- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Single-Molecule Studies of Membrane Receptors from Brain Region Specific Nanovesicles
Published: Vol 11, Iss 10, May 20, 2021 DOI: 10.21769/BioProtoc.4018 Views: 5965
Reviewed by: Shalini Low-NamAnca Flavia SavulescuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Conjugation of Fab’ Fragments with Fluorescent Dyes for Single-molecule Tracking on Live Cells
I-Ting Teng [...] Inhee Chung
Sep 20, 2019 8005 Views

FRET-based Microscopy Assay to Measure Activity of Membrane Amino Acid Transporters with Single-transporter Resolution
Didar Ciftci [...] Olga Boudker
Apr 5, 2021 6756 Views

Single Protein Detection and Imaging with Evanescent Scattering Microscopy
Pengfei Zhang [...] Shaopeng Wang
Oct 20, 2022 1885 Views
Abstract
Single molecule imaging and spectroscopy are powerful techniques for the study of a wide range of biological processes including protein assembly and trafficking. However, in vivo single molecule imaging of biomolecules has been challenging because of difficulties associated with sample preparation and technical challenges associated with isolating single proteins within a biological system. Here we provide a detailed protocol to conduct ex vivo single molecule imaging where single transmembrane proteins are isolated by rapidly extracting nanovesicles containing receptors of interest from different regions of the brain and subjecting them to single molecule study by using total internal reflection fluorescence (TIRF) microscopy. This protocol discusses the isolation and separation of brain region specific nanovesicles as well as a detailed method to perform TIRF microscopy with those nanovesicles at the single molecule level. This technique can be applied to study trafficking and stoichiometry of various transmembrane proteins from the central nervous system. This approach can be applied to a wide range of animals that are genetically modified to express a membrane protein-fluorescent protein fusion with a wide range of potential applications in many aspects of neurobiology.
Graphic abstract:
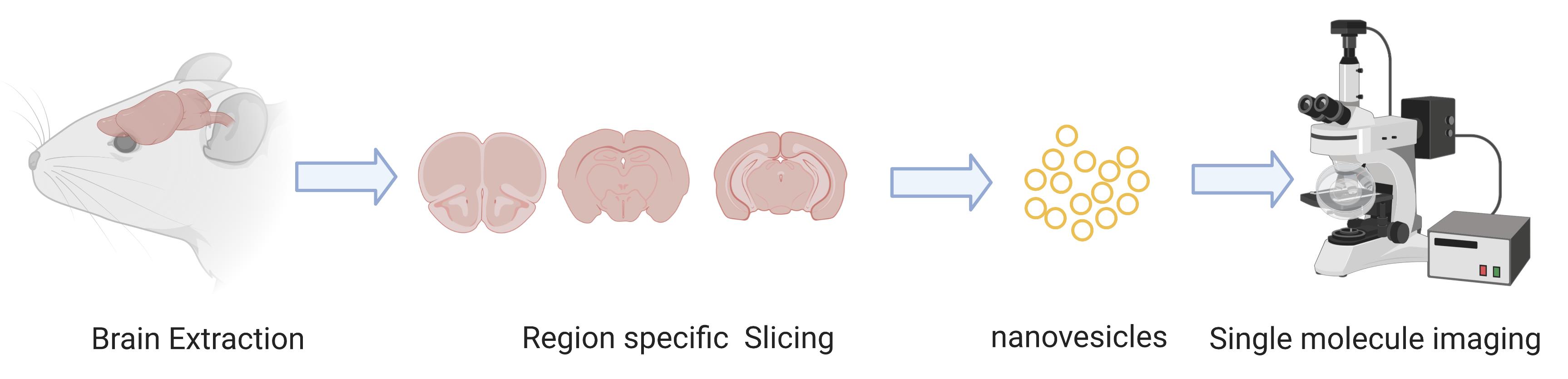
EX vivo single molecue imaging of membrane receptors
Background
Single molecule imaging techniques have emerged as powerful methods to study the structure, function, and interactions of biomolecules (Joo et al., 2008; Choi et al., 2019). While ensemble measurements provide important information about the average population, single molecule methods can be used to evaluate the heterogeneity within this population revealing differences in structural and functional states (Lee et al., 2020). Single molecule methods have already been used in a diverse range of applications including measurements of protein and ligand interaction, conformational changes in proteins, and ion channel function (Xia et al., 2013; Martinac, 2017; Fu et al., 2019). These methods have provided powerful ways to characterize individual cellular constituents and single molecules in vitro.
Despite their successful implementations in vitro, single molecule methods for in vivo studies have been particularly challenging. Major challenges for in vivo studies include the heterogenous nature of cellular components, the difficulties associated with light penetration in thick tissues, and the need to isolate a single species from the complex environment within a living system (Fu et al., 2019). In addition, the concentration of protein of interest in some cases is much higher in the native environment than the optimum concentration required for single molecule imaging (Richards et al., 2012). Additionally, membrane proteins are not spatially isolated in a single location within the cellular environment and instead diffuse across a wide region, complicating single molecule studies. This makes isolation and separation of the molecule of interest a key aspect of any ex vivo single molecule imaging protocol. This protocol details an approach to address these challenges, by utilizing nanoscale vesicles generated from primary tissue as a way of isolating and immobilizing membrane proteins. We demonstrate this ex vivo single molecule imaging approach using knock-in mice containing a fluorescent protein labeled nicotinic acetylcholine receptor (nAChRs). Nicotinic acetylcholine receptors (nAChRs) are transmembrane proteins which respond to and can be upregulated during nicotine exposure (Cano et al., 2020). Their trafficking and stoichiometry have also shown to be altered by nicotine (Srinivasan et al., 2011). In this protocol, we used a mouse model where the nAChR α4 subunit is labelled with green fluorescence protein (GFP).
We believe this protocol can be widely applied to virtually any mouse model that focuses on a transmembrane protein that is genetically labeled with a fluorescent protein. This protocol will enable researchers to extend single molecule studies beyond common in vitro approaches by performing ex vivo characterization of physiological changes occurring in live animals at the individual protein level. Specifically, this protocol describes a detailed method to isolate nanovesicles from specific regions of the mouse brain in order to perform single molecule imaging to characterize membrane receptors. This technique can be used to understand brain region-specific structural and functional changes in various animal models. An advantage of this approach is that during the isolation of the vesicles, transmembrane receptors maintain their structural integrity because they remain in their endogenous membrane in the nanovesicles. This allows one to characterize their structural assembly as well as their functional properties as in their native physiological environment (Fu et al., 2019). This single molecule protocol which involves sample preparation and labelling along with Total Internal Reflection Fluorescence (TIRF) Microscopy should be immediately applicable to a wide range of existing mouse models already in use for other neuroscience applications.
Materials and Reagents
Tygon tubing (Garinger, Item number: 9MH86)
1 ml plastic syringe (Grainger, Item number: 19G334, Thermo Fisher Scientific, catalog number: NC0786233)
Syringe needles (Air-Tite, catalog number: 161028)
50 ml centrifuge tubes (VWR, catalog number: 89039-658)
5 ml Serological pipettes (VWR, catalog number: 89130-896)
15 ml centrifuge tubes (VWR, catalog number: 89039-666)
1.5 ml Flex tubes Eppendorf (VWR, catalog number: 20901-421)
Ultra-Clear ultracentrifuge tubes (Beckman Coulter, catalog number: 344061)
Dounce Tissue Grinder 7ml (Omni International, catalog number: 07-357542)
Glass bottom dish (MatTek, catalog number: P35G-1.5-14-C)
Fatal Plus Solution (Drugs.com, product number: V.P.L. 9373, NDC number: 0298-9373-68)
Protease inhibitor mini tablet (Thermo Fisher Scientific, catalog number: A32955)
Sucrose (VWR, catalog number: 97063-788)
HEPES (Fisher Scientific, catalog number: BP310-500)
Ice
Milli-Q Water (Fisher Scientific, catalog number: QGARD00D2)
1× PBS buffer (VWR, catalog number: 97062-948)
HCl (VWR, catalog number: 470301-260)
NaOH ((VWR, catalog number: BDH7225-4)
Biotin-PEG-Silane (Laysan Bio, Item number: Biotin-PEG-SIL-3400-500mg)
Neutravidin protein (Thermo Fisher Scientific, catalog number: 31000)
Biotinylated anti-GFP antibody (Rockland antibodies and assays, item number: 600-406-215)
Sucrose buffer/Homogenization buffer, pH 7.4 (see Recipes)
Fatal Plus Working Solution (see Recipes)
Equipment
Smooth fine forceps (Electron Microscopy Sciences, 1209K43, catalog number: 72911-6)
Surgical scissors (Medline, catalog number: MDS0838410)
Operating scissors (Stoelting, catalog number: 52138-51)
Flat tip forceps (Fisher Scientific, catalog number: 12-000-123)
Ice bucket (VWR, catalog number: 10146-202)
Adult Mouse Brain Slicer Matrix (Zivic Instruments, catalog number: BSMAS002-1)
1 ml pipettor (Gilson, catalog number: F123602)
Pipet controller (VWR, catalog number: 613-4442)
Autoclave (Steris Amsco Eagle, serial no. 096899 SAW)
Perfusion pump (Grainger, catalog number: 2688)
Centrifuge (Beckman Coulter, model: Allergra® X-22R; Rotor: Beckman Coulter, model: SX4250)
Ultracentrifuge (Beckman Coulter, model: L-60)
Swing bucket rotor (Beckman Coulter, model: SW 28)
Fixed angle rotor (Beckman Coulter, model: 70 Ti)
Belly dancer or orbital shaker (Fisher Scientific, catalog number: 15-453-211)
Sonicator (Amsco Reliance Sonic 550)
Fume Hood (Supreme Air LV, Kewaunee Scientific, VWR, catalog number: 97006-002) or an air compression system
Oxygen plasma cleaner (Harrick Plasma, catalog number: PDC-32G)
Vortex (VWR, catalog number: 10153-836)
4 °C fridge
-20 °C freezer
A TIRF-capable microscope (Olympus, model: IX2-ZDC2, serial number: 0F83094)
Software
Metamorph (Available on Olympus Microscope)
ImageJ/FIJI (Freely available from NIH)
Procedure
There are animal experiments involved in the following procedures. All animal experiments were conducted within the guidelines set forth by the National Institutes of Health (NIH) and were approved by the University of Kentucky’s Institutional Animal Care and Use Committee (IACUC). In our case, the α4-GFP knock-in mouse (2-4 months old) which has green fluorescence protein (GFP) expressed in the α4 subunit of α4 β2 nicotinic acetylcholine receptor (nAChR) were used throughout. The strains of α4-GFP knock-in mice were obtained from Dr. Jerry Stitzel’s Lab (Institute for Behavioral Genetics University of Colorado). These mice express a green fluorescent protein CHRNA4 fusion protein in replacement of the endogenous nicotinic receptor subunit.
Perfusion and Brain Extraction
Prepare the required buffers (see the Recipes section), rinse the surgical tools with water, dry them and autoclave in cycle 2 (Gravity cycle) prior to animal perfusion.
Perform humane euthanasia to a mouse by injecting 200 μl of Fatal plus working solution (see Recipe 2). Wait until the animal becomes nonresponsive by checking foot pinch reflex. Usually, the mouse will become nonresponsive 2-3 min after the injection of fatal plus.
Note: Intravenous injection is the most preferred method, however other injection method such as subcutaneous could be used when it is impractical to do so.
Once nonresponsive, lay the mouse keeping ventral side up atop a foam piece and immobilize it with tape or metal pins. Using small scissors and forceps, pinch a piece of skin and make a small cut in the lower thorax. Make a parabolic, sagittal cut that runs up the length of each side of abdomen. Pull back the loosened tissue. Make a transverse cut across the pectoral region whilst tearing through the diaphragm. Anchor the newly loosened tissue to ensure it is out of the way (Gage et al., 2012).
Note: You will require 3-4 mice for region specific nanovesicles preparation.
Set the perfusion pump to Level 3. Run tygon tubing from the PBS buffer to the pump and then to a 1ml plastic syringe. Insert the needle into the left ventricle (LV). Turn the pump on to perfuse with PBS. Immediately make an incision in the aorta. Exsanguinate until the liver turns tan-colored and the blood is nearly translucent.
Note: A flow rate of around 0.72 ml/min is good. It is important that all the blood is cleared, and liver is tan colored after perfusion otherwise there might still be some blood in the brain vasculature which can interfere with single molecule imaging. Usually, 100 ml PBS is enough to clear blood from a mouse.
For the brain extraction, remove the head by decapitation using large, 6” scissors. Locate the spinal column and remove excess cervical vertebrae. Make an approximately 4 mm cut from the base of the magnum foramen (MF) towards the nose and from the MF laterally towards the ears. Using regular forceps, peel back loosened bone tissue. Make a midsagittal cut from the base of the skull all the way up to the nose being sure to insert the blunt edge of the scissors in first and dragging on the underside of the brain and then making the cut. Make a transverse cut from one eye to the other eye. Using tweezers peel back newly loosened skull tissue. Gently pull the brain out and put in a conical tube containing ice-cold homogenization buffer (Gage et al., 2012).
Brain region specific nanovesicles isolation
Put the brain in a 2 mm Zivic mouse brain matrix and slice the brain (Figure 1). Remove the olfactory bulb and the region of spinal cord if any is left. The first slice which comes after the olfactory bulb will contain the prefrontal cortex and striatum. Separate them and collect in 15 ml centrifuge tubes. The second slice will contain the cortex, hippocampus, thalamus, and hypothalamus. Isolate the regions and collect them in 15 ml centrifuge tubes. The slice posterior to the second slice will contain the cortex, hippocampus, and midbrain. Separate the regions and collect them in 15 ml centrifuge tubes. The remaining region will mostly contain cerebellum. Take out the slices and with the help of forceps, isolate the regions and combine the identical brain regions from multiple mice together. Be careful not to contaminate one region by another.

Figure 1. Set up for Brain Slicing and Region Identification. A. A mouse brain kept in a 2 mm Zivic brain matrix. B. Blades kept on the brain matrix to make 2 mm thin mouse brain slices. C. An illustration of mouse brain to identify various brain regions from the slices. The anterior part contains the olfactory bulb, and the posterior part is the cerebellum attached to the spinal cord. Slices shown at the bottom contain parts between the olfactory bulb and the cerebellum.Use a Dounce homogenizer to homogenize the region-specific fresh tissues by using 2 ml of cold homogenization buffer (Recipe 1 below). Add additional 3 ml of buffer to the mixture and centrifuge at 200 × g for 15 min at 4 °C. Remove the pellet carefully and proceed to the next step with the supernatant.
Centrifuge the supernatant brain lysate at 1,000 × g for 15 min at 4 °C to remove the pellet which contain nuclear fraction.
Collect the supernatant and perform ultracentrifugation at 10,000 × g for 20 min at 4 °C to remove mitochondria. Remove the supernatant and centrifuge it at 100,000 × g for 2 h at 4 °C. This step will yield pellets containing vesicles. Resuspend the vesicles in 300-400 μl of 1× PBS buffer. The vesicles can be stored for few weeks at -80 °C.
Note: Store 50-100 μl per vial to make more convenient while performing TIRF imaging.
Single molecule labelling for TIRF imaging
To perform glass cleaning, take 4-5 gamma irradiated glass bottom dishes and sonicate them in 5 M NaOH solution for 1 h at 45 °C. Rinse them 3 times with DI water and sonicate them again in 0.1 M HCl solution for 1 h at 45 °C. Finally rinse the dishes 3 times with water followed by rinsing with ethanol 3 times.
Dry the cleaned dish by using compressed air and perform oxygen plasma cleaning. Oxygen plasma cleaning should be performed at high plasma level for 5 min. This helps to remove some organic impurities which are not removed from previous cleaning steps (Raiber et al., 2005).
Note: It is strongly recommended to check background fluorescence of the dishes during this step. If the dishes have a large background characterized by the presence of >10-20 isolated background molecules in a field of view discard them or clean them again.
Perform a functionalization of each dish using 1 mg/ml solution of Biotin-PEG-silane in 95% ethanol for 30 min. After 30 min, wash the dishes 3 times with DI water.
Note: Functionalization of glass bottom dishes and immobilization of nanovesicles is carried out at room temperature.
Add 0.1 mg/ml of NeutrAvidin solution in PBS to the dishes for 5 min. After 5 min, rinse the dishes with 1× PBS 3 times.
Add 1 μg/ml of biotinylated anti-GFP antibody in 1× PBS and incubate the dishes for 30 min. Rinse the dishes with 1× PBS 3 times.
The dishes should be imaged before adding vesicles, some dishes could also be kept filled with 1.5 ml of 1× PBS (without vesicles) to compare against the dishes where vesicles are added.
Immobilize the spatially isolated nanovesicles on the functionalized dishes by incubating at room temperature for 30 min (The nanovesicles will be immobilized as shown in the Figure 2).
Note: Usually 50 μl of nanovesicles in 200 μl of 1× PBS is good for TIRF imaging. If the vesicle concentration is too high (resulting in very bright fluorescence but no isolated molecules) or too low (resulting in very low or no single molecule fluorescence), change the concentration accordingly.
Rinse the dish again with 1 ml of 1× PBS 3 times. Add 1 ml 1× PBS to the dish to perform TIRF imaging.
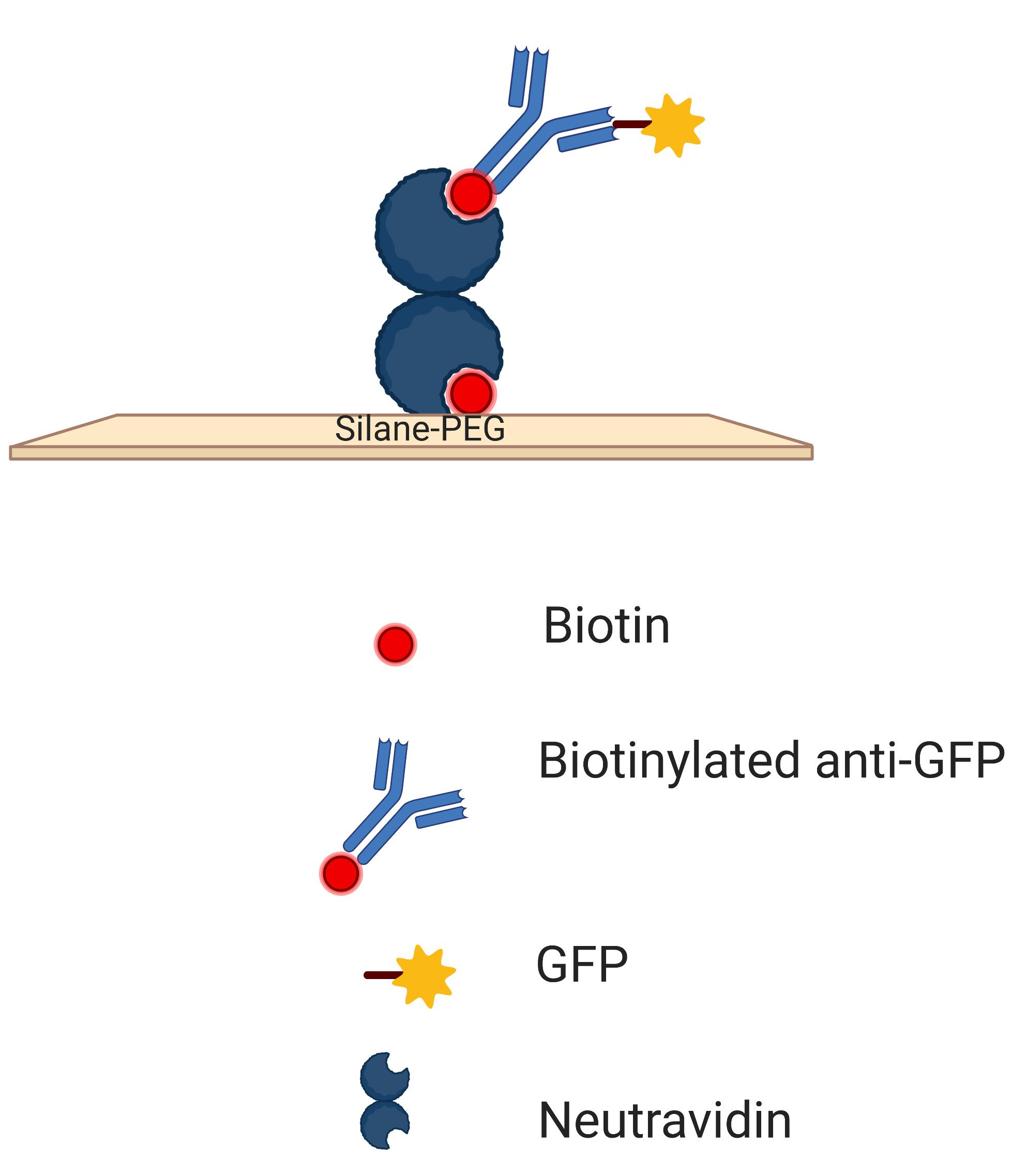
Figure 2. Schematic Diagram of GFP containing vesicles immobilized in a dish. A clean glass bottom dish is coated with biotin-peg-silane, the silane will bind with the glass and biotin will be available to bind with neutravidin. Neutravidin has binding sites to bind with biotin. During washing steps, unbound neutravidin will be washed away while the leftover sites of the bound neutravidin will be used to bind biotinylated anti-GFP antibody. This antibody will bind to the GFP molecule present in the nicotinic acetylcholine receptor of the nanovesicles.
TIRF Imaging of single molecules and data analysis
Turn on the Microscope system including laser, stage controller and accompanying software (e.g., Metamorph for Olympus microscope). Wait for 30 min to allow software, camera, and other parts in the microscope system to equilibrate.
Note: The microscope camera should be sufficiently cooled (~ -80 °C) before conducting the imaging. Please do not turn on the camera at higher temperatures.
Enter into the epifluorescence mode by using the appropriate software in the microscope system (Metamorph software in the Olympus Microscope). Set the laser power to approximately 3 mW/cm2, and make sure the laser is focused and aligned. Ideally the laser should be focused on a point or a small circle at a distance from the objective while in epifluorescence mode. Set up an exposure time to 100 ms. Emission light is detected by an electron multiplying charge coupled device (EMCCD, Andor).
Put a drop of oil on the 60× oil objective. Place a clean glass bottom dish containing PBS above the objective and use eyepiece and focus knob to bring the objective close to the bottom of the dish. Be careful not to hit the dish by objective. The oil drop should stay between the base of the dish and the objective.
Note: You can visualize the fluorescence by using the eyepiece or the software accompanying the microscope. Once the molecules are in focus, you can enter into TIRF mode.
Adjust the angle to gain total internal reflection. You can visualize the laser beam to ensure it is in TIRF mode (as shown in Figure 3). In addition, the nanovesicles will also be spatially isolated in TIRF mode (as shown in Figure 4A). Acquire 800 frames with 100 ms exposure time for each field of view. Store the collected frame by using the stream acquisition mode in the software.
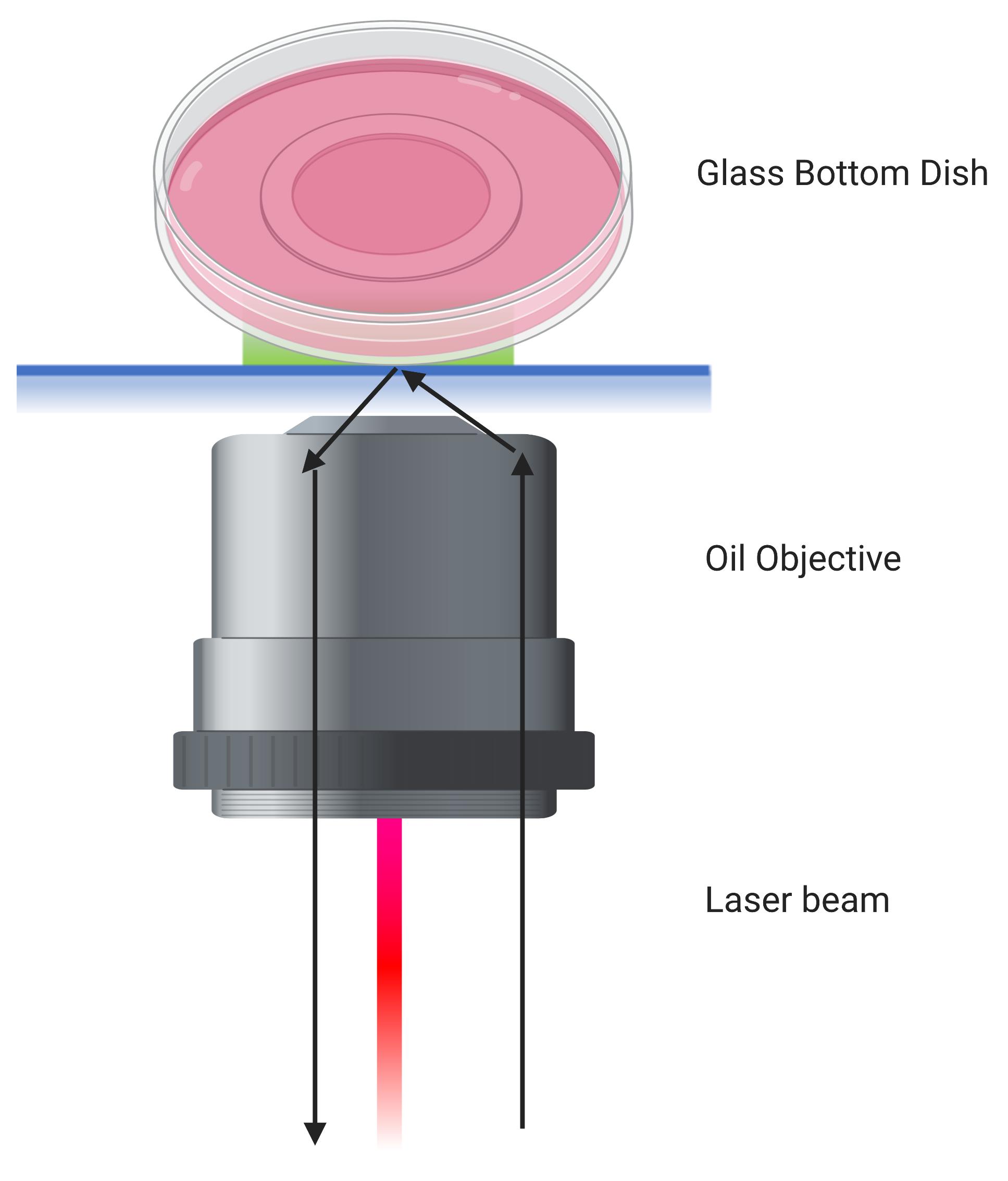
Figure 3. Schematic Diagram of TIRF Microscopy on a Glass Bottom Dish. An oil objective is used where the laser beam is adjusted such that it returns by total internal reflection. The bottom ~200 nm thickness of the dish is illuminated by the evanescent field that originates from total internal reflection on the glass side. This filed allows to image fluorescent single molecule nanovesicles on the bottom surface of the glass bottom dish.Use ImageJ/FIJI software to open the frames (Figure 4) and select regions of interest to perform analysis of change in fluorescence intensity (Rueden et al., 2017).
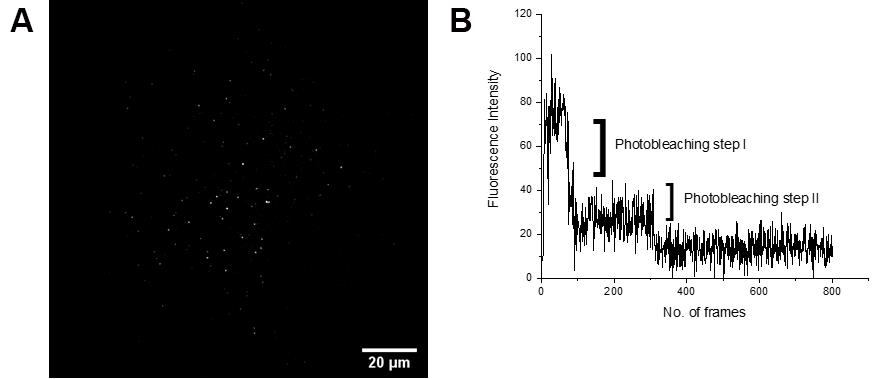
Figure 4. Example data from the single molecule imaging of alpha-4 GFP nanovesicles. A. A TIRF image of spatially isolated nanovesicles containing GFP tethered membrane receptors (scale bar = 5 µm). B. Fluorescence intensity pattern of an individual single molecule from the nanovesicle showing two photobleaching steps (a single molecule region of interest (ROI) is shown in (A)). These photobleaching steps give information about how the alpha-4 beta-2 nAChRs are distributed in the brain.
Recipes
Sucrose buffer/Homogenization buffer, pH 7.4
0.32 M sucrose
10 mM HEPES
2 mM EDTA
Protease inhibitor (One protease inhibitor tablet per 10 ml buffer)
Fatal Plus Working Solution
1 ml Fatal Plus
10 ml PBS (1×)
Acknowledgments
This work was adapted from Fu et al. Anal. Chem. 2019, 91(15): 10125-10131. Biorender was used to construct most figures.
Competing interests
We declare no conflict of interest or competitive interest related to this publication.
Ethics
All the experiments were conducted within the guidelines set forth by the National Institutes of Health and were approved by the University of Kentucky’s Institutional Animal Care and Use Committee.
References
- Cano, M., Reynaga, D. D., Belluzzi, J. D., Loughlin, S. E. and Leslie, F. (2020). Chronic exposure to cigarette smoke extract upregulates nicotinic receptor binding in adult and adolescent rats. Neuropharmacology 181: 108308.
- Choi, J., Grosely, R., Puglisi, E. V. and Puglisi, J. D. (2019). Expanding single-molecule fluorescence spectroscopy to capture complexity in biology. Curr Opin Struct Biol 58: 233-240.
- Fu, X., Moonschi, F. H., Fox-Loe, A. M., Snell, A. A., Hopkins, D. M., Avelar, A. J., Henderson, B. J., Pauly, J. R. and Richards, C. I. (2019). Brain Region Specific Single-Molecule Fluorescence Imaging. Anal Chem 91(15): 10125-10131.
- Gage, G. J., Kipke, D. R. and Shain, W. (2012). Whole animal perfusion fixation for rodents. J Vis Exp(65): e3564.
- Joo, C., Balci, H., Ishitsuka, Y., Buranachai, C. and Ha, T. (2008). Advances in single-molecule fluorescence methods for molecular biology. Annu Rev Biochem 77: 51-76.
- Lee, H., Kang, H., Kang, M., Han, C., Yi, J., Kwon, Y. and Park, J. (2020). Heterogeneous Subcellular Origin of Exosome-Mimetic Nanovesicles Engineered from Cells. ACS Biomater Sci Eng 6(11): 6063-6068.
- Martinac, B. (2017). Single-molecule FRET studies of ion channels. Prog Biophys Mol Biol 130(Pt B): 192-197.
- Raiber, K., Terfort, A., Benndorf, C., Krings, N. and Strehblow, H. H. (2005). Removal of self-assembled monolayers of alkanethiolates on gold by plasma cleaning.Surface Science 595(1-3): 56-63.
- Richards, C. I., Luong, K., Srinivasan, R., Turner, S. W., Dougherty, D. A., Korlach, J. and Lester, H. A. (2012). Live-cell imaging of single receptor composition using zero-mode waveguide nanostructures. Nano Lett 12(7): 3690-3694.
- Rueden, C. T., Schindelin, J., Hiner, M. C., DeZonia, B. E., Walter, A. E., Arena, E. T. and Eliceiri, K. W. (2017). ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18(1): 529.
- Srinivasan, R., Pantoja, R., Moss, F. J., Mackey, E. D., Son, C. D., Miwa, J. and Lester, H. A. (2011). Nicotine up-regulates α4β2 nicotinic receptors and ER exit sites via stoichiometry-dependent chaperoning. 137(1): 59-79.
- Xia, T., Yuan, J. and Fang, X. (2013). Conformational dynamics of an ATP-binding DNA aptamer: a single-molecule study. J Phys Chem B 117(48): 14994-15003.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Aryal, S. P., Fu, X., Masud, A. A., Neupane, K. R. and Richards, C. I. (2021). Single-Molecule Studies of Membrane Receptors from Brain Region Specific Nanovesicles. Bio-protocol 11(10): e4018. DOI: 10.21769/BioProtoc.4018.
Category
Biophysics > Force spectroscopy
Neuroscience > Basic technology > Receptor-receptor interactions
Biochemistry > Protein > Single-molecule Activity > Imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








