image from this study is shown in Figure 1.
While the experimental approach described above was developed to explore kinase interaction with, and activation by, a viral protein in a cell-based setting, the overall concept should be readily adaptable to any combination of kinases and interacting partners. The cell-based approach has important advantages over older in vitro methods such as immune-complex kinase assays, which do not provide information about the subcellular localization of the active kinase complex or the range of responses across a population of individual cells. However, one important caveat includes consideration of where to add the fragments of Venus for the BiFC assay. For example, both Nef and Tec family kinases localize to the cell membrane by virtue of N-terminal signals. Nef is myristoylated on its N-terminus, while Tec kinases have an N-terminal Pleckstrin homology (PH) domain that binds to membrane phosphoinositides. To avoid interference with these membrane-targeting signals, we were careful to fuse the Venus fragments to the C-terminus of each protein. Control experiments are also essential to verify that Venus fragment fusion does not influence basal kinase activity or localization, which is readily accomplished by comparing unfused with fused versions of each kinase in transfected cells and staining with kinase and phosphospecific antibodies. Finally, it should be noted that the Venus fluorophore, once reconstituted via BiFC, is irreversible. While this feature of BiFC may help to stabilize transient interactions for endpoint assessment by microscopy as described here, other techniques are more appropriate to assess the kinetics of interaction, such as the split-FAST reversible complementation system (Tebo and Gautier, 2019).
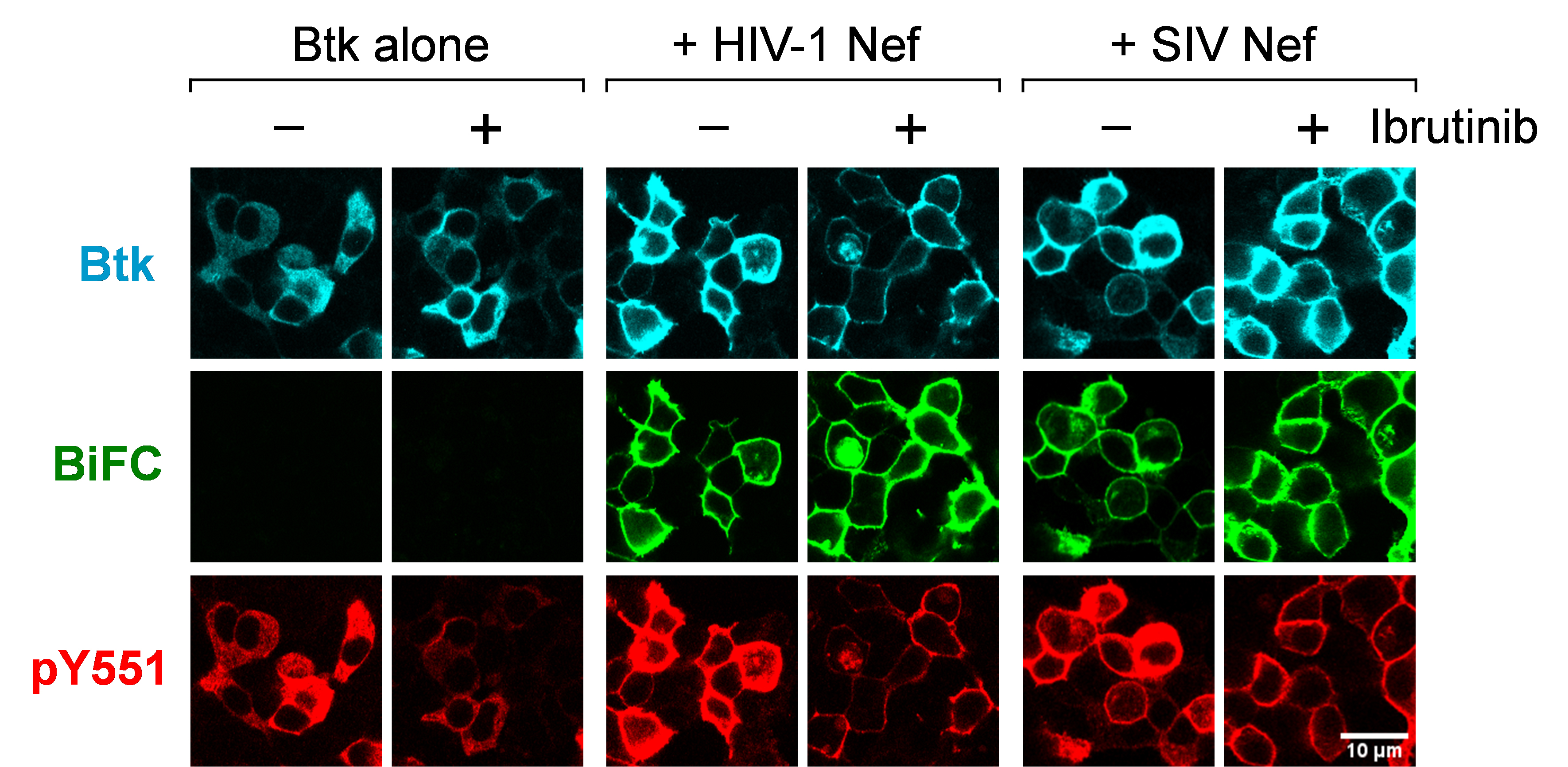
Figure 1. HIV-1 and SIV Nef proteins induce constitutive Btk activation loop autophosphorylation at the cell membrane. Btk was expressed in 293T cells either alone or together with HIV-1 Nef (SF2 isolate) or SIV Nef (mac239) as BiFC pairs in the absence or presence of the Btk/Itk inhibitor ibrutinib (1 μM). Cells were fixed and stained for confocal microscopy with phosphospecific antibodies against the Btk activation loop phosphotyrosine (pY551; red) and the Btk protein (V5 epitope; cyan). Nef interaction with Btk is observed as fluorescence complementation of the YFP variant, Venus (BiFC; green). Note that interaction and kinase activation occur at the plasma membrane.
Materials and Reagents
Molecular biology reagents
Phusion high-fidelity DNA polymerase (New England Biolabs, catalog number: M0530S)
Venus template (gift from Dr. Atsushi Miyawaki, RIKEN Brain Science Institute, Saitama, Japan)
HIV-1 (SF2 allele) and SIV (mac239) Nef clones (NIH AIDS Reagent Program, HIV #11431; SIV #2476)
Full-length human Tec family kinase cDNA clones (Dana-Farber/Harvard Cancer Center PlasmID DNA Resource Core, Btk # HsCD00346954; Itk # HsCD00021352)
Mammalian expression vector, pCDNA3.1(−) (Thermo Fisher, catalog number: V79520)
Anti-V5 tag mouse monoclonal antibody (Thermo Fisher, catalog number: R960-25)
Anti-V5 tag rabbit polyclonal antibody (Sigma, catalog number: AB3792)
BTK anti-pY551 rabbit monoclonal antibody (Abcam, catalog number: ab40770)
Anti-pTyr antibody pY99 (Santa Cruz, catalog number: sc-7020)
Anti-HIV-1 Nef monoclonal antibody 6.2 (NIH AIDS Reagent Program, catalog number: 1539)
Goat anti-rabbit IgG (H+L), mouse/human ads-TXRD (Texas Red conjugate; cross-adsorbed to mouse and human immunoglobulins; Southern Biotech, catalog number: 4050-07)
Goat anti-mouse IgG (H+L), human ads-TXRD (Texas Red conjugate; cross-adsorbed to human immunoglobulins; Southern Biotech, catalog number: 1031-07)
Pacific Blue goat anti-mouse IgG antibody (Thermo Fisher/Molecular Probes, catalog number: P31582)
Pacific Blue goat anti-rabbit IgG antibody (Thermo Fisher/Molecular Probes, catalog number: P10994)
35 mm microwell dishes (MatTek, catalog number: P35G-1.5-14-C)
Human embryonic kidney 293T cells (American Type Culture Collection, catalog number: CRL-11268)
Dulbecco’s modified Eagle’s medium (DMEM; ThermoFisher/Invitrogen, catalog number: 11965-118)
Fetal bovine serum (FBS; Gemini Bio-Products, catalog number: 900-108)
Trypsin-EDTA, 0.05% (ThermoFisher/Invitrogen catalog number: 25300054)
X-tremeGENE 9 DNA transfection reagent (Sigma-Aldrich, catalog number: 06365787001)
Paraformaldehyde, 16% aqueous solution (Fisher, catalog number: 50980487)
Triton X-100 (Sigma, catalog number: X100-1L)
Bovine serum albumin (BSA, Sigma, catalog number: A3059-500G)
Itk inhibitor, BMS-509744 (Calbiochem, catalog number: 41-982-05MG)
Itk/Btk inhibitor, ibrutinib (SelleckChem, catalog number: S2680)
Equipment
Olympus FluoView FV1000 Confocal Microscope
Software
Prism v. 8.0 (GraphPad Software, Inc.; www.graphpad.com)
ImageJ (National Institutes of Health; https://imagej.net/Welcome)
Olympus FluoView Software (https://www.olympus-lifescience.com/en/)
Procedure
Construction of expression vectors for BiFC based on Venus. This procedure is based on our published work with BiFC vectors for lentiviral Nef alleles and the Tec family kinases Btk and Itk (Tarafdar et al., 2014; Li et al., 2020) but can be easily adapted to virtually any combination of interacting protein partners. While our approach uses pcDNA3-based expression vectors that drive strong protein expression in 293T cells, other vectors can be substituted depending on the target cell type and application. Additional details related to primer design, subcloning strategy, and sequences of the final fusion constructs are provided in the Appendix.
PCR-amplify the coding sequence for the Venus N-terminal (VN: residues Val2 to Asp173) and C-terminal (VC: residues Ala154 to Lys238) fragments containing the appropriate restriction sites and a stop codon, and subclone the PCR product into the mammalian expression vector, pcDNA3.1(–).
PCR-amplify the coding sequence for HIV-1 Nef (SF2) and SIV Nef (mac239) in the same manner and subclone the product into the pcDNA3.1(–)/VN construct.
PCR-amplify the full-length human Tec family kinase cDNA clones containing the appropriate restriction sites and a C-terminal V5 epitope tag for ligation in-frame into the pcDNA3.1(–)/VC construct.
293T cell culture and transfection
Culture 293T cells in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum in a 37°C humidified incubator with a 5% CO2 atmosphere. Use 10 ml DMEM per 10-cm culture dish and maintain cells at a density of 2-5 × 104 cells/cm2. When cultures reach 80% confluence, remove the spent medium and split 1:10 by incubating with 1.0 ml 0.05% trypsin-EDTA solution for 1 min at 37°C to generate a single cell suspension. Add 10 ml fresh medium and spin at 500 × g for 5 min. Resuspend the cell pellet in 10 ml fresh medium and transfer 1.0 ml cell suspension to new 10-cm dishes with an additional 10 ml fresh medium. For transfection, seed 2.5 × 105 cells per MatTek plate and culture overnight.
Transfect cells with BiFC expression vectors using X-tremeGene 9 DNA transfection reagent according to the manufacturer’s protocol. Briefly, mix 3 µl X-tremeGene 9 with 97 µl DMEM, add each BiFC expression plasmid (0.5 µg each VN and VC construct), and incubate the mixture at room temperature for 15 min. After incubation, add the transfection complex dropwise to the MatTek culture plate. Treat the cells with inhibitors or the DMSO carrier solvent (0.1% final concentration) 4 h after transfection as needed.
Immunofluorescence staining
Forty hours post-transfection, fix the cells by replacing the medium with 2 ml 4% paraformaldehyde and incubate for 10 min at room temperature. Wash cells with 2 ml PBS (pH 7.4) for 5 min with occasional gentle shaking by hand.
Permeabilize the cells with 2 ml 0.2% Triton X-100 in PBS for 15 min. Wash cells twice with 2.0 ml PBS for 5 min each.
Block the cells with 2 ml 2% BSA in PBS for 1 h at room temperature or overnight at 4°C.
Incubate the cells with anti-V5 (kinase tag) and anti-Nef or anti-pTyr (or anti-Btk pY551/anti-Itk pY511) antibodies diluted 1:1,000 in 250 µl PBS with 2% BSA for 1 h at room temperature. Wash cells three times with 2 ml PBS for 5 min each.
Incubate the cells with the appropriate secondary antibodies conjugated to Texas Red or Pacific Blue at a dilution of 1:500 or 1:1,000, respectively. Wash the cells three times with 2 ml PBS for 5 min each. Keep cells in 2 ml PBS for imaging.
Note: This protocol was developed for 3-color imaging to allow simultaneous detection of interaction (Venus complementation) and protein expression of each interacting partner (kinase and Nef), or interaction, kinase expression, and kinase activity (overall cellular phosphotyrosine content or activation loop autophosphorylation).
Fluorescence imaging and analysis
Acquire images using multi-color confocal microscopy with a 60× objective using x-y scan mode. On the Olympus FluoView1000, we used the violet laser to detect Pacific Blue (405 nm), the green laser to detect fluorescence complementation (Venus; 543 nm), and the red laser to detect Texas Red (633 nm).
Perform single-cell image analysis using the Java-based image processing program, ImageJ, as described under Data analysis.
Data analysis
As an example, the following protocol uses ImageJ to quantitate the mean fluorescence intensities (MFI) of cells stained with anti-phosphotyrosine (anti-pTyr) and anti-Btk antibodies. The ratio of the pTyr to Btk MFIs is then calculated for a minimum of 100 cells as a measure of the kinase activity within the cell population. The same approach can be used to quantitate other MFI ratios such as interaction (BiFC fluorescence) normalized to partner protein expression.
File/Open: Open the Btk protein expression and confocal image pair (Figure 2). Minimize the pTyr image and do not click on it again. Click on the Btk protein image window.
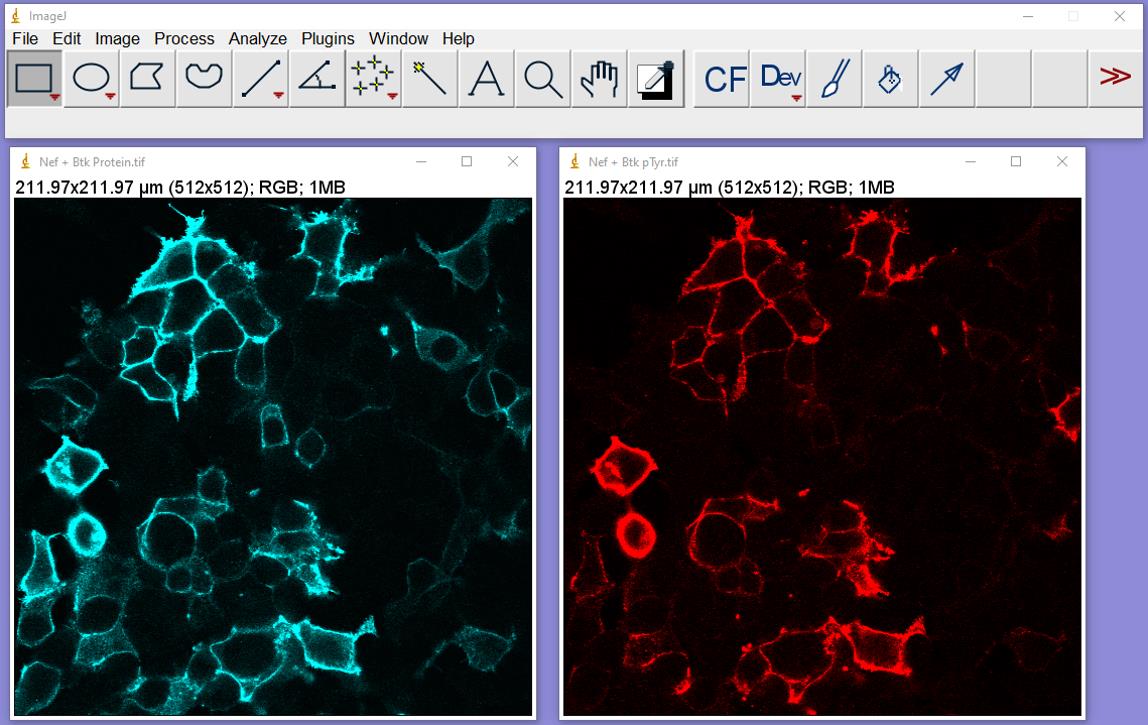
Figure 2. Confocal images of 293T cells stained for Btk protein expression and pTyr opened in ImageJ. Cells stained for Btk protein are shown on the left (cyan), with pTyr shown on the right (red). These images are from cells co-expressing Btk and HIV-1 Nef.Image/Type: Change to 8-bit, which will convert the image to black and white (Figure 3).
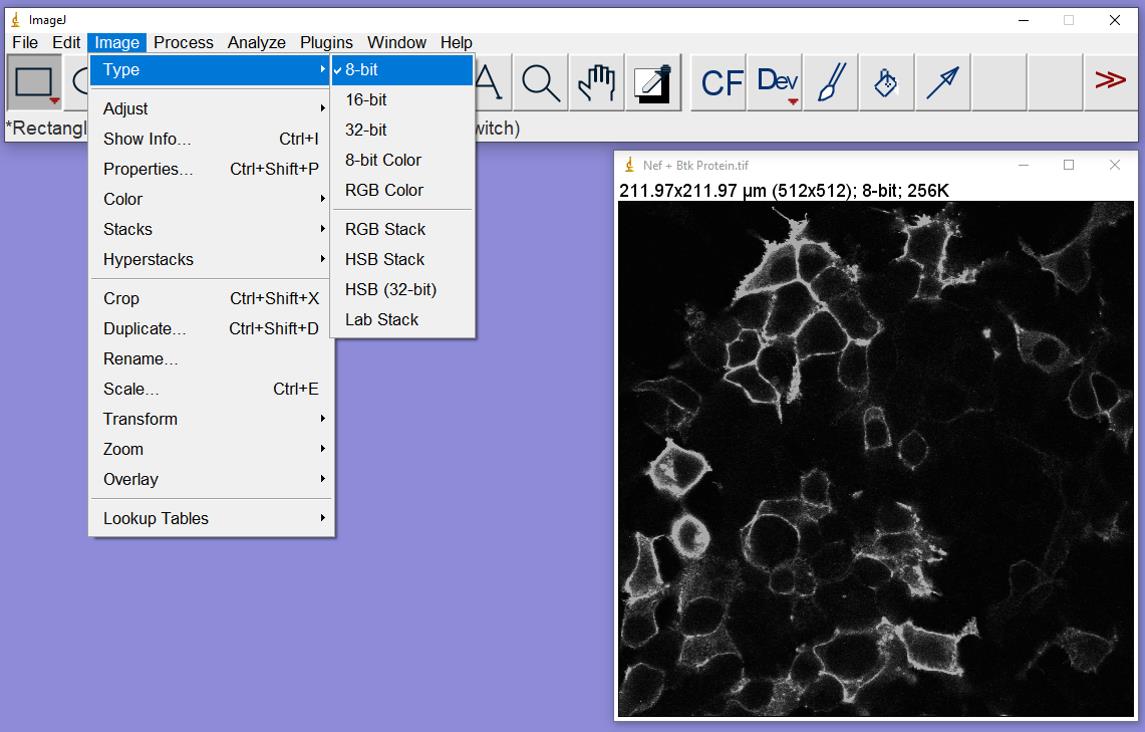
Figure 3. Conversion of Btk protein image to an 8-bit greyscale. In the control bar, click on Image and then 8-bit, which will convert the image to an 8-bit grey scale image (right).Image/Adjust/Threshold: Set levels by moving the two sliders to set the min/max to include staining but minimize the background and overexposure. Check the “Dark Background” box. Click “Set” and “OK.” Close the Threshold box. The red highlighted areas show the pixels that will be included in the analysis (Figure 4).
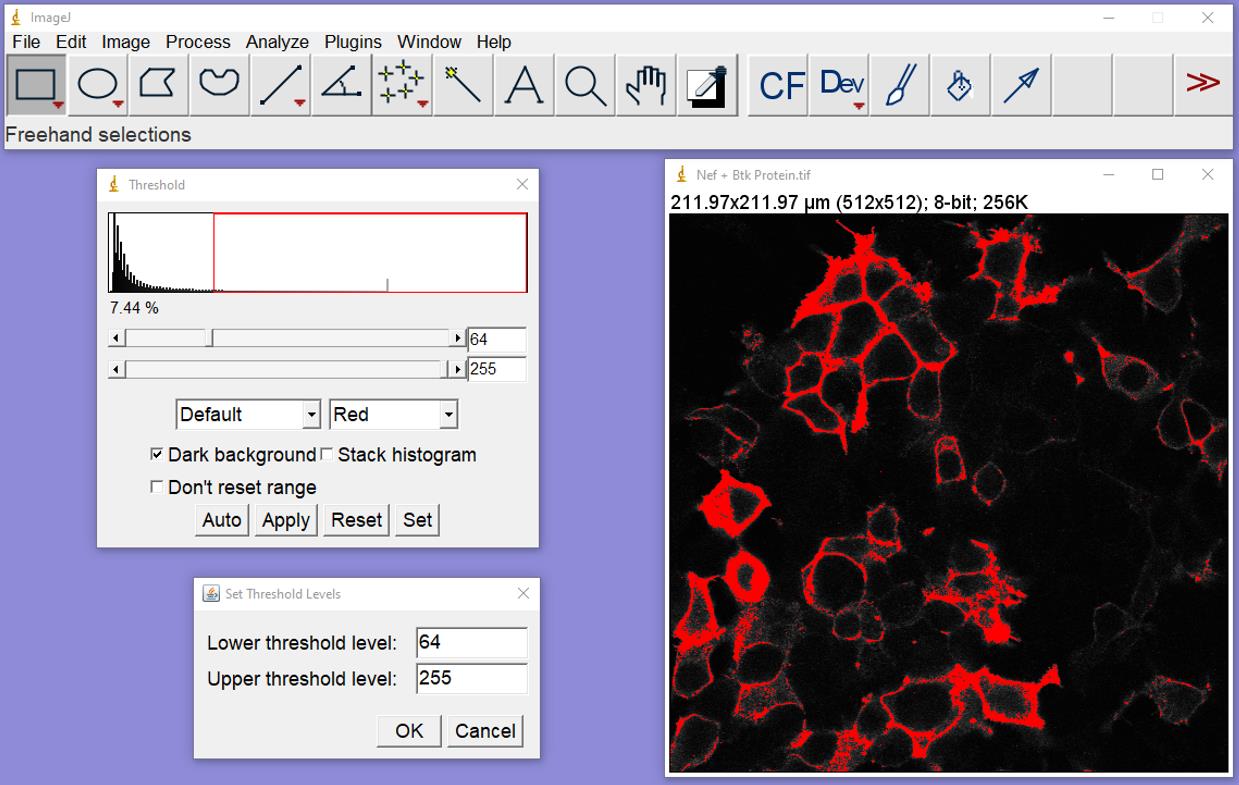
Figure 4. Setting the threshold. Click on Image in the control bar, select Adjust, and then Threshold. The box on the upper left will appear, and the pixels that will be included in the analysis will be shown in red on the image (right). Click Set; the lower box shown will appear. Click OK to finish.Analyze/Set Measurements: Select these options (Figure 5):
Area, Min & Max Gray Value, and Mean Gray Value.
Set the “Redirect to” box to “None.”
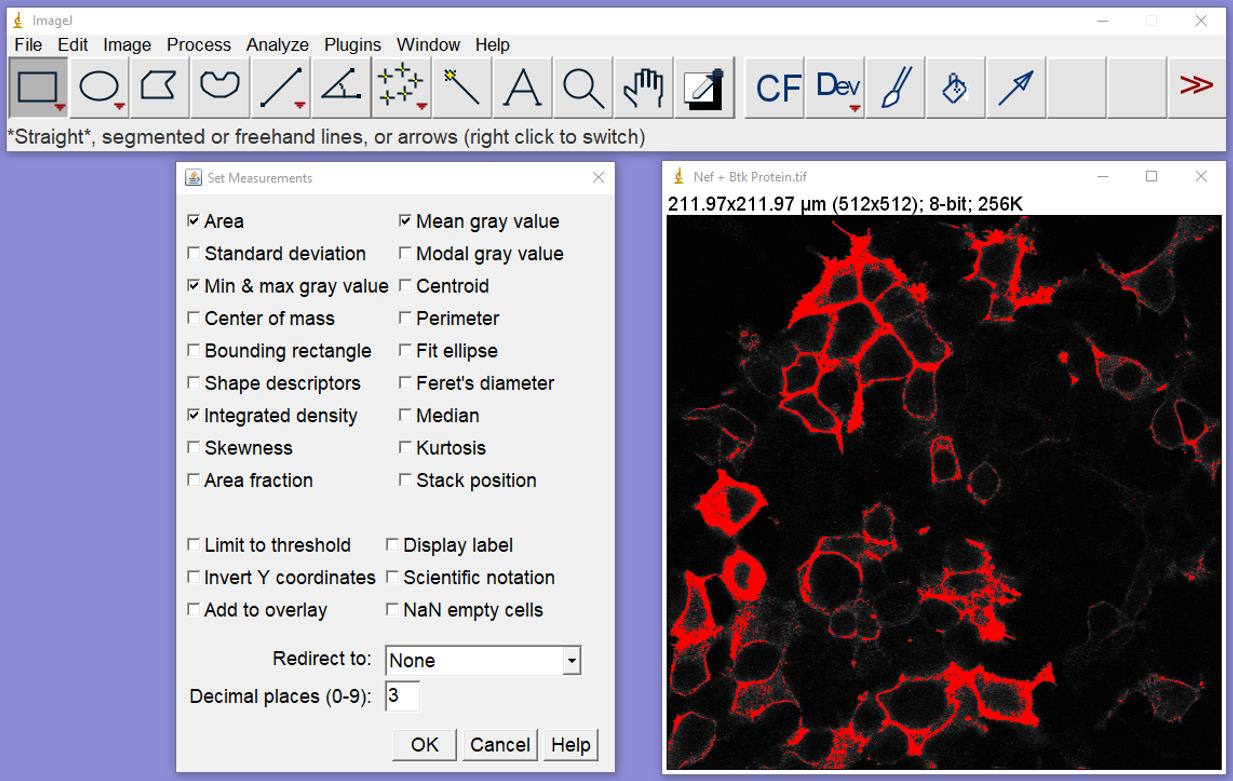
Figure 5. Setting the analysis parameters. From the control bar, click on Analyze and then Set Measurements; the box shown on the left will appear. Select the parameters shown.
Analyze/Analyze Particles (Figure 6):
Size: Determines the cell area that will be analyzed. Typically, [20-infinity] and [50-infinity] works best for 400× and 100× images of 293T cells, respectively.
Check the “pixel units” box.
Circularity: Start with [0.0-1.0] to exclude background specks and noise.
Show: Chose the “Outlines” option to reveal the location and size of the areas selected for measurement (controlled by adjusting the size/circularity values above).
Check the “Display Results” box and uncheck the “Clear results” box.
Click “OK.” Two additional windows will be generated: One (Figure 7) shows outlines of the cell areas used in the analysis of the thresholded image (left), and the second (right) is the results table window containing MFI data (‘Mean’ column).
Copy the data in the Results window into an Excel file and close the Results window only.
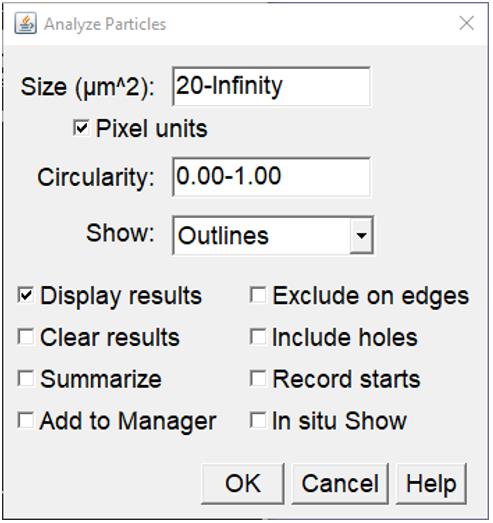
Figure 6. Particle (pixel) analysis settings. From the control bar, select Analyze and then analyze particle, which will open this box. Select the parameters shown.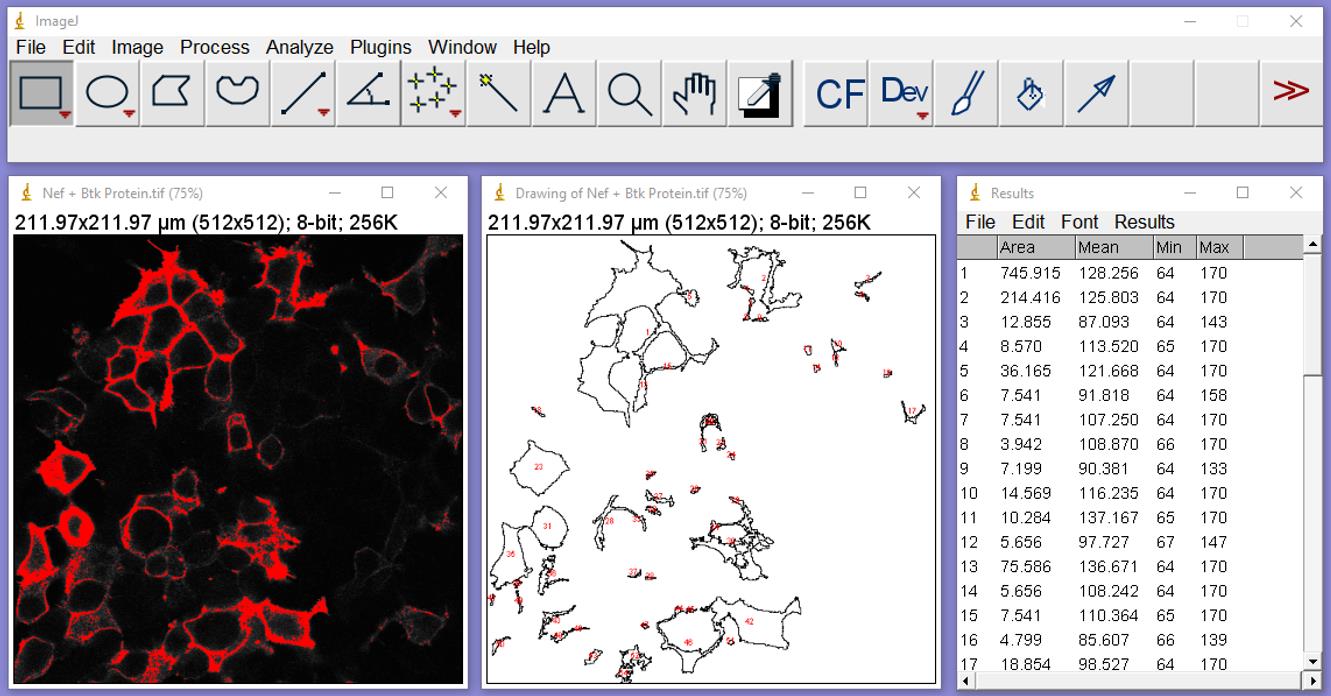
Figure 7. Results of particle analysis. The center image shows outlines of the areas where pixel intensities have been analyzed and numbered. The table on the right shows the resulting data including the number of the analyzed feature and its area, followed by the mean and minimum and maximum fluorescence intensities. Note that cells will be analyzed individually, as groups, or as fragments, with several fragments from the same cell.The next step will use the same masks to analyze the anti-pTyr image. Be sure the “Results” box from the protein analysis is closed. Click on the ‘Nef + Btk’ protein box to activate it.
Analyze/Set Measurements: Set the “Redirect to” box to the corresponding pTyr image file name (previously minimized and not modified) while keeping the other parameters the same. Click “OK.” The selected (outlined) fields established for the Btk protein image are now directly transposed onto the pTyr image and will analyze the pTyr levels relative to the identical cell locations (pixels) provided by the Btk immuno-stain.
Analyze/Analyze Particles: Use the previous settings and click “OK.” Select all and copy the data in the Results window into the Excel file. The Results windows from the Btk protein and pTyr analyses are shown side-by-side below (Figure 8). Note that the number of rows in each are the same (54; not shown) and the areas used for analysis of each region are also identical. However, the MFI values differ (Mean columns) as expected, since the intensities are different.
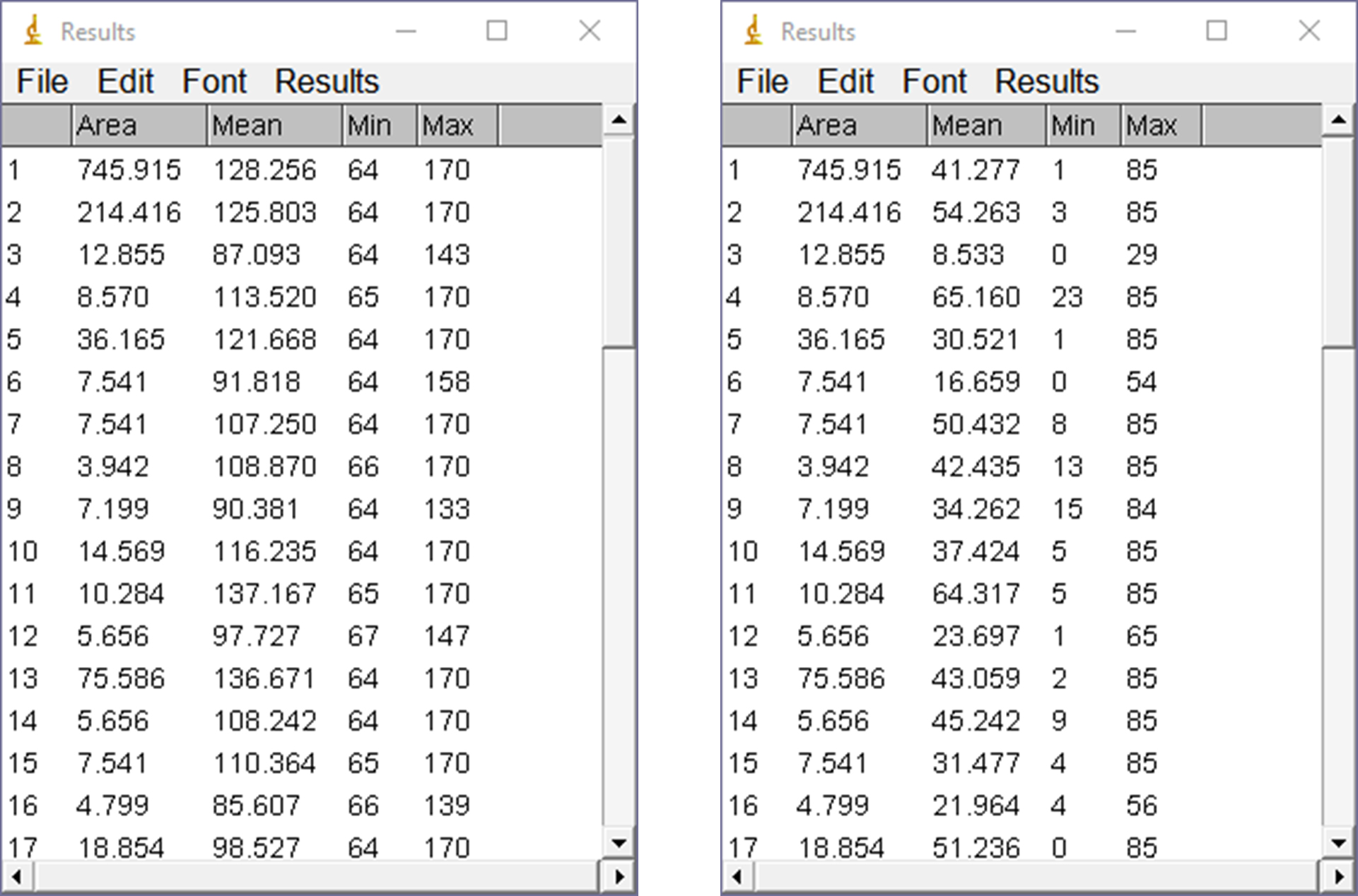
Figure 8. Results of particle analysis of the Btk pTyr image. The Results tables from the Btk protein (from Figure 7, left) and pTyr (right) analyses are shown side-by-side for comparison. Note that areas of the features in each image are identical, as are the number of features (54 in each case, not shown). Both conditions must be true for the subsequent calculations to be valid.Using Excel, calculate the ratio of pTyr to Btk mean pixel densities (“Mean” columns).
Analyze at least 100 cells per condition, which may require several fields of cells depending on the cell density, size, and magnification used. Significant differences between groups can be analyzed using an unpaired Student’s t-test (GraphPad Prism v.8.0). For added rigor, perform three biological replicates of each experiment.
The data can be presented in several different ways using Prism. We prefer showing the results for individual “cells” (really from masks of individual cells or cell fragments as identified by ImageJ) as a series of bars, which gives a view of the distribution of ratios across the cell population. Alternatively, box-and-whisker plots or violin plots can be used. Figure 9 shows the distribution of ratios obtained using the above approach following ImageJ analysis of the results shown in Figure 1.
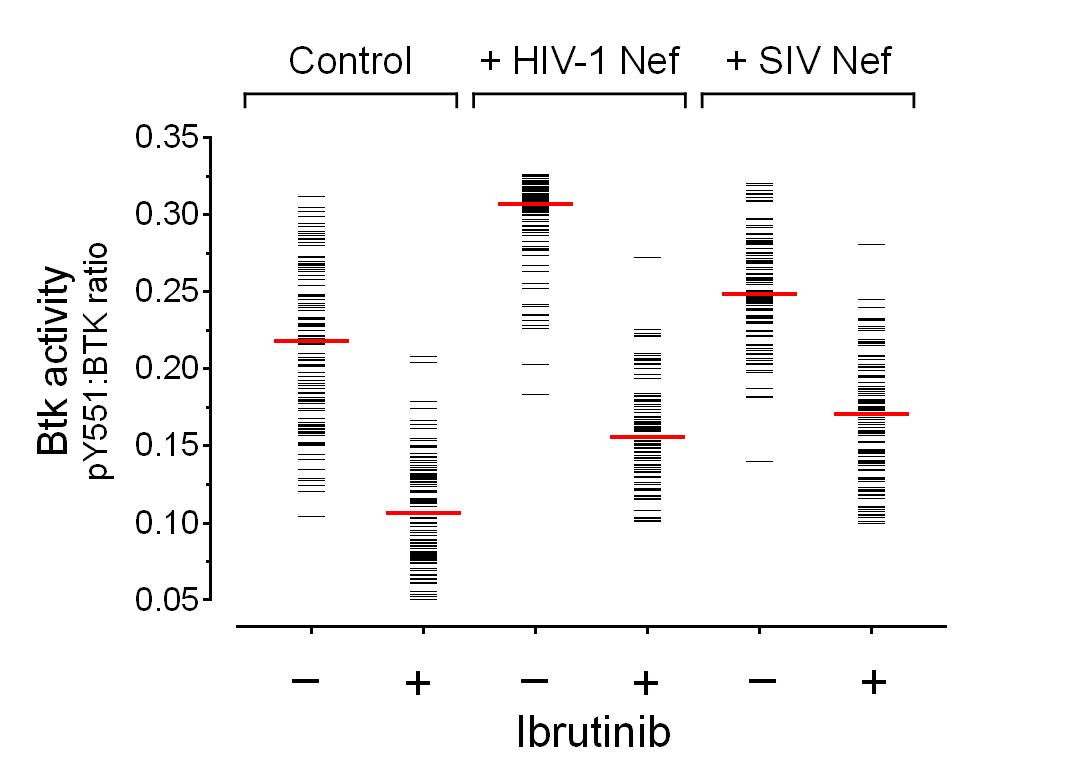
Figure 9. Image analysis of Btk autophosphorylation in the presence and absence of lentiviral Nef proteins. Mean fluorescence intensities for Btk activation loop autophosphoryla-tion (pY551) and Btk protein expression signals were determined for ≥100 cells from each condition using ImageJ and the data shown in Figure 1. The fluorescence intensity ratio (pY551: Btk expression) for each cell (or cell fragment as determined by ImageJ) is shown as a horizontal bar, with the median value indicated by the red bar. Student’s t-test shows significant increases in Btk activation loop phosphorylation in the presence of both HIV-1 and SIV Nef (P < 0.0001 in each case). When Btk is expressed alone, a wide range of pY551:Btk ratios are observed because the extent of Btk activity increases in a non-linear fashion as the amount of protein increases. When expressed with HIV-1 Nef, note that most of the black bars shift to the top of the stack, consistent with maximal activation of Btk by Nef in almost all the cells imaged. Co-expression with SIV Nef produces a more subtle, albeit statistically significant, shift in the pY551:Btk ratio. Ratios from all three populations shift downward in the presence of the Btk kinase inhibitor, ibrutinib.
Notes
Do not allow the dishes to dry out prior to imaging. Drying will seriously affect staining and create false positive artifacts.
Protect fluorophores from light to avoid bleaching by wrapping dishes in aluminum foil.
Always include positive and negative BiFC pairs as controls, especially when developing the assay for a new protein-protein interaction.
293T cells should be maintained at a low passage number to ensure cell integrity (i.e., less than 10 passages).
Keep all confocal image acquisition and ImageJ analysis settings the same throughout an experiment. Results should be reproducible when all conditions are kept constant.
When testing a new phosphospecific antibody, titrate a range of antibody concentrations during the staining procedure to identify the optimal concentration.
After staining, the cells can be imaged up to 2 days later, and as long as one week in some cases. When storing plates, protect from light and keep at 4°C.
During imaging, adjust the laser power to avoid oversaturating the signal in the image, as this may affect subsequent image analysis.
Acknowledgments
This work was funded by the National Institutes of Health grants AI152677 and AI057083 to T.E.S. These protocols were originally reported by Li et al. (2020) (doi: 10.1074/jbc.RA120.012536). The following reagent was obtained through the NIH HIV Reagent Program, Division of AIDS, NIAID, NIH: Anti-Human Immunodeficiency Virus 1 (HIV-1) Nef Monoclonal (6.2), ARP-1539, contributed by Drs. Kai Krohn and Vladimir Ovod.
Competing interests
The authors have no competing interests.
References
- Amatya, N., Lin, D. Y. and Andreotti, A. H. (2019). Dynamic regulatory features of the protein tyrosine kinases. Biochem Soc Trans 47(4): 1101-1116.
- Andreotti, A. H., Schwartzberg, P. L., Joseph, R. E. and Berg, L. J. (2010). T-cell signaling regulated by the Tec family kinase, Itk. Cold Spring Harb Perspect Biol 2(7): a002287.
- Basmaciogullari, S. and Pizzato, M. (2014). The activity of Nef on HIV-1 infectivity. Front Microbiol 5: 232.
- Briggs, S. D., Sharkey, M., Stevenson, M. and Smithgall, T. E. (1997). SH3-mediated Hck tyrosine kinase activation and fibroblast transformation by the Nef protein of HIV-1. J Biol Chem 272: 17899-17902.
- Deacon, N. J., Tsykin, A., Solomon, A., Smith, K., Ludford-Menting, M., Hooker, D. J., McPhee, D. A., Greenway, A. L., Ellett, A., Chatfield, C., Lawson, V. A., Crowe, S., Maerz, A., Sonza, S., Learmont, J., Sullivan, J. S., Cunningham, A., Dwyer, D., Dowton, D. and Mills, J. (1995). Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science 270: 988-991.
- Emert-Sedlak, L., Kodama, T., Lerner, E. C., Dai, W., Foster, C., Day, B. W., Lazo, J. S. and Smithgall, T. E. (2009). Chemical library screens targeting an HIV-1 accessory factor/host cell kinase complex identify novel antiretroviral compounds. ACS Chem Biol 4(11): 939-947.
- Emert-Sedlak, L. A., Narute, P., Shu, S. T., Poe, J. A., Shi, H., Yanamala, N., Alvarado, J. J., Lazo, J. S., Yeh, J. I., Johnston, P. A. and Smithgall, T. E. (2013). Effector Kinase Coupling Enables High-Throughput Screens for Direct HIV-1 Nef Antagonists with Antiretroviral Activity. Chem Biol 20(1): 82-91.
- Foster, J. L. and Garcia, J. V. (2008). HIV-1 Nef: at the crossroads. Retrovirology 5: 84.
- Joseph, R. E., Kleino, I., Wales, T. E., Xie, Q., Fulton, D. B., Engen, J. R., Berg, L. J. and Andreotti, A. H. (2013). Activation loop dynamics determine the different catalytic efficiencies of B cell- and T cell-specific tec kinases. Sci Signal 6(290): ra76.
- Kestler, H., Ringler, D. J., Mori, K., Panicali, D. L., Sehgal, P. K., Daniel, M. D. and Desrosiers, R. C. (1991). Importance of the nef gene for maintenance of high viral loads and for development of AIDS. Cell 65: 651-662.
- Kirchhoff, F., Greenough, T. C., Brettler, D. B., Sullivan, J. L. and Desrosiers, R. C. (1995). Absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N Engl J Med 332: 228-232.
- Li, W. F., Aryal, M., Shu, S. T. and Smithgall, T. E. (2020). HIV-1 Nef dimers short-circuit immune receptor signaling by activating Tec-family kinases at the host cell membrane. J Biol Chem 295(15): 5163-5174.
- Lin, T. A., McIntyre, K. W., Das, J., Liu, C., O'Day, K. D., Penhallow, B., Hung, C. Y., Whitney, G. S., Shuster, D. J., Yang, X., Townsend, R., Postelnek, J., Spergel, S. H., Lin, J., Moquin, R. V., Furch, J. A., Kamath, A. V., Zhang, H., Marathe, P. H., Perez-Villar, J. J., Doweyko, A., Killar, L., Dodd, J. H., Barrish, J. C., Wityak, J. and Kanner, S. B. (2004). Selective Itk inhibitors block T-cell activation and murine lung inflammation. Biochemistry 43(34): 11056-11062.
- Mujib, S., Saiyed, A., Fadel, S., Bozorgzad, A., Aidarus, N., Yue, F. Y., Benko, E., Kovacs, C., Emert-Sedlak, L., Smithgall, T. E. and Ostrowski, M. A. (2017). Pharmacologic HIV-1 Nef Blockade Enhances the Recognition and Elimination of Latently HIV-1 Infected CD4 T cells by Autologous CD8 T cells. J Clin Invest Insight 2(17): e93684.
- Nagai, T., Ibata, K., Park, E. S., Kubota, M., Mikoshiba, K. and Miyawaki, A. (2002). A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 20(1): 87-90.
- Narute, P. S. and Smithgall, T. E. (2012). Nef alleles from all major HIV-1 clades activate Src-family kinases and enhance HIV-1 replication in an inhibitor-sensitive manner. PLoS One 7(2): e32561.
- Pawlak, E. N. and Dikeakos, J. D. (2015). HIV-1 Nef: a master manipulator of the membrane trafficking machinery mediating immune evasion. Biochim Biophys Acta 1850(4): 733-741.
- Readinger, J. A., Schiralli, G. M., Jiang, J. K., Thomas, C. J., August, A., Henderson, A. J. and Schwartzberg, P. L. (2008). Selective targeting of ITK blocks multiple steps of HIV replication. Proc Natl Acad Sci U S A 105(18): 6684-6689.
- Romei, M. G. and Boxer, S. G. (2019). Split Green Fluorescent Proteins: Scope, Limitations, and Outlook. Annu Rev Biophys 48: 19-44.
- Roskoski, R., Jr. (2016). Ibrutinib inhibition of Bruton protein-tyrosine kinase (BTK) in the treatment of B cell neoplasms. Pharmacol Res 113(Pt A): 395-408.
- Schneider, C. A., Rasband, W. S. and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9(7): 671-675.
- Staudt, R. P., Alvarado, J. J., Emert-Sedlak, L. A., Shi, H., Shu, S. T., Wales, T. E., Engen, J. R. and Smithgall, T. E. (2020). Structure, function, and inhibitor targeting of HIV-1 Nef-effector kinase complexes. J Biol Chem 295 (44): 15158-15171.
- Tarafdar, S., Poe, J. A. and Smithgall, T. E. (2014). The Accessory Factor Nef Links HIV-1 to Tec/Btk Kinases in an Src Homology 3 Domain-dependent Manner. J Biol Chem 289(22): 15718-15728.
- Tebo, A. G. and Gautier, A. (2019). A split fluorescent reporter with rapid and reversible complementation. Nat Commun 10(1): 2822.
- Trible, R. P., Emert-Sedlak, L. and Smithgall, T. E. (2006). HIV-1 Nef selectively activates SRC family kinases HCK, LYN, and c-SRC through direct SH3 domain interaction. J Biol Chem 281: 27029-27038.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Shu, S. T., Li, W. F. and Smithgall, T. E. (2021). Visualization of Host Cell Kinase Activation by Viral Proteins Using GFP Fluorescence Complementation and Immunofluorescence Microscopy. Bio-protocol 11(13): e4068. DOI: 10.21769/BioProtoc.4068.
- Li, W. F., Aryal, M., Shu, S. T. and Smithgall, T. E. (2020). HIV-1 Nef dimers short-circuit immune receptor signaling by activating Tec-family kinases at the host cell membrane. J Biol Chem 295(15): 5163-5174.
Category
Biochemistry > Protein > Imaging
Microbiology > Microbe-host interactions > Virus
Microbiology > Microbial signaling > Phosphorylation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








