- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Characterising Plant Deubiquitinases with in vitro Activity-based Labelling and Ubiquitin Chain Disassembly Assays
Published: Vol 11, Iss 9, May 5, 2021 DOI: 10.21769/BioProtoc.4015 Views: 4585
Reviewed by: Agnieszka ZienkiewiczKomuraiah MyakalaNoelia Foresi

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Semi-throughput Procedure for Assaying Plant NADP-malate Dehydrogenase Activity Using a Plate Reader
Kevin Baudry and Emmanuelle Issakidis-Bourguet
Aug 20, 2023 1465 Views

An in vitro Assay to Probe the Formation of Biomolecular Condensates
Yu Zhang and Shen Lisha
Sep 5, 2023 3196 Views

Immunofluorescence for Detection of TOR Kinase Activity In Situ in Photosynthetic Organisms
Ana P. Lando [...] Giselle M. A. Martínez-Noël
Dec 20, 2024 1812 Views
Abstract
Post-translational modification of proteins by ubiquitin is an essential cellular signaling mechanism in all eukaryotes. Ubiquitin is removed from target proteins by a wide range of deubiquitinase (DUB) enzymes with different activities and substrate specificities. Understanding how DUBs function in vitro is a vital first step to uncovering their cellular roles. Here, we provide protocols for the rapid analysis of DUB activity in vitro by activity-based labelling with the suicide probe, HA-ubiquitin vinyl sulfone (HA-UbVS), and ubiquitin chain disassembly assays. We have previously used these methods to analyse the activity of the Arabidopsis thaliana DUB, UBP6, but in principle, these protocols are applicable to any DUB of interest.
Keywords: UbiquitinBackground
Regulation of cellular protein homeostasis (proteostasis), the intricate balance between protein synthesis, modification, and degradation, is essential for the survival of all organisms (Balch et al., 2008). Proteostasis is maintained by a diverse range of post-translational modifications (PTMs) that increase the size and functionality of proteomes (Skelly et al., 2016). Ubiquitination, the covalent attachment of the small regulatory protein ubiquitin to target proteins, is a prevalent PTM involved in numerous cellular processes in eukaryotes. Ubiquitin is conjugated predominantly to lysine residues of target substrates via its C-terminus, which requires the sequential actions of three types of enzyme; E1 ubiquitin-activating enzymes, E2 ubiquitin-conjugating enzymes, and E3 ubiquitin ligases. Ubiquitin itself contains seven lysine residues that can also be modified, facilitating formation of polyubiquitin chains of various linkage types. Different ubiquitin chain topologies have specific signalling roles and are associated with different cellular processes. For example, K48-linked chains generally target proteins for degradation by the proteasome, a large multi-protein complex that is responsible for the majority of proteolytic activity in eukaryotic cells. However, ubiquitin modifications can also be reversed by the action of a wide range of deubiquitinase (DUB) enzymes with diverse specificities for different ubiquitin linkage types (Mevissen and Komander, 2017). Since deubiquitination determines the stability, activity, or function of proteins, entire proteomes can be shaped by the activities of specific DUBs. Many human diseases, including cancers, are associated with dysregulated DUB activity, demonstrating the physiological and pathological importance of these enzymes (Harrigan et al., 2018; Clague et al., 2019).
To understand DUB function, their activity can be analysed by various in vitro methods to determine substrate specificity and regulation of activity (Cho et al., 2020). These methods include activity-based protein profiling using synthetic ubiquitin suicide probes that irreversibly label DUB active sites (Borodovsky et al., 2002; Gong et al., 2018; Schauer et al., 2020), as well as assays in which purified ubiquitin chains are incubated with DUBs and chain disassembly is monitored by western blotting or gel staining. Recently, we used these methods to uncover the signalling roles of the Arabidopsis thaliana proteasome-associated deubiquitinases, UBP6 and UBP7, in plant immunity (Skelly et al., 2019). Here, we describe protocols for analysing recombinant UBP6 activity by HA-UbVS labelling and in vitro deubiquitination assays. Importantly, these methods can be applied to any DUB of interest and in the case of HA-UbVS, labelling can also be applied to protein extracts from any organism to uncover proteome-wide DUB activity. Characterising the activity and regulation of DUBs is vital to understand how these enzymes act in vivo and will advance the fields of ubiquitin signalling and proteostasis across eukaryotes.
Materials and Reagents
HA-ubiquitin-vinyl sulfone (Boston Biochem, catalog number: U-212)
26S proteasome (Ub-VS treated) (Ubiquigent, catalog number: 65-1020-010)
Recombinant purified His6-T7-UBP6 (Skelly et al., 2019) or other DUB of interest
Ubiquitin subtrates for deubiquitination assays:
Poly-ubiquitin (Ub3-7) K48-linked (Boston Biochem, catalog number: UC-220)
Poly-ubiquitin (Ub3-7) K63-linked (Boston Biochem, catalog number: UC-320)
Di-ubiquitin K48-linked (Boston Biochem, catalog number: UC-200B)
Di-ubiquitin K63-linked (Boston Biochem, catalog number: UC-300B)
UltraPure Tris (Invitrogen, catalog number: 15504-020)
MgCl2 hexahydrate (Sigma-Aldrich, catalog number: M2670)
Adenosine triphosphate (ATP) (Sigma-Aldrich, catalog number: A26209)
Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: 1114740005)
Glycerol (Sigma-Aldrich, catalog number: G5516)
Sodium dodecyl sulfate (SDS) (Sigma-Aldrich, catalog number: L3771)
Bromophenol blue (Sigma-Aldrich, catalog number: B8026)
Anti-HA antibody (ThermoFisher, catalog number: 26183)
Anti-ubiquitin antibody (Santa Cruz Biotechnology, catalog number: sc-8017)
Antibody against the DUB being analysed, e.g., anti-T7 for His6-T7-UBP6 (Millipore, catalog number: 69522)
Double-distilled H2O (ddH2O)
15% SDS-PAGE gel (e.g., compatible with Bio-Rad Mini-PROTEAN system)
SuperSignal West Pico PLUS Chemiluminescent Substrate (ThermoFisher, catalog number: 34577)
10× DUB labelling buffer (store at -20°C) (see Recipes)
10× deubiquitination buffer (store at -20°C) (see Recipes)
2× SDS-PAGE loading buffer (see Recipes)
DUBs, proteasomes, HA-UbVS, and ubiquitin substrates (see Recipes)
Equipment
1.5 ml microcentrifuge tubes (e.g., Starlab, catalog number: S1615-5500)
Pipettes and tips (e.g., Gilson Pipettman, various volumes)
Heat block (e.g., ThermoFisher, catalog number: 88870004)
Protein gel electrophoresis and western blotting apparatus (e.g., Bio-Rad Mini-PROTEAN system)
Procedure
HA-UbVS labelling
Set up 10-μl reactions including the following: 1 μl 10× DUB labelling buffer, 5 μl 700 nM DUB, 1.25 μl 80 nM 26S proteasome (if required), 1 μl 7 μM HA-UbVS, and 1.75 μl ddH2O. Control samples should be included in which components are omitted and replaced with ddH2O.
Incubate for 30 min at room temperature or in a heat block at 22°C.
Add 10 μl 2× SDS-PAGE loading buffer to each sample.
Incubate for 10 min in a heat block at 70°C.
Perform SDS-PAGE with a 15% gel and western blotting using standard methods with:
Anti-HA antibodies (at 1:5000 dilution) to detect free HA-UbVS (~9.8 kDa) and HA-UbVS-labelled DUB (Figure 1).
Antibodies (at antibody-specific dilutions) against the appropriate DUB or epitope tag to detect both unlabelled and labelled DUB. Any standard imaging system can be used depending on the secondary antibodies (e.g., film or CCD imaging system for chemiluminescence).
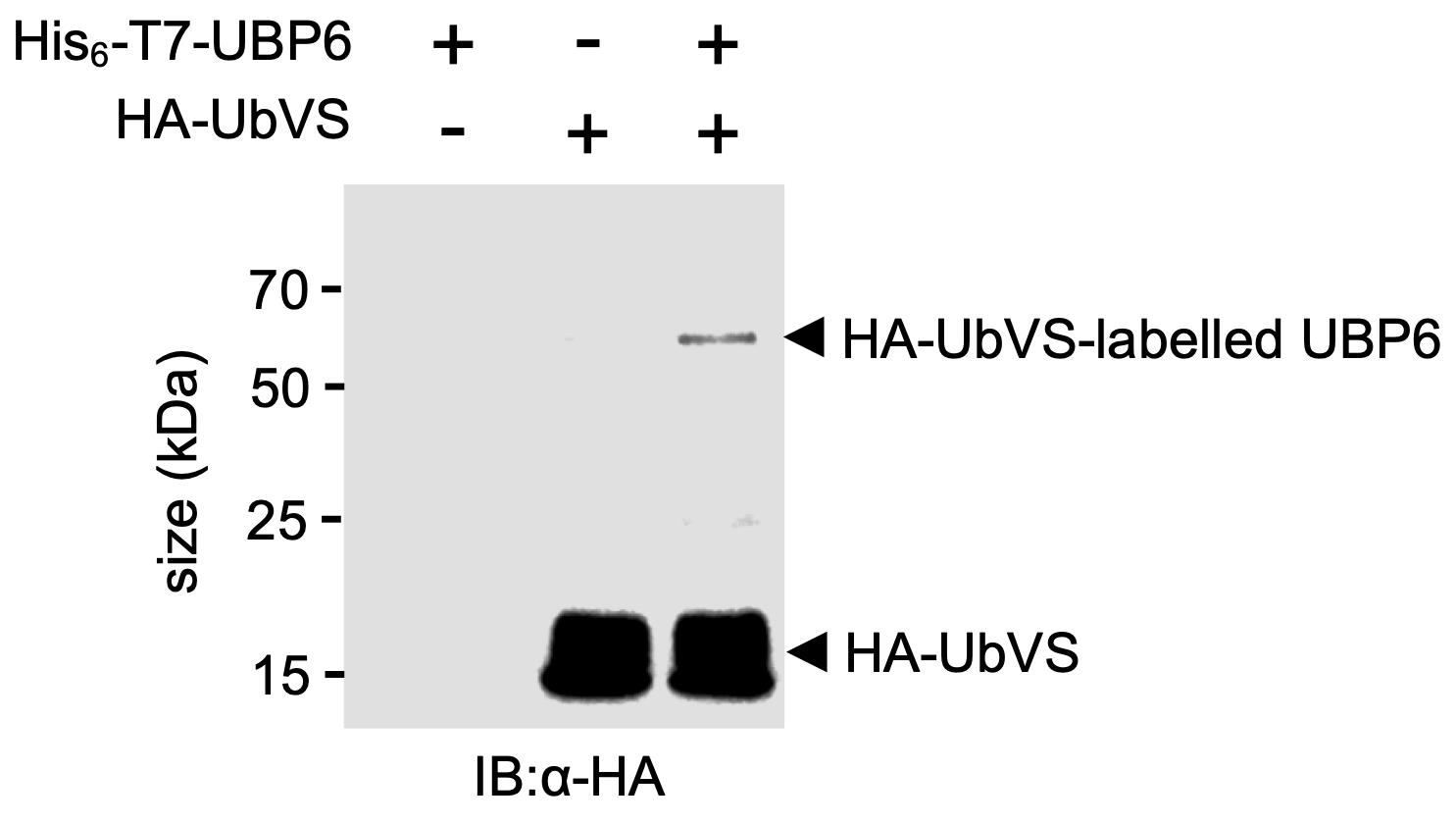
Figure 1. HA-UbVS labelling of recombinant purified His6-T7-UBP6. A typical western blot showing free and HA-UbVS-labelled recombinant His6-T7-UBP6. Adapted from Figure 6B of Skelly et al. (2019).
In vitro deubiquitination assays
Set up 10 μl reactions including the following: 1 μl 10× deubiquitination buffer, 1 μl 200 nM DUB, 1 μl 12.5 nM 26S proteasome (if required), 1 μl 4 μM ubiquitin substrate, and 6 μl ddH2O. Control samples should be included in which components are omitted and replaced with ddH2O.
Incubate in a heat block at 30°C for the desired time [2-16 h for His6-T7-UBP6 (Skelly et al., 2019), but dependent on the DUB and substrate].
Add 10 μl 2× SDS-PAGE loading buffer to each sample.
Incubate for 10 min in a heat block at 70°C.
Perform SDS-PAGE with a 15% gel and western blotting using standard methods with anti-ubiquitin antibodies to detect the original ubiquitin chain substrate and observe any DUB-mediated chain disassembly as lower-order ubiquitin species. Any standard imaging system can be used depending on the secondary antibodies (e.g., film or CCD imaging system for chemiluminescence).
Recipes
10× DUB labelling buffer (store at -20°C)
500 mM Tris pH 7.4
50 mM MgCl2
10 mM DTT
10 mM ATP
10× deubiquitination buffer (store at -20°C)
500 mM Tris pH 7.4
50 mM MgCl2
10 mM DTT
50 mM ATP
2× SDS-PAGE loading buffer (store at room temperature, add DTT fresh before use)
20% glycerol
120 mM Tris pH 6.8
4% SDS
0.02% Bromophenol Blue
100 mM DTT
DUBs, proteasomes, HA-UbVS, and ubiquitin substrates
Diluted in ddH2O from stocks to the required concentrations described above
Acknowledgments
This work was supported by a Royal Society University Research Fellowship (UF090321), a BBSRC grant (BB/L006219/1), and the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No. 678511). This protocol was derived from Skelly et al. (2019).
Competing interests
The authors declare that no competing interests exist.
References
- Balch, W. E., Morimoto, R. I., Dillin, A. and Kelly, J. W. (2008). Adapting proteostasis for disease intervention. Science 319(5865): 916-919.
- Borodovsky, A., Ovaa, H., Kolli, N., Gan-Erdene, T., Wilkinson, K. D., Ploegh, H. L. and Kessler, B. M. (2002). Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family.Chem Biol 9(10): 1149-1159.
- Cho, J., Park, J., Kim, E. E. and Song, E. J. (2020). Assay Systems for Profiling Deubiquitinating Activity. Int J Mol Sci 21(16): 5638.
- Clague, M. J., Urbe, S. and Komander, D. (2019). Breaking the chains: deubiquitylating enzyme specificity begets function. Nat Rev Mol Cell Biol 20(6): 338-352.
- Gong, P., Davidson, G. A., Gui, W., Yang, K., Bozza, W. P. and Zhuang, Z. (2018). Activity-based ubiquitin-protein probes reveal target protein specificity of deubiquitinating enzymes. Chem Sci 9(40): 7859-7865.
- Harrigan, J. A., Jacq, X., Martin, N. M. and Jackson, S. P. (2018). Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat Rev Drug Discov 17(1): 57-78.
- Mevissen, T. E. T. and Komander, D. (2017). Mechanisms of Deubiquitinase Specificity and Regulation. Annu Rev Biochem 86: 159-192.
- Schauer, N. J., Magin, R. S., Liu, X., Doherty, L. M. and Buhrlage, S. J. (2020). Advances in Discovering Deubiquitinating Enzyme (DUB) Inhibitors. J Med Chem 63(6): 2731-2750.
- Skelly, M. J., Frungillo, L. and Spoel, S. H. (2016). Transcriptional regulation by complex interplay between post-translational modifications. Curr Opin Plant Biol 33: 126-132.
- Skelly, M. J., Furniss, J. J., Grey, H., Wong, K. W. and Spoel, S. H. (2019). Dynamic ubiquitination determines transcriptional activity of the plant immune coactivator NPR1. Elife 8: e47005.
Article Information
Copyright
Skelly and Spoel. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Skelly, M. J. and Spoel, S. H. (2021). Characterising Plant Deubiquitinases with in vitro Activity-based Labelling and Ubiquitin Chain Disassembly Assays. Bio-protocol 11(9): e4015. DOI: 10.21769/BioProtoc.4015.
- Skelly, M. J., Furniss, J. J., Grey, H., Wong, K. W. and Spoel, S. H. (2019). Dynamic ubiquitination determines transcriptional activity of the plant immune coactivator NPR1. Elife 8: e47005.
Category
Biochemistry > Protein > Activity
Plant Science > Plant biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









