- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A New Method for Studying RNA-binding Proteins on Specific RNAs
(*contributed equally to this work) Published: Vol 11, Iss 10, May 20, 2021 DOI: 10.21769/BioProtoc.4022 Views: 10582
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Normalized Ribo-Seq for Quantifying Absolute Global and Specific Changes in Translation
Katharina Hoerth [...] Johanna Schott
Feb 20, 2022 5211 Views

Preparation of Cardiac Extracts from Embryonal Hearts to Capture RNA–protein Interactions by CLIP
Giulia Buonaiuto [...] Monica Ballarino
Oct 20, 2023 2184 Views

Flow Cytometry Analysis of Microglial Phenotypes in the Murine Brain During Aging and Disease
Jillian E. J. Cox [...] Sarah R. Ocañas
Jun 20, 2024 3568 Views
Abstract
Proximity-based protein labeling has been developed to identify protein-nucleic acid interactions. We have reported a novel method termed CRUIS (CRISPR-based RNA-United Interacting System), which captures RNA-protein interactions in living cells by combining the RNA-binding capacity of CRISPR/Cas13 and the proximity-tagging activity of PUP-IT. Enzymatically deactivated Cas13a (dCas13a) is fused to the proximity labeling enzyme PafA. In the presence of a guide RNA, dCas13a binds specific target RNA region, while the fused PafA mediates the labeling of biotin-tagged Pup on proximal proteins. The labeled proteins can be enriched by streptavidin pull-down and identified by mass spectrometry. Here we describe the general procedure for capturing RNA-protein interactions using this method.
Keywords: Proximity-based protein labelingBackground
RNA-binding proteins (RBPs) play important roles in regulating cellular processes and function. Mapping RNA-protein interactions is a key endeavor to address how proteins regulate the location and function of target RNAs. The traditional methods are mostly protein-centric methods, including RNA immunoprecipitation (RIP) and crosslinking and immunoprecipitation (CLIP). They rely on specific antibodies to immunoprecipitate RNA-protein complexes and identify the RNAs by sequencing. Although some RNA-centric methods have been developed, they focused on systematic labeling of all RNAs, rather than one specific RNA, for the purification of RNA-protein complexes (Chu et al., 2015; McHugh et al., 2015). Therefore, there is an urgent need to develop a method that can be widely used to study RNA-protein interactions of the specific RNA in living cells. Since the CRISPR-based RNA-targeting Cas nucleases have been reported, it provides us a new powerful tool to bind and cleave specific RNAs to regulate RNA function (Abudayyeh et al., 2016; Cox et al., 2017). In addition, the enzymatically deactivated Cas13a provides a new strategy for tracking the specific RNA. The proximity labeling system PUP-IT has been widely used to identify protein-protein interactions. It employs the proximity ligase PafA to mediate the ligation of a small protein (PupE) to lysines on the surrounding proteins (Liu et al., 2018). Combining PUP-IT with the RNA-targeting Cas nucleases (Figure 1), we have developed a new class of RNA-centric methods for studying RNA-protein interactions in living cells (Zhang et al., 2020).
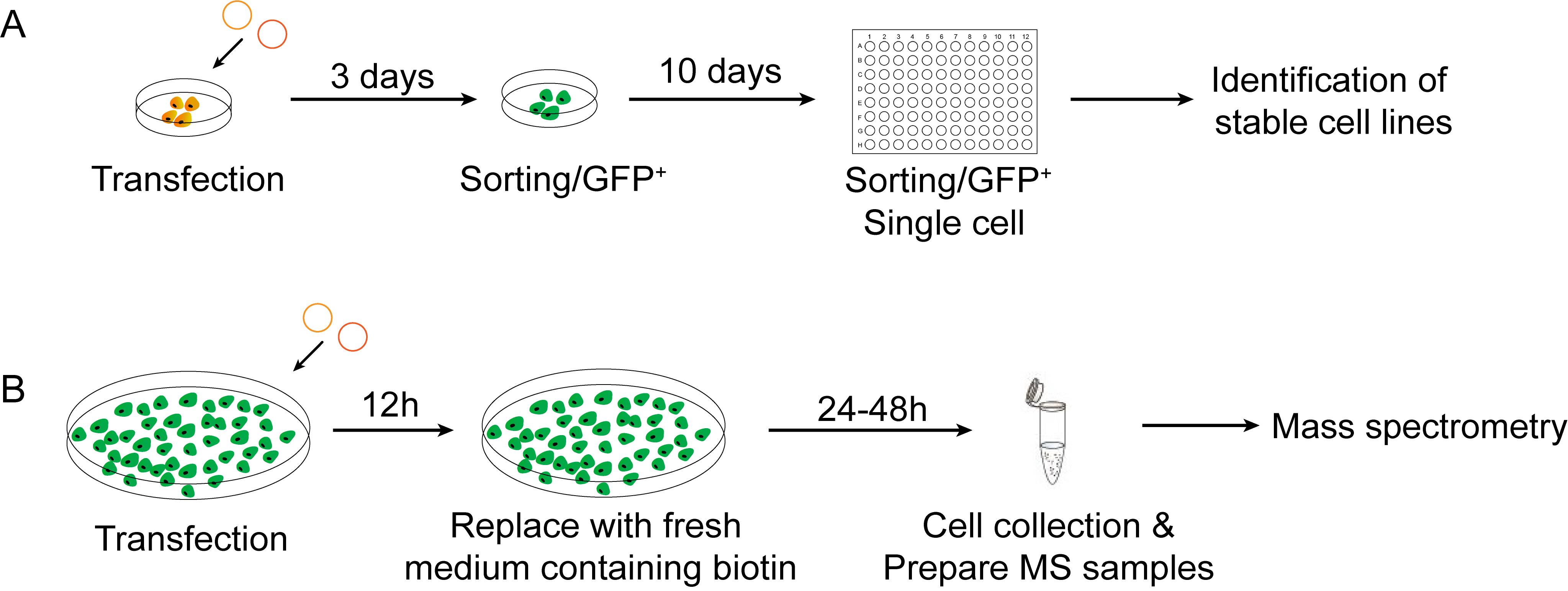
Figure 1. The workflow of capturing RNA-protein interactions. A. The workflow for isolating HEK293T cell line stably expressing CRUIS. B. The workflow of mass spectrometry sample preparation.
Materials and Reagents
Pipette tips (Filtered) (Thermo, catalog numbers: TF112-1000-Q, T104RS-Q, TF140-200-Q)
0.2 ml PCR tubes (Thermo, catalog number: 431-MIXED-Q)
1.5 ml Microtubes (AXYGEN, catalog number: MCT-150-L-C)
50 ml Tubes (Falcon, catalog number: 352098)
96-well plates (Corning, catalog number: 3599)
24-well plates (Corning, catalog number: 3524-ND)
6-well plates (Corning, catalog number: 3516)
10 cm dishes (Corning, catalog number: 430167)
pC0040-LwaCas13a crRNA backbone (Addgene, catalog number: 103851)
pB-CAGGS-dCas9 (Addgene, catalog number: 110823)
Super PiggyBac Transposase (SBI, catalog number: PB210PA-1)
DMEM (Life, catalog number: C11995500CP)
FBS (Gemini, catalog number: 100-307)
Penicillin-Streptomycin (Life, catalog number: 15140122)
Opti-MEM (Gibco, catalog number: 31985062)
PEI (Polysciences, catalog number: 23966)
PBS (Life, catalog number: C20012500CP)
0.25% Trypsin-EDTA (Life, catalog number: 25200072)
Biotin (SCRC, catalog number: 67000260)
Triton® X-100 (Sangon Biotech, catalog number: T0694-100 ml)
1 M Tris pH 7.5, sterile (Sangon Biotech, catalog number: B548127-0500)
100× protease inhibitor cocktail (Biotool, catalog number: B14001)
Urea (Sangon Biotech, catalog number: UT0907)
DTT (MDBio, catalog number: D023-5G)
Iodoacetamide (SIGMA, catalog number: I1149-5G)
Streptavidin magnetic beads (PIERCE, catalog number: 88816)
1 M Tris pH 8.0, sterile (Sangon Biotech, catalog number: B548127-0500)
NaCl (Sangon Biotech, catalog number: A501218-0001)
SDS (MDBio, catalog number: S001-100)
EDTA (Sangon Biotech, catalog number: E0105-500g)
Ammonium bicarbonate reagent plus(R) (Sigma, catalog number: A6141-500G)
RapiGest SF (Waters, catalog number: 186008090)
Sequencing Grade Modified Trypsin (Promega, catalog number: V5113)
Formic acid (Sigma, catalog number: 94318-250ML)
Acetonitrile (Merck Chemicals, catalog number: 1.00030.4008)
Trifluoroacetic acid LC-MC Ultra (Sigma, catalog number: 14264-50ML)
Amino acid sequence information of CRUIS and Bio-PupE are available at https://doi.org/10.1093/nar/gkaa143
ZIPTIP C18 (Millipore,catalog number: ZTC18S960)
Anti-myc antibody (Cell Signaling, catalog number: 2276s)
Goat anti-mouse IgG Antibody (H&L) [HRP] (GenScript, catalog number: A00160)
Streptavidin-HRP (Cell Signaling, catalog number: 3999s)
BSA (AMRESCO, catalog number: 0903-5G)
SurePAGE (GenScript, catalog number: M00657)
PVDF Western blotting membranes (Roche, catalog number: 30100400)
BbsI-HF (NEB, catalog number: R3539)
Lysis buffer (see Recipes)
Buffer 1 (see Recipes)
Buffer 2 (see Recipes)
Buffer 3 (see Recipes)
Buffer 4 (see Recipes)
Equipment
Standard molecular biology lab equipment
Eppendorf mixer (Eppendorf, catalog number: 5382000074)
Magnetic separator(Bimake, catalog number: B23803)
-80°C freezer (Therom, catalog number: 905)
FACSAriaTM III (BD)
Trans-blot turbo (Biorad, catalog number: 1704150)
Chemiluminescence imaging system (GE, Amersham Imager 600)
Vacuum concentrator (Thermo, SpeedVac)
Spectrometer (Thermo, Orbitrap Fusion)
Procedure
Molecular cloning: design and assembly of sgRNA expression vector for target gene, subclone CRUIS into the plasmid for generating the stable cell line
For the sgRNA expression vectorDesign sgRNAs for the target gene. We recommend design with CRISPR-RT. The parameters can be set as the length of the target complementarity region of crRNA at 28 nt, the length of the seed region at 10 nt (default parameters of CRISPR-RT). The system will give a series of crRNAs, if you are interested in a specific region of the target RNA, you can choose the crRNA targeted to the corresponding position. For more information and guidelines, please follow the link to CRISPR-RT (http://bioinfolab.miamioh.edu/CRISPR-RT/interface/C2c2.php) (Zhu et al., 2018).
The sgRNA expression plasmids cloned by inserting annealed oligos into the U6 promoter-based expression vector between LwaCas13a-DR and poly-T that digested by BbsI (For the forward oligo should add AAAC in the 5’, reverse oligo should add AAAA at 5’ as an overhand to insert the LwaCas13a crRNA backbone digested by BbsI). For cloning the gRNAs into the plasmid by annealed oligo cloning, we recommend the protocol provided by Addgene as a reference (http://www.addgene.org/protocols/annealed-oligo-cloning).
Note: Any plasmid with LwaCas13a crRNA is suitable, here we take pC0040-LwaCas13a crRNA backbone (Addgene #103851) as an example.
For the plasmid used to construct stable cell line
Clone CRUIS-P2A-EGFP into the backbone of pB-CAGGS-dCas9 vector between the restriction enzyme sites NheI and HpaI.
Notes:
Any PiggyBac expression vector is suitable, here we take pB-CAGGS-dCas9 (Addgene #110823) as an example, the sequence information of CRUIS-P2A-EGFP is available at https://doi.org/10.1093/nar/gkaa143.
In order to facilitate the following description, we use CRUIS, which is the method’s name to represent the dCas13-PafA fusion gene; and use 293T-CRUIS to represent the 293T cells stably expressing dCas13-PafA.
Preparation of endotoxin-free plasmids for subsequent transfection with PEI.
Generation of 293T-CRUIS stable cell line
Note: Any method that can be used to construct a stable cell line is suitable. We prefer to recommend the lentivirus transfection system. The size of the CRUIS is about 5k bp, which is close to the upper limit of lentiviral vectors. If you need to introduce some selection marker, it is recommended to apply the PiggyBac system.
Culture 293T cells in DMEM with 10% FBS at 37°C in 5% CO2.
The day before transfection, seed 5 × 105 cells/well in one well of a 6-well dish with 2.5 ml medium.
Co-transfect 293T cells with PB-CRUIS-P2A-EGFP and Super PiggyBac Transposase (SBI: PB210PA-1) by PEI follow Steps B4-B6 when cells grow to 70% cell confluency.
Dilute 1.5 μg PB-CRUIS-P2A-EGFP plasmids and 1.5 μg Super PiggyBac Transposase plasmids in 125 μl Opti-MEM, dilute 7.5 μl of 1 mg/ml PEI in 125 μl Opti-MEM.
Mix the diluted plasmids and PEI in a 1.5 ml tube, incubate the mixture at room temperature for 20 min.
Add PEI-DNA complex to the cells dropwise.
Three days after transfection, EGFP positive cells were sorted by flow cytometry for continuous culture.
Note: This step is to sort out the successfully transfected cells; For the sorting, wild-type 293T cells were used as control.
Ten days after transfection, EGFP positive cells were sorted into 96-well plates with 1 cell/well for single clone selection.
Note: The purpose of this step is to sort out the cells with CRUIS-P2A-EGFP stable expression.
Three weeks after sorting, 10 EGFP positive clones were selected for further amplification.
Western Blot is used to test the expression of CRUIS in each clone. 1 million cells were harvested, washed with cold PBS, and lysed in 200 μl lysis buffer supplemented with 1× protease inhibitor for 1 h on ice. After centrifugation at 13,000 rcf, 150 μl supernatant was mixed with 30 μl 6× protein loading buffer and denatured at 100°C for 10 min. 20 μl sample was loaded on 4-20% SDS-PAGE gels, followed by immune-blotting with anti-myc antibody (1:3,000 dilute) to detect the expression of CRUIS.
To test the proximity labeling activity of CRUIS in those clones stably expressing CRUIS. Cells were transfected with pcDNA3.1-Bio-PupE in a 6-well plate by PEI.
Twelve hours after transfection, supplement fresh media with biotin at the final concentration of 20 μM (For convenience, biotin is prepared as 100× concentrated stock in DMEM, and stored at 4°C).
After adding biotin for 24-48 h, cells are harvested and further examined for PafA activity, indicated by biotin signals on western blot. The cell clone with high enzymatic activity (with strong biotin signals) will be selected for future experiments.
Note: The blocking reagent used for detecting biotin by western is 5% BSA, since most commonly used milk based blocking reagents contain biotin and result in background.
CRUIS labeling in cells
Prepare 20 million cells in a 150 mm dish as the experimental group and another one as the control group.
Note: The number of required cells depends on the abundance of target RNA. More cells generally produce better results.
When the CRUIS cells reach about 70% confluence, co-transfect the cells with sgRNA and pcDNA3.1-Bio-PupE plasmids by PEI. A co-transfection of non-targeting sgRNA and pcDNA3.1-Bio-PupE are used as a control (the ratio of sgRNA and pcDNA3.1-Bio-PupE is 1:1, totally 15 μg).
Twelve hours after transfection, replace with fresh medium containing biotin at the final concentration of 20 μM.
After adding biotin for 24-48 h, cells are harvested, washed with cold PBS 3 times, stored or lysed for the mass spectrometry sample preparation.
Mass spectrometry preparation
Day 1Lyse about 30 million cells by 2 ml lysis buffer with 1× protease inhibitor at 4°C with shaking for one hour.
Clarify the lysates by centrifuging at 13,000 rcf for 10 min at 4°C.
Transfer 900 μl supernatant into 1.5 ml microtubes, with the addition of 576 mg urea to a final concentration of about 8 M (The final volume is about 1.2 ml).
Add 12 μl 1 M DTT to a final concentration of 10 mM, then incubate lysate for 1 h at 56°C.
Treat lysate with 30 μl 1 M iodoacetamide to a final concentration of 25 mM, incubate in dark at room temperature for 45 min to aminocarbonyl modify the Cys site of proteins.
Add 30 μl 1 M DTT to a final concentration of 25 mM, and incubate at room temperature for 30 min to terminate the modification.
Take 50 μl streptavidin magnetic beads (stock in PBS containing 0.1% BSA, 0.05% NaN3 and 0.05% Tween 20) into 1.5 ml microtubes, put microtubes on a magnetic stand for 3 min at room temperature, then remove the supernatant with pipette and wash beads three times with 500 μl PBS, then resuspend beads with 50 μl lysis buffer.
Transfer beads into the lysate and incubated overnight on a rotator at 4°C.
After incubation, the microtubes were placed into magnetic stand for 3 min, then aspirate out lysate and wash beads on Eppendorf mixer with the following four buffers step by step.
Add 1 ml buffer 1 to each microtube and shake on Eppendorf mixer at 600 rpm for 5 min at room temperature.
Place the microtube into magnetic band for 3 min and aspirate out buffer 1.
Repeat the Steps D10 and D11 for once.
Add 1 ml buffer 2 to each microtube and shake on Eppendorf mixer at 600 rpm for 5 min at room temperature.
Place the microtube into magnetic band for 3 min and aspirate out buffer 2.
Add 1 ml buffer 3 to each microtube and shake on Eppendorf mixer at 600 rpm for 5 min at room temperature.
Place the microtube into magnetic band for 3 min and aspirate out buffer 3.
Repeat Steps D15 and D16 for once.
Add 1 ml buffer 4 to each microtube and shake at room temperature for 5 min on Eppendorf mixer at 600 rpm.
Place the microtube into magnetic band for 3 min and aspirate out buffer 4.
Repeat Steps D18 and D19 for twice.
Resuspend beads by 100 μl buffer 5 with addition of 6 μg trypsin, transfer the mixture into new PCR tubes and shake samples at 37°C for overnight.
After incubation, place the tubes into magnetic stand and transfer the supernatant to a new PCR tube, adjust sample pH to a lower than 3 with addition of 8 μl 10% formic acid (Under normal circumstances, the pH of the sample will be below 3, which can pass the PH paper test).
Wash Ziptip with 100 μl 100% ACN (Acetonitrile) with pipette.
Aspirate out ACN slowly with pipette.
Balance Ziptip with 100 μl 0.1% TFA (Trifluoroacetic acid) with pipette.
Repeat Step D25 for once.
Bind peptides to Ziptip carefully for three times with pipette.
Desalt peptides with 100 μl 0.1% TFA.
Repeat Step D28 for once.
Elute peptides into new 1.5 ml microtubes by addition of 50 μl 70% ACN-0.1% TFA.
Repeat step D30 for twice.
Dry peptides in the SpeedVac concentrator. The samples can be stored in -80°C or analyzed on an Orbitrap Fusion.
Recipes
Lysis buffer
1% TritonX-100
50 mM Tris pH 7.5, sterile
150 mM NaCl
Note: Prepared in advance and store at 4°C.
Buffer 1
50 mM Tris pH 8.0, sterile
8 M urea
200 mM NaCl
0.2% SDS
Note: Prepared in advance and stored at room temperature).
Buffer 2
50 mM Tris 8.0
200 mM NaCl
8 M urea
Note: Prepared in advance and store at room temperature.
Buffer 3
50 mM Tris 8.0
0.5 mM EDTA
1 mM DTT(store at -80°C)
Note: Prepared in advance and store at room temperature,add DTT to a final concentration of 1 mM before used.
Buffer 4
50 mM Ammonium bicarbonate
Note: Prepared in advance and store at room temperature.
Buffer 5
100 mM Ammonium bicarbonate with 1 mg/ml RapiGest
Note: Prepared 100 mM Ammonium bicarbonate in H2O in advance and store at room temperature,add RapiGest to a final concentration of 1 mg/ml before used.
Acknowledgments
We thank the Molecular Imaging Core Facility (MICF), the Molecular and Cell Biology Core Facility (MCBCF) and the Multi-Omics Core Facility (MOCF) at the School of Life Science and Technology, ShanghaiTech University for providing technical support. This work has been supported by Shanghai Municipal Science and Technology Commission (19JC1413700 to M.Z.) and National Natural Science Foundation of China (31922038 to M.Z.; 31771490 to J.L.L).
Competing interests
None declared.
References
- Abudayyeh, O. O., Gootenberg, J. S., Konermann, S., Joung, J., Slaymaker, I. M., Cox, D. B., Shmakov, S., Makarova, K. S., Semenova, E., Minakhin, L., Severinov, K., Regev, A., Lander, E. S., Koonin, E. V. and Zhang, F. (2016). C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353(6299): aaf5573.
- Chu, C., Zhang, Q. C., da Rocha, S. T., Flynn, R. A., Bharadwaj, M., Calabrese, J. M., Magnuson, T., Heard, E. and Chang, H. Y. (2015). Systematic discovery of Xist RNA binding proteins. Cell 161(2): 404-416.
- Cox, D. B. T., Gootenberg, J. S., Abudayyeh, O. O., Franklin, B., Kellner, M. J., Joung, J. and Zhang, F. (2017). RNA editing with CRISPR-Cas13. Science 358(6366): 1019-1027.
- Liu, Q., Zheng, J., Sun, W., Huo, Y., Zhang, L., Hao, P., Wang, H. and Zhuang, M. (2018). A proximity-tagging system to identify membrane protein-protein interactions. Nat Methods 15(9): 715-722.
- McHugh, C. A., Chen, C. K., Chow, A., Surka, C. F., Tran, C., McDonel, P., Pandya-Jones, A., Blanco, M., Burghard, C., Moradian, A., et al. (2015). The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521: 232-236.
- Zhang, Z., Sun, W., Shi, T., Lu, P., Zhuang, M. and Liu, J. L. (2020). Capturing RNA-protein interaction via CRUIS. Nucleic Acids Res 48(9): e52.
- Zhu, H., Richmond, E. and Liang, C. (2018). CRISPR-RT: a web application for designing CRISPR-C2c2 crRNA with improved target specificity. Bioinformatics 34(1): 117-119.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Sun, W., Zhang, Z., Liu, J. and Zhuang, M. (2021). A New Method for Studying RNA-binding Proteins on Specific RNAs. Bio-protocol 11(10): e4022. DOI: 10.21769/BioProtoc.4022.
Category
Molecular Biology > RNA > RNA-protein interaction
Neuroscience > Cellular mechanisms
Biochemistry > Protein > Labeling
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









