- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
FACS Enrichment of Total Interstitial Cells and Fibroblasts from Adult Mouse Ventricles
(*contributed equally to this work) Published: Vol 11, Iss 10, May 20, 2021 DOI: 10.21769/BioProtoc.4028 Views: 5109
Reviewed by: Dongsheng JiangPhilipp WörsdörferAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Selective Enrichment and Identification of Cerebrospinal Fluid-Contacting Neurons In Vitro via PKD2L1 Promoter-Driven Lentiviral System
Wei Tan [...] Qing Li
Nov 20, 2025 1337 Views

Revisiting Primary Microglia Isolation Protocol: An Improved Method for Microglia Extraction
Jianwei Li [...] Guohui Lu
Dec 5, 2025 1466 Views

FLARE: A Flow Cytometry–Based Fluorescent Assay for Measuring HSV-1 Nuclear Egress
Bing Dai [...] Ekaterina E. Heldwein
Jan 5, 2026 591 Views
Abstract
Besides cardiomyocytes, the heart contains numerous interstitial cell types, including cardiac fibroblasts, endothelial cells, immune (myeloid and lymphoid) cells, and mural cells (pericytes and vascular smooth muscle cells), which play key roles in heart repair, regeneration, and disease. We recently published a comprehensive map of cardiac stromal cell heterogeneity and flux in healthy and infarcted hearts using single-cell RNA sequencing (scRNA-seq) (Farbehi et al., 2019). Here, we describe the FACS (Fluorescent Activated Cell Sorting)-based method used in that study for isolation of two cardiac cell fractions from adult mouse ventricles: the total interstitial cell population (TIP; non-cardiomyocytes) and enriched (Pdgfra-GFP+) cardiac fibroblasts.
Keywords: Adult mouse heartBackground
Cardiovascular disease, including myocardial infarction (MI), is the leading cause of death worldwide. As adult mammalian hearts have a very limited capacity to repair damaged myocardium following injury, considerable efforts have been made to augment endogenous repair processes and stimulate heart regeneration (Tzahor and Poss, 2017). However, the structural and cellular complexity of the heart presents challenges to obtaining a complete mechanistic understanding of its functional and repair processes.
Cardiomyocytes are the primary functional cell type in the heart; however, they coexist within a complex support network of interstitial cells, which include fibroblasts, vascular, immune, and nerve cells. Cardiac interstitial cells constitute 60%-70% of total hearth cells and have essential regulatory functions in tissue homeostasis and repair (Frangogiannis, 2019). Ischaemic injuries, such as MI, induce a complex cascade of molecular and cellular events to clear necrotic cardiomyocytes and replace the affected area with a fibrotic scar (Tallquist and Molkentin, 2017). Thus, characterizing the cellular heterogeneity of the heart and its flux during the post-injury stages of inflammation and repair is necessary to both advance insights into disease mechanisms and facilitate the discovery of novel therapeutic targets.
Recent advances in single-cell technologies have enabled the systematic investigation of cellular heterogeneity in a wide range of tissues, yielding fresh insights into regulatory mechanisms underpinning cell states in development and disease (Marioni and Arendt, 2017). A comprehensive understanding of cellular complexity in the heart using single-cell methods requires developing reliable and efficient protocols to isolate the heavily interconnected and heterogeneous cardiac cell types in a relatively intact and healthy state. Here, we present a mechanical and enzymatic digestion protocol that relies on FACS to generate large numbers of high-quality, viable cardiac interstitial cells, as well as enriched fibroblasts, using lineage tracing of GFP+ cells from the ventricles of Pdgfra+/GFP mice. For details on the use and execution of this protocol, please refer to Chong et al., 2011 and Farbehi et al., 2019.
Materials and Reagents
DNA LoBind 1.5 ml tubes (Eppendorf, catalog number: 022431005)
60 mm cell culture dish (Corning, catalog number: CLS430166)
100 mm cell culture dish (Corning, catalog number: CLS430167)
50 ml and 15 ml Falcon conical centrifuge tubes (Thermo Fisher Scientific, catalog numbers: 14-959-53A; 14-432-22)
5 ml and 10 ml Stripette serological pipettes (Corning, catalog numbers: CLS4487 and CLS4488)
Sterile plastic transfer pipettes (Thermo Scientific Samco, catalog number: SAM-336-1S)
10 μl, 200 μl and 1,000 μl ART pipette tips (Sigma-Aldrich, catalog numbers: A2598, A3098, and A3948)
5 ml round bottom FACS tubes (Corning, catalog number: 352052)
LS columns (Miltenyi Biotec, catalog number: 130-042-401)
Surgical scalpel blades No. 22 (Swann-Morton, catalog number: 0208)
0.2 μm syringe filters (Thermo Fisher Scientific, catalog number: NC9103939)
40 μm cell strainers (Corning, catalog number: CLS431750)
BD ultra-fine insulin syringe (Becton Dickinson, catalog number: 326719)
10 ml and 50 ml BD syringes (Becton Dickinson, catalog numbers: 305482 and 309653)
Cannula (blunt-end 22-G needle) (Becton Dickinson, catalog number: 305156)
Silk suture (Ethicon, catalog number: 768G)
70 ml specimen Jar (Sarstedt, catalog number: SAR00002)
Dead cell removal Kit (Miltenyi Biotec, catalog number: 130-090-101)
C57BL/6J mice (The Jackson Laboratory, JAX: 000664)
PdgfraGFP/+ mice (The Jackson Laboratory, MGI: 2663656)
Bovine Serum Albumin (BSA) (Sigma-Aldrich, catalog number: A2153)
1× Phosphate Buffered Saline (PBS), pH 7.4 (Thermo Fisher Scientific, catalog number: 10010023)
Fetal Bovine Serum (FBS) (Thermo Fisher Scientific, catalog number: 10100147)
Red blood cell (RBC) lysis buffer (Sigma-Aldrich, catalog number: R7757)
Collagenase type II (Worthington Biochemical Corporation, catalog number: LS004177)
DAPI (4',6-diamidino-2-phenylindole, dihydrochloride) (Thermo Fisher Scientific, catalog number: D1306)
APC-coupled anti-PDGFRA (CD140a) antibody (Thermo Fisher Scientific, catalog number: 17-1401-81)
Heparin (Pfizer Australia Pty Ltd, catalog number: AUST R 49232)
Ketamine (Provet (NSW) Pty Ltd)
Xylazine (Provet (NSW) Pty Ltd)
FACS buffer (see Recipes)
Collagenase type II in PBS (see Recipes)
Equipment
Timer (Thermo Fisher Scientific, catalog number: 06664251)
Hemocytometer (NEUBAUER, catalog number: HL-8100204 I)
Fine surgical tools:
Surgical scissors, Sharp (Fine Science Tools, catalog number: 14002-14)
Dumont Forceps, Micro-Blunted Tips (Fine Science Tools, catalog number: 11253-25)
Spring Scissors, 8 mm Cutting Edge (Fine Science Tools, catalog number: 15024-10)
Graefe Forceps (Fine Science Tools, catalog number: 11051-10)
Extra Fine Graefe Forceps (Fine Science Tools, catalog number: 11150-10)
Delicate Suture Tying Forceps (Fine Science Tools, catalog number: 11063-07)
Leica DFC295 digital microscope
Refrigerated centrifuge (Thermo Fisher Scientific, catalog number: 75004270)
Refrigerated microcentrifuge (Thermo Fisher Scientific, catalog number: 75002402)
FACS Aria II (5 lasers, 100 μm nozzle) (BD Bioscience, catalog number: SCR_018091)
QuadroMACS separator (Miltenyi Biotec, catalog number: 130-090-976)
Shaker water bath (Analytical Instruments, catalog number: 7746-22220)
Procedure
Before you start
Set all centrifuges to 4°C.
Set the water bath to 37°C and adjust the water level to cover half of the specimen jar.
Place the collagenase type II stock bottle in a desiccator at room temperate (RT) for at least 30 min before opening.
Note: Collagenase II is supplied as a lyophilized powder and is very hygroscopic. The hygroscopicity significantly affects the enzymatic activity if the powder is exposed to moisture. Thus, desiccation of the stock bottle helps ensure the long-term storage of the collagenase II stock. The stock bottle should not be opened in a humid environment.
Pre-warm the red cell lysis buffer to RT.
Note: Perform all steps on ice unless otherwise indicated. A simplified workflow of this protocol is illustrated in Figure 1A-1J.Method
Inject the PdgfraGFP/+ mouse with heparin (200 IU/mouse) to prevent microthrombus formation in the coronary circulation.
Note: In the original eLife paper (Farbehi et al., 2019), experimental mice were sacrificed by cervical dislocation without heparin administration, and the heart cavity was flushed manually after excision. Here, we present a revised method in which mice are heparinized and the heart removed, allowing flushing of the coronary arteries by ex vivo retrograde perfusion to minimize contamination of blood cells. In this method, euthanasia occurs by exsanguination under anesthesia.
Ten minutes after heparin injection, anesthetize the mouse with an intraperitoneal injection of ketamine (100 mg/kg body weight) and xylazine (20 mg/kg body weight).
After sedation, check for the lack of pain using the toe-pinch reflex before proceeding to the thoracotomy.
Incise the skin along the mid-sternal axis using fine surgical scissors and expose the rib cage. Cut the rib cage and separate the ribs to expose the thoracic cavity. Using fine forceps and surgical scissors, free the heart and ascending aorta from connective tissue. Cut the aorta 2-3 mm from the aortic valve, freeing the heart from the thoracic cage (Figure 1A).
Immediately place the heart in a 100 mm Petri dish containing cold PBS.
Trim excess soft tissue from the aorta. Cannulate the aorta and secure with silk suture as previously documented in video form Li et al. (2014) (Figure 1B). Make an incision in the right atrium to permit coronary sinus effluent to flow freely.
Attach a 10-ml syringe filled with PBS to the cannula, taking care not to introduce any air bubbles.
Initiate perfusion by gently pressing the syringe plunger, maintaining a steady flow rate until the heart is cleared of blood (the fluid should be running clear).
Note: Alternative cardiac perfusion methods, e.g., Langendorff perfusion or whole body in situ perfusion, extend the protocol time significantly and may cause additional ischemic stress.
After perfusion, remove the cannula and transfer the heart to a new 100 mm Petri dish containing PBS. Remove atria, great vessels, and top one-quarter of ventricles to avoid contaminating the samples with atrioventricular mesenchymal tissue (Figure 1C).
Transfer the heart to a 60 mm Petri dish and mince it with a scalpel to obtain ~1-3 mm3 pieces. Cover the tissue in a small volume of collagenase solution (~500 µl) to keep it moist during mincing.
Note: This step should be performed within a 5 min timeframe. Small pieces of tissue (~1-3 mm3) should be visible (Figure 1D). Excessive mincing generates a cell slurry which will negatively affect cell viability.
Add 5 ml of collagenase solution to the dish and transfer the minced tissue into a 70 ml specimen jar using a plastic transfer pipette. Mix by gently pipetting the sample up and down to break down the clumps.
Incubate the specimen jar at 37°C in a water bath with shaking (60 rpm) for 10 min (Figure 1E). Break down clumps by intermittent pipetting while avoiding bubble formation.
After 10 min of incubation, remove the sample from the water bath and let the pieces settle down to the bottom of the specimen jar. Transfer the supernatant (cell suspension) to a 50 ml Falcon tube by filtering through a 40 μm sterile cell strainer to remove cardiomyocytes and undigested debris (size of adult mouse cardiomyocytes ranges 100-200 μm) (Figure 1F).
Add fresh 5 ml collagenase solution to the remaining undigested tissue in the specimen jar, mix by gently pipetting up and down, and repeat Steps B12-B13 further two times further. Pool the cell suspension from each heart in the same designated 50 ml Falcon tube. Centrifuge the cell suspension at 300 × g for 5 min and discard the supernatant.
Note: At this stage, most of the tissue should be completely digested; however, if larger pieces are visible, the incubation period should be extended to ~15 min, and/or another round of digestion with 5 ml collagenase solution should be done.
Resuspend the pellet in 1 ml of red cell lysis buffer and incubate at RT for 1 min (Figure 1G).
Centrifuge the tube at 300 × g for 5 min and discard the supernatant.
Gently resuspend the pellet in 1 ml of ice-cold FACS buffer and transfer the contents into 1.5 ml Eppendorf tubes. Remove the supernatant carefully by aspiration without disturbing the cell pellet.
Centrifuge the tube at 300 × g for 5 min after washing, discard supernatant, and resuspend the pellet in 200 µl of dead cell removal microbeads; mix well and incubate for 15 min at RT (Figure 1H).
During the incubation, prepare 15 ml (per heart) of 1× column binding buffer from the 20× stock solution (included in the dead cell removal kit), place MACS LS columns in the magnetic field of a QuadroMACS Separator, and precondition the column with 3 ml of 1× binding buffer (Figure 1I). Discard the flow-through.
Load the cell suspension into the LS column and slowly add the remaining 12 ml of 1× binding buffer to each column, collecting the flow-through into a 15 ml tube placed on ice. The flow-through contains live cells.
Centrifuge the tubes at 300 × g for 5 min. After discarding the supernatant, resuspend the pellet in 1 ml of ice-cold FACS buffer and transfer the contents into a 1.5 ml Eppendorf tube.
Centrifuge at 300 × g for 5 min and resuspend the pellet in 500 µl of ice-cold FACS buffer.
Transfer 100 µl of the cell suspension into a 5 ml FACS tube and dilute with ice-cold FACS buffer to achieve a cell density of approximately 1 million cells per ml. Add DAPI (final concentration: 100 ng/ml) to the cell suspension and proceed to FACS sorting (Figure 1J).
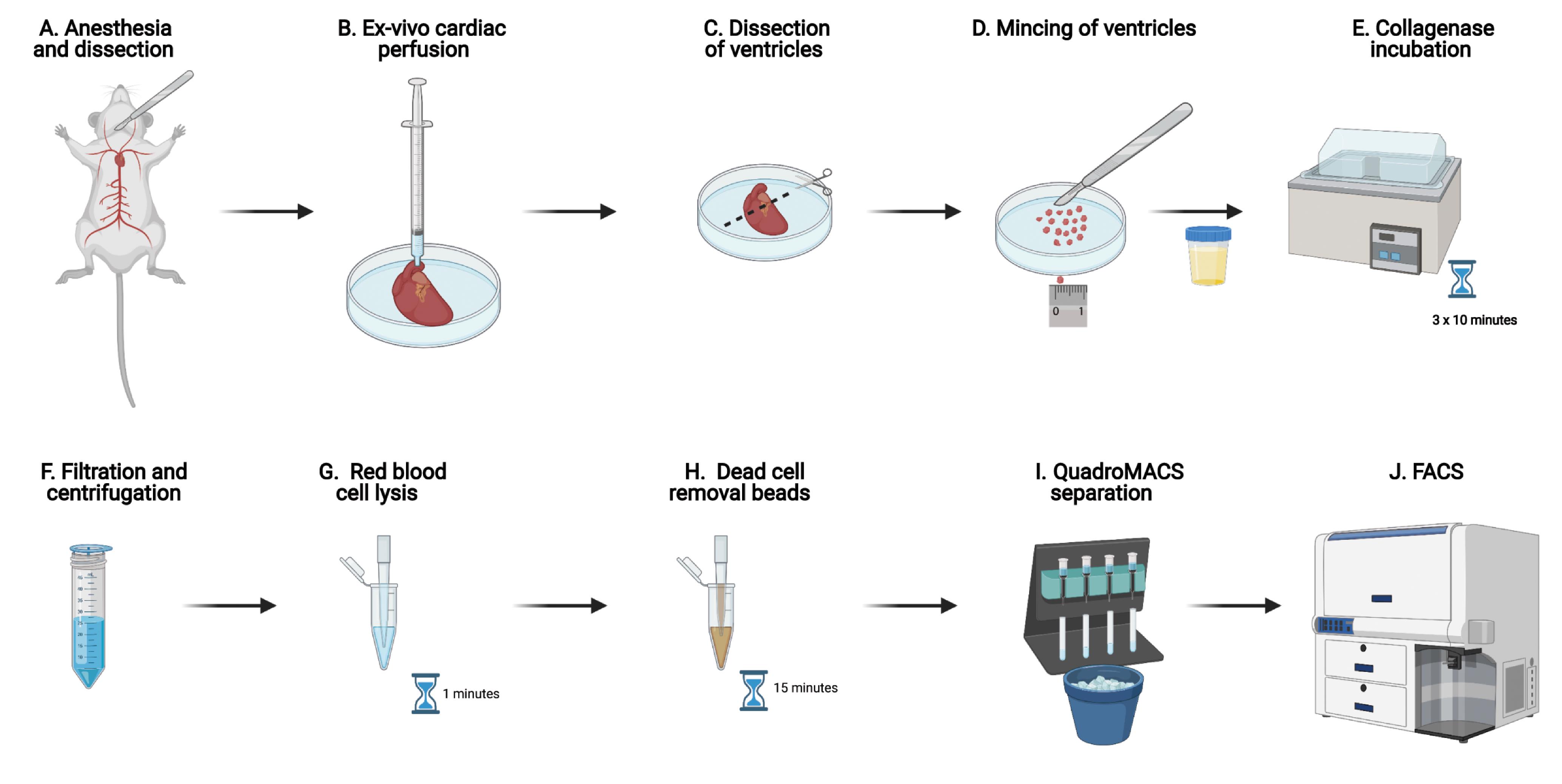
Figure 1. Experimental workflow in graphical formAcquire cells on the BD FACS Aria II sorter fitted with a 100 μm nozzle (30 kHz, 20 PSI). Adjust the flow rate to achieve ~1,000 events/second. After establishing the gates (see below), sort TIP cells (Figure 1D) with 4-way purity precision mode in a 1.5 ml Eppendorf tube containing 50 μl FACS buffer. Once the desired number of TIP cells are sorted, sort GFP+ cells (Figure 1E).
Gating strategy:
Set FSC-A vs SSC-A gate to include TIP cells and exclude the remaining cardiomyocytes, debris, and cell aggregates (Figure 2A).
Gate single cells and exclude cell aggregates based on FSC gating (Figure 2B).
Gate single cells and exclude cell aggregates based on SSC gating (Figure 2C).
Gate the DAPI- fraction to sort for “Live TIP” cells (Figure 2D).
From the “Live TIP” fraction, gate the GFP+ fraction to sort cardiac fibroblasts (Figure 2E) (Chong et al., 2011).
Notes:
Both unstained and single-stained samples should be prepared from the same cell suspension obtained from wild-type (C57BL/6J) mouse hearts. This will adjust the compensation parameter settings to set background fluorescence levels and establish the gate for GFP+ cells.
If PdgfraGFP/+ mice are not available, a cell suspension obtained from wild-type (C57BL/6J) mouse hearts (at Step B22) should be stained with a fluorophore-conjugated anti-PDGFRA antibody (e.g., APC-conjugated anti-PDGFRA (CD140a) antibody) to sort cardiac fibroblasts as the “APC” positive fraction from the “Live TIP” fraction. TIP cells can be sorted as explained above for PdgfraGFP/+ mice (Step B24 and Figures 2A-2D).
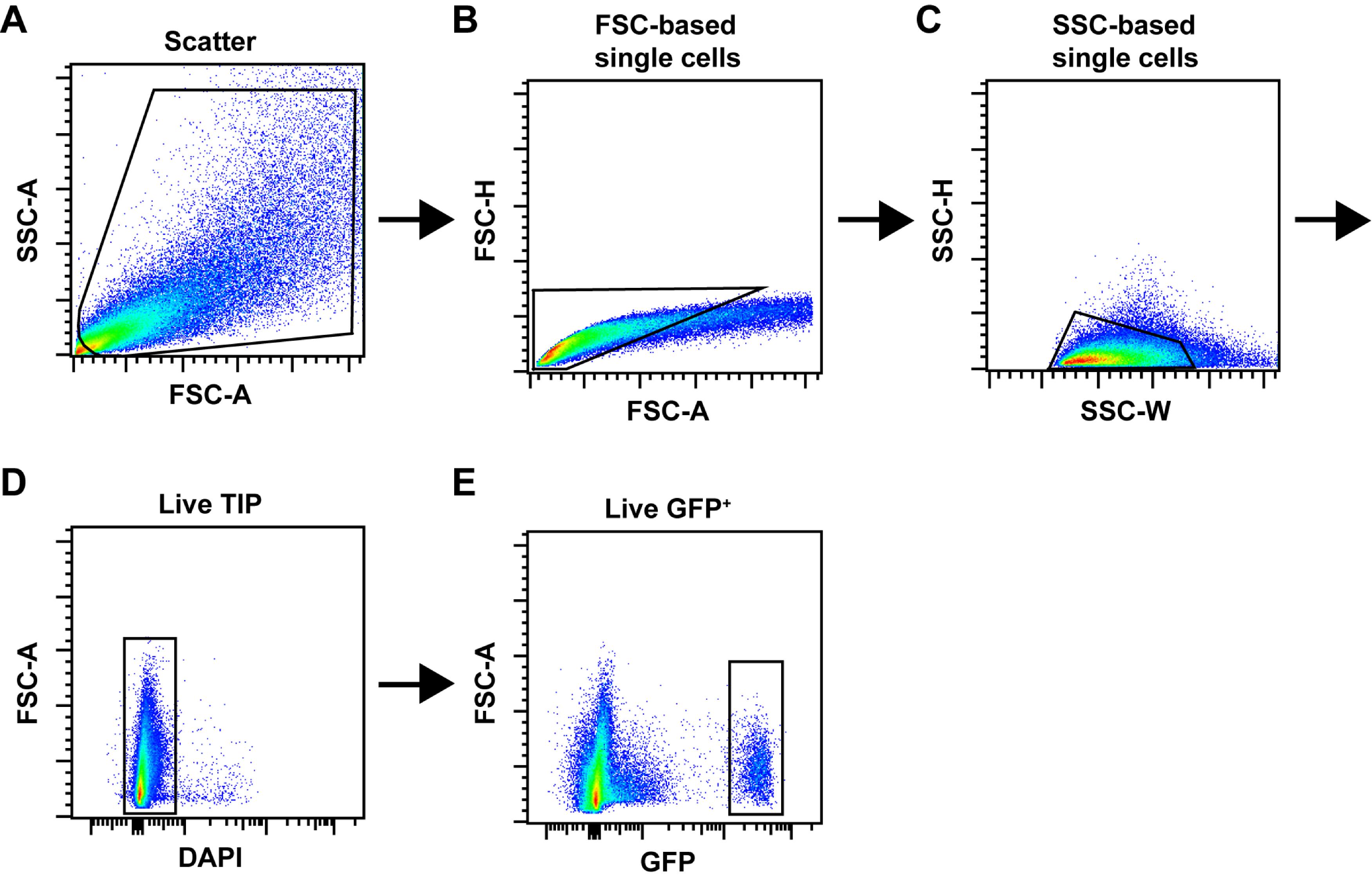
Figure 2. FACS gating strategy. (A-C) A typical workflow of sequential gating for doublet exclusion is shown. Total events were gated in a scatter plot (A) and subjected to FSC-based (B) and SSC-based (C) single-cell gating. (D) Live TIP cells were then gated as the DAPI- fraction. (E) Example of gating for GFP+ cells isolated from PdgfraGFP/+ mice.
Recipes
FACS buffer
Prepare 2% FBS in PBS and filter through a 0.2 µm syringe filter.
Collagenase type II in PBS (working concentration: 265 Unit/ml)
Prepare fresh solution each time. Dissolve collagenase powder in PBS and filter through a 0.2 µm syringe filter. Prepare 15 ml of collagenase solution for each heart sample.
Note: The enzymatic activity per unit mass (Unit/mg) of the lyophilized powder and weigh as required (Unit/ml) considering working concentration and number of samples.
Acknowledgments
This work was supported by funding from the National Health and Medical Research Council (1118576, 573707, 1105271, 1074286), Leducq Foundation (13CVD01, 15CVD03), Stem Cells Australia (SR110001002), St. Vincent’s Clinic Foundation (100711), Victor Chang Cardiac Research Institute and UNSW Sydney.
Competing interests
The authors declare no conflict of interest.
Ethics
Animal experiments were performed following the guidelines and with the approval of the Garvan Institute of Medical Research/St. Vincent’s Animal Ethics Committee, research approvals 16/03 and 16/10 (Validity period: 2016-2019).
References
- Chong, J. J., Chandrakanthan, V., Xaymardan, M., Asli, N. S., Li, J., Ahmed, I., Heffernan, C., Menon, M. K., Scarlett, C. J., Rashidianfar, A., Biben, C., Zoellner, H., Colvin, E. K., Pimanda, J. E., Biankin, A. V., Zhou, B., Pu, W. T., Prall, O. W. and Harvey, R. P. (2011). Adult cardiac-resident MSC-like stem cells with a proepicardial origin. Cell Stem Cell 9(6): 527-540.
- Farbehi, N., Patrick, R., Dorison, A., Xaymardan, M., Janbandhu, V., Wystub-Lis, K., Ho, J. W., Nordon, R. E. and Harvey, R. P. (2019). Single-cell expression profiling reveals dynamic flux of cardiac stromal, vascular and immune cells in health and injury. Elife 8: e43882.
- Frangogiannis, N. G. (2019). Cardiac fibrosis: Cell biological mechanisms, molecular pathways and therapeutic opportunities. Mol Aspects Med 65: 70-99.
- Li, D., Wu, J., Bai, Y., Zhao, X. and Liu, L. (2014). Isolation and culture of adult mouse cardiomyocytes for cell signaling and in vitro cardiac hypertrophy. J Vis Exp(87): 51357.
- Marioni, J. C., and Arendt, D. (2017). How Single-Cell Genomics Is Changing Evolutionary and Developmental Biology. Annu Rev Cell Dev Biol 33: 537-553.
- Tallquist, M. D. and Molkentin, J. D. (2017). Redefining the identity of cardiac fibroblasts. Nat Rev Cardiol 14(8): 484-491.
- Tzahor, E. and Poss, K. D. (2017). Cardiac regeneration strategies: Staying young at heart. Science 356(6342): 1035-1039.
Article Information
Copyright
Farbehi et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Farbehi, N., Janbandhu, V., Nordon, R. E. and Harvey, R. P. (2021). FACS Enrichment of Total Interstitial Cells and Fibroblasts from Adult Mouse Ventricles. Bio-protocol 11(10): e4028. DOI: 10.21769/BioProtoc.4028.
- Farbehi, N., Patrick, R., Dorison, A., Xaymardan, M., Janbandhu, V., Wystub-Lis, K., Ho, J. W., Nordon, R. E. and Harvey, R. P. (2019). Single-cell expression profiling reveals dynamic flux of cardiac stromal, vascular and immune cells in health and injury. Elife 8: e43882.
Category
Cell Biology > Cell isolation and culture > Cell isolation
Cell Biology > Cell-based analysis > Flow cytometry
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











