- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Analysis of Monocyte Cell Fate by Adoptive Transfer in a Murine Model of TLR7-induced Systemic Inflammation
Published: Vol 11, Iss 9, May 5, 2021 DOI: 10.21769/BioProtoc.4007 Views: 5928
Reviewed by: Chiara AmbrogioNaushaba HasinAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Participant-Derived Xenograft Mouse Model to Decode Autologous Mechanisms of HIV Control and Evaluate Immunotherapies
Emma Falling Iversen [...] R. Brad Jones
Apr 5, 2025 2516 Views

PBMC-Humanized Mouse Model for Multiple Sclerosis: Studying Immune Changes and CNS Involvement
Anastasia Dagkonaki [...] Lesley Probert
May 20, 2025 3976 Views

Advancing EAE Modeling: Establishment of a Non-Pertussis Immunization Protocol for Multiple Sclerosis
Shruti Gupta [...] Kayla L. Nguyen
Feb 5, 2026 29 Views
Abstract
Myeloid plasticity is a hallmark of the innate immune response to Toll-like receptor (TLR) activation. Here, we provide a protocol for monocyte cell fate tracking by adoptive transfer in the context of systemic inflammation induced by TLR7 activation, the principal innate immune receptor sensing viral RNA in mice. Defined monocyte subsets are isolated from the bone marrow of donor mice by cell sorting and adoptively transferred into the systemic circulation of congenic hosts, with or without concurrent activation of TLR7 via the topical application of the small molecule agonist, imiquimod, in a cream formulation that induces a systemic inflammatory response. Advantages are the precise definition of donor cell populations and resulting cell fate without the need for host conditioning in a model that recapitulates key aspects of the systemic inflammatory response to TLR7 stimulation.
Keywords: Aldara/ImiquimodBackground
The diversity of myeloid differentiation responses is an integral part of the adaptive immune response and is critical for tissue restoration and maintenance during inflammation and inflammatory dysregulation. Infectious agents, such as viruses, or tissue injury trigger an inflammatory response involving monocytes of the major (classic) subtype in mice, Ly6Chi monocytes (Bonnardel and Guilliams, 2018). These monocytes originate from progenitor cells in the bone marrow (BM), circulate in peripheral blood (PB), and respond to tissue signals by differentiating into a spectrum of effector phagocytes: macrophages (MF), dendritic cells (DC), and monocytes with patrolling behavior (Hettinger et al., 2013; Gamrekelashvili et al., 2016; Bonnardel and Guilliams, 2018; Arazi et al., 2019; Chakarov et al., 2019). Monocyte lineage studies are facilitated by Cx3cr1GFP/+ reporter mice, in which monocytes and macrophages, but not granulocytes, express distinct GFP signals (Jung et al., 2000).
Toll-like receptor 7 (TLR7) is a member of the family of pathogen sensors expressed on myeloid cells. TLR7 is the principal innate immune receptor sensing viral RNA in mice, a function homologous to TLR7/8 in humans (Diebold et al., 2004). It is highly expressed in endosomes of myeloid cells, where viral RNA triggers the production of pro-inflammatory cytokines and type I interferons, causing local and systemic inflammation and the induction of an antiviral state (Iwasaki and Pillai, 2014). Importantly, myeloid TLR7 signaling is required for the efficient protection of mice against virulent RNA viruses (Kaminski et al., 2012), and it is involved in immune responses to the influenza A virus and SARS-CoV2. The activation of TLR7 can also be induced by the small molecule TLR7 agonist, imiquimod (IMQ), an imidazoquinoline derivative used clinically in a cream formulation as antiviral and antitumor therapies (van der Fits et al., 2009). TLR7 stimulation induces cytokine production in both mouse and human patrolling monocytes and mediates the sensing and disposal of damaged endothelial cells by Ly6Clo monocytes (Cros et al., 2010; Carlin et al., 2013); however, chronic TLR7 stimulation drives the differentiation of Ly6Chi monocytes into specialized macrophages and causes the development of anemia (Akilesh et al., 2019). Furthermore, systemic stimulation with TLR7 agonists induces the conversion of Ly6Chi monocytes into patrolling Ly6Clo monocytes in wildtype mice (Santiago-Raber et al., 2011; Gamrekelashvili et al., 2020).
Here, we provide a protocol to study monocyte cell fate under inflammatory conditions using adoptive transfer of monocyte subsets from genetically defined mouse strains, with the concurrent stimulation of host TLR7 via the topical application of IMQ in a cream formulation (Aldara). Ly6Chi monocytes from the bone marrow of Cx3cr1GFP/+ CD45.2+ mice are FACS-sorted and transferred into congenic CD45.1+ mice by tail-vein injection, and donor-derived cells are identified by GFP and CD45.2 expression. The main distinguishing features in comparison with conventional bone-marrow reconstitution experiments are the precise definition of donor cell populations and resulting cell fate without the need for host conditioning. The protocol is transferrable to other progenitor or mature cell populations if suitable numbers can be retrieved from the donors. Furthermore, key aspects of the systemic response to TLR7 stimulation seen in viral models are recapitulated in a virus-free model.
Materials and Reagents
LS columns (Miltenyi Biotec, catalog number: 130-042-401)
0.5 ml and 1.5 ml microfuge tubes (Sarstedt, catalog numbers: 72.699 and 72.690.001)
15 ml tubes (Greiner Bio-One, CELLSTAR, catalog number: 188261)
1 ml syringes with 25 G and 27 G needles (B. Braun, Injekt-F 9166033V, Sterican 4657705)
K3EDTA-containing blood collection tubes (Microvette 100 µl; Sarstedt, catalog number: 20.1278)
96-well V-bottomed plates (Greiner Bio-One, catalog number: 651101)
6-well plates (Merck, Biochrom, catalog number: TPP92406)
Cell strainers (Greiner Bio-One, catalog numbers: 542040 [40 µm], 542000 [100 µm])
Nylon mesh (Heidland, catalog number: 03-70/41)
Anti-biotin microbeads (Miltenyi Biotec, catalog number: 130-090-485)
Propidium iodide (Merck, Sigma-Aldrich, catalog number: 81854-25MG, Stock 1 mg/ml in PBS, 1:12,000 final)
Histopaque 1083 (Merck, Sigma-Aldrich, catalog number: 10831-100ML)
Red blood cell lysis buffer (RBC lysis buffer; BioLegend, catalog number: 420301)
Imiquimod (Meda, Aldara, 5% cream, catalog number: PZN-00111981)
Vaseline (Winthrop GmbH, catalog number: PZN-02726847)
Depilating cream (Silk & Fresh depilating cream)
Bepanthen eye ointment (Bayer vital GmbH, catalog number: PZN-01578675)
Ketamine (Ketamine, 100 mg/ml; CP-Pharma, catalog number: 1202)
Rompun (Xylavet, 20 mg/ml; CP-Pharma, catalog number: 1205)
Midazolam (Midazolam-Actavis 1 mg/ml; Actavis, catalog number: PZN-03831807)
Isofluran (Isofluran CP 1 ml/ml; CP-Pharma, catalog number: 1214)
Heparin 5000 U/ml (Heparin-Natrium, 25000; Ratiopharm, catalog number: PZN-03029843)
Cell counting slides (Thermo Fisher Scientific, catalog number: C10228)
PBS (Merck, Sigma-Aldrich, catalog number: D8537)
Antibodies (Tables 1 and 2)
Table 1. Antibodies and fluorescent dyes for the sorting of inflammatory monocytes
Antibody/Dye Clone Company Cat. No. Final Dilution CD16/32 (TruStain FcX) 93 BioLegend 101319 1:200 CD3 PE 17A2 BioLegend 100205 1:400 Ter119 PE Ter119 BioLegend 116208 1:800 CD45R/B220 PE RA3-6B2 BD Pharmingen 553089 1:800 Ly6G PE 1A8 BioLegend 127608 1:800 CD19 PE 6D5 BioLegend 115507 1:400 CD49b PE DX5 BD Pharmingen 553858 1:200 NK1.1 PE PK136 BioLegend 108707 1:200 CD90.2 PE Thy1.2 BD Pharmingen 553005 1:800 CD11b Pacific Blue M1/70 BioLegend 101224 1:200 Ly6C PE-Cy7 HK1.4 BioLegend 128018 1:1400
Table 2. Antibodies and fluorescent dyes for donor cell enrichment and flow cytometryAntibody/Dye Clone Company Cat. No. Final Dilution CD16/32 (TruStain FcX) 93 BioLegend 101319 1:200 CD3 Biotin 17A2 BioLegend 100243 1:400 Ter119 Biotin Ter119 BioLegend 116203 1:400 CD45R/B220 Biotin RA3-6B2 BioLegend 103203 1:400 Ly6G Biotin 1A8 BioLegend 127603 1:400 CD19 Biotin 6D5 BioLegend 115504 1:400 CD45.1-Biotin A20 BioLegend 110703 1:200 CD117 APC-Cy7 2B8 BioLegend 105825 1:100 CD11b Pacific Blue M1/70 BioLegend 101224 1:200 Ly6C PE-Cy7 HK1.4 BioLegend 128018 1:2800 F4/80 BV650 BM8 BioLegend 123149 1:200 CD11c BV605 N418 BioLegend 117334 1:400 I-A/I-E BV510 M5/114.15.2 BioLegend 107635 1:400 NK1.1 Biotin PK136 BioLegend 108704 1:400 CD45.2 Alexa700 104 BioLegend 109821 1:200 CD43 PerCP-Cy5.5 S7 BD Pharmingen 562865 1:400 Streptavidin-PE-Dazzled594 BioLegend 405247 1:400 Flow cytometry buffer (FCS-EDTA-PBS, see Recipes)
Cell collection buffer (20% FCS-PBS, see Recipes)
Heparin-PBS (250 U/ml) for collection of peripheral blood (see Recipes)
RBC lysis buffer (see Recipes)
Equipment
Hair shaver (Aesculap Isis GT420, Type HS61)
Caliper (Pariere, 8030/R)
Mouse heat pad (Beurer, HK25, Type P10)
Centrifuge (Hermle, Z383K)
Table-top centrifuge (VWR, Micro Star 17R)
Flow cytometer (Beckton Dikinson, LSR-II)
Cell sorter (Beckton Dikinson, FACSAria Fusion)
Quadro MACS separation unit (Miltenyi Biotec, catalog number: 130-090-976)
Automated cell counter (Thermo-Fisher Scientific, Invitrogen, Countless II FL, AMQAF1000)
Automated blood counter (Scil Animal Care Company, Scil Vet ABC)
Software
FlowJo (version 10.6.2, FLOWJO LLC, https://www.flowjo.com)
Prism (version 7.04, GraphPad, https://www.graphpad.com)
Adobe Illustrator (version 25.0, https://www.adobe.com)
Procedure
Induction of systemic inflammation using imiquimod
Notes:
Experimental procedures can only be performed after approval by the local Animal Care Committee.
Perform experiments with 8-12-week-old mice. Reporter mice are B6.129P-Cx3cr1tm1Litt/J, obtained from the Jackson Laboratory (Jung et al., 2000; Gamrekelashvili et al., 2016 and 2020).
IMQ-treated mice must be kept separately from the Vaseline (Sham) group.
Mix blood thoroughly with Heparin-PBS or K3EDTA to avoid coagulation.
Pre-warm RBC lysis buffer at room temperature to achieve efficient lysis.
Induction of systemic inflammation using IMQ (Figure 1)
On Day -1, weigh the mice, record the weight, and intraperitoneally inject Ketanest (Esketamin 80 mg/kg), Rompun (Xylazin 2.5 mg/kg), and Dormicum (Midazolam 2.5 mg/kg) in 0.9% NaCl solution to anesthetize the mice (injection volume 10 µl/g mouse weight, using a 1-ml syringe and 27 G needle).
Apply a drop of Bepanthen eye ointment from the tube to the eye to avoid dry eyes, and test paw reflexes to make sure the mouse is sleeping.
Shave the back (appr. 3 cm2) of the mice with an electric shaver and apply the depilating cream.
Keep the mice for appr. 2 min on a heat pad and wipe away the depilating cream with a spadel.
Wash and rinse the back of the mice with warm water to remove the remaining depilating cream.
Keep the mice on the heat pad until awake and then transfer to cages with ad libitum access to food and water.
Two days later (Day 1), weigh the mice, record the weight, and place each mouse separately in a glass jar with appr. 200-300 µl isofluran for 10-15 s.
Place each dizzy/sleepy mouse on the heat pad, measure the thickness of the ear using a caliper, and quickly apply 50 mg IMQ (Aldara) cream or an equal amount of Vaseline to the back and the right ear using a stainless-steel spadel.
Repeat the same procedure for 3-4 consecutive days (Figure 1).
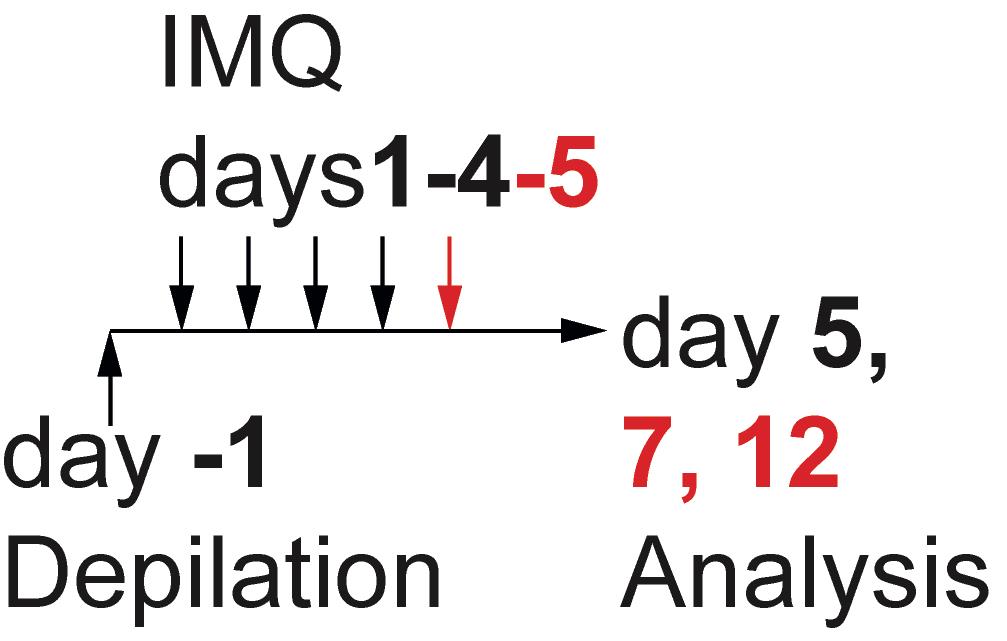
Figure 1. Experimental set-up for the induction of inflammationMouse preparation and analysis of systemic inflammation
On Day 5, 7, or 12, anesthetize the mice by intraperitoneal injection with Ketanest (Esketamin 80 mg/kg), Rompun (Xylazin 2.5 mg/kg), and Dormicum (Midazolam 2.5 mg/kg) in 0.9% NaCl solution (injection volume 10 µl/g mouse weight, using a 1-ml syringe and 27 G needle).
Check the footpad reflex to confirm that the mouse is sleeping.
Fix the sleeping mice to the pad, open the abdomen, and collect the blood from the inferior vena cava using a 1-ml syringe with a 25 G ×5/8’’ needle (Braun Melsungen) (Figure 2):
300 µl blood (PB) in 100 µl Heparin-PBS (250 U/ml) for cell analysis.
50 µl blood in K3EDTA-containing tubes for blood counts.
The remaining blood without heparin for serum collection and analysis.
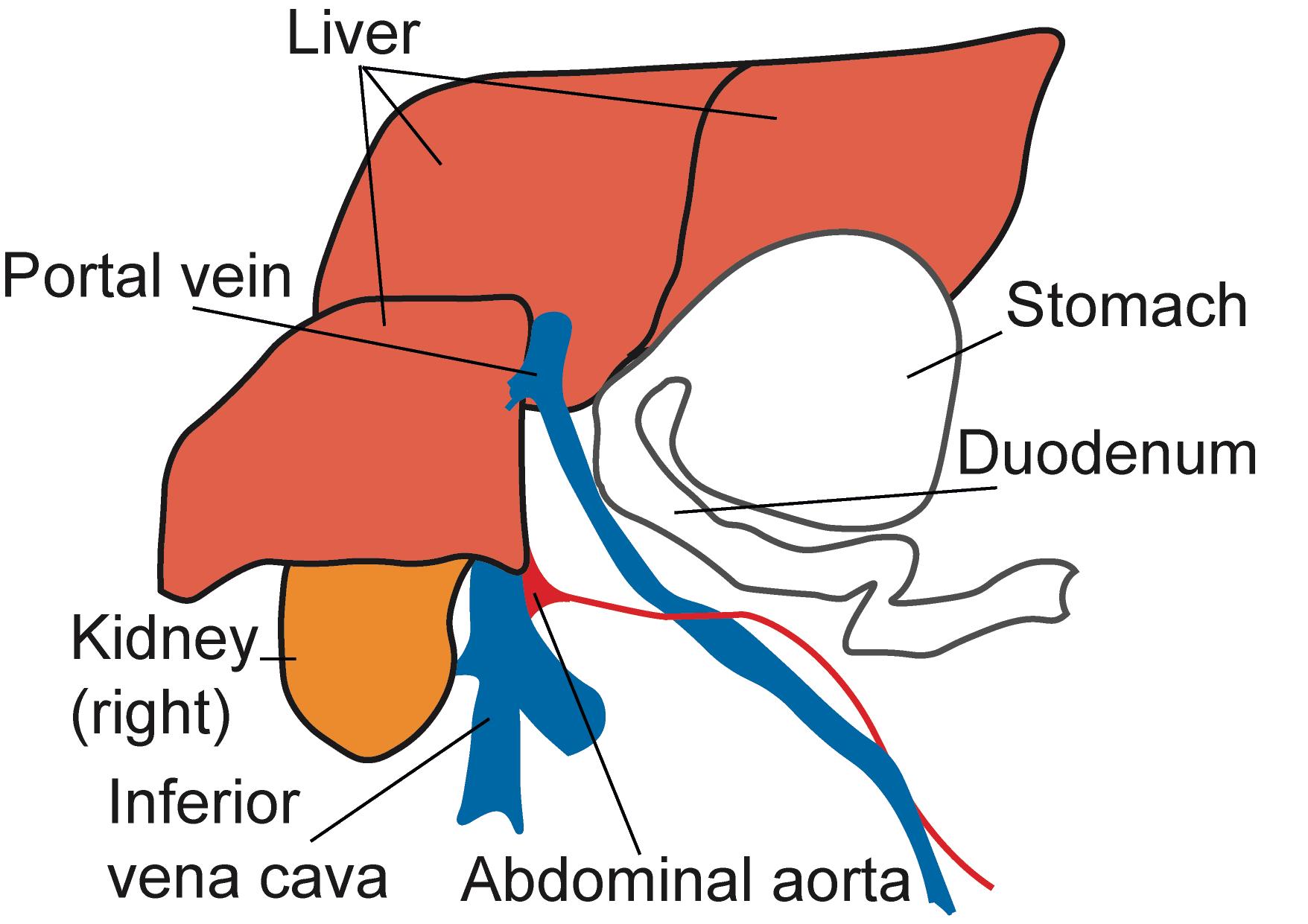
Figure 2. Schematic representation of the mouse abdominal situs, ventral view. After opening the abdominal cavity of deeply anesthetized mice, collect blood from the inferior vena cava.Euthanize the mice, open the abdomen, excise the spleen (Spl), and place in PBS on ice.
Cut and remove the skin from the hind limb and collect the hind limb with the muscles on a paper towel.
Carefully remove the muscles using scissors. Clean the rest of the muscles and soft tissue with a paper towel and isolate the bones [one tibial (Tibia) and one femoral (Femur) bone (Figure 3)].
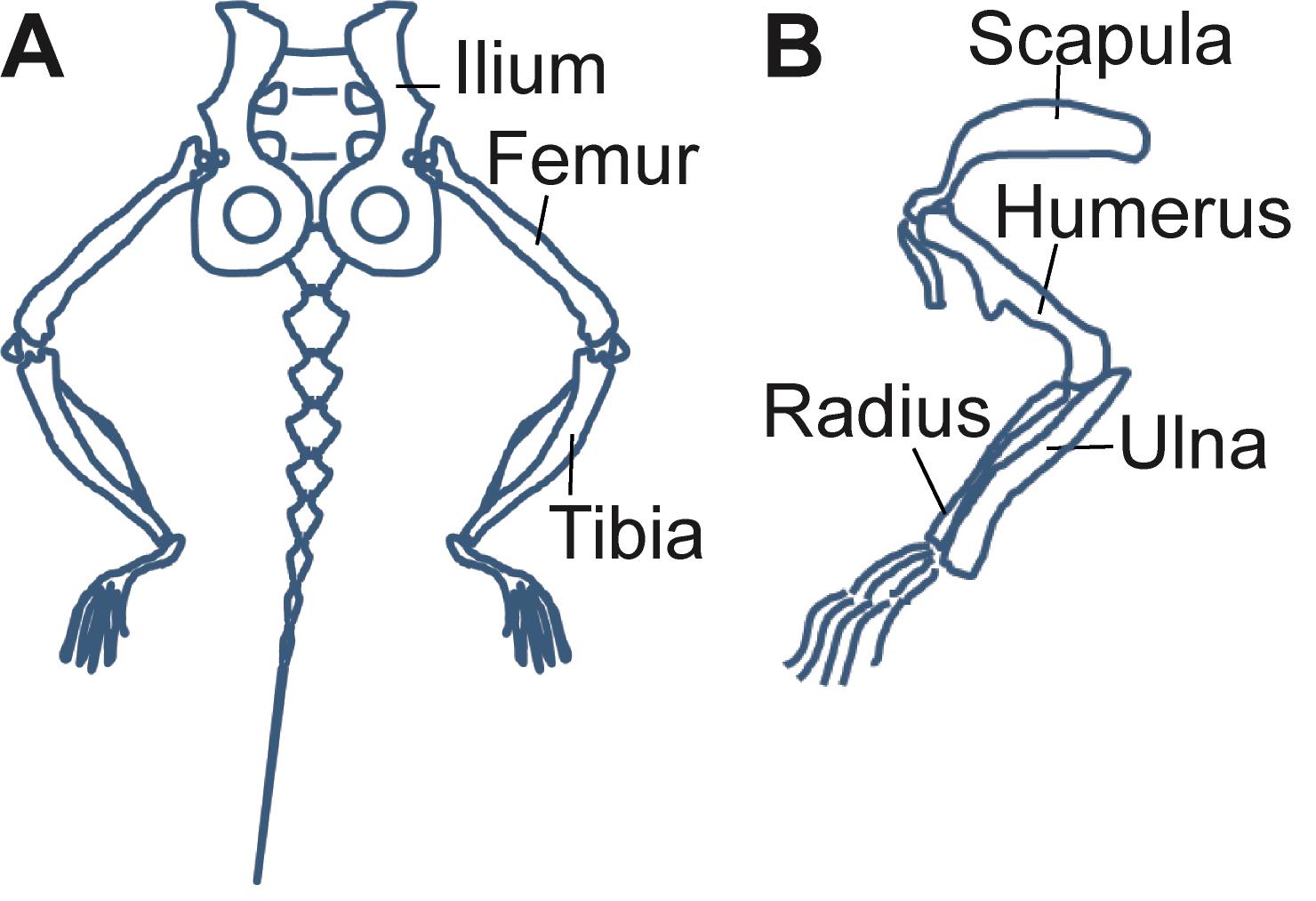
Figure 3. Schematic representation of the bones of the mouse hindlimb (A) and forelimb (B). The Ilium, Femur, Tibia, and Humerus are collected for bone-marrow isolation and analysis.Pre-weigh the 1.5-ml Eppendorf collection tube of the custom-made bone-marrow (BM) isolating device (Figure 4). Cut one end of the bone, place in a 0.5-ml tube with a hole in the bottom, and collect the BM by centrifugation of the isolating device in a table-top centrifuge at 5,000 × g for 20 s.
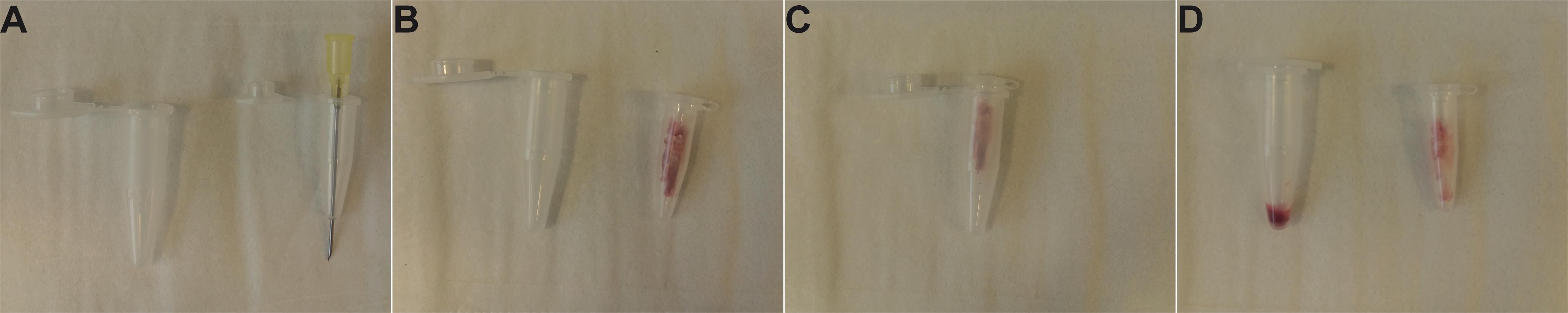
Figure 4. Assembly and use of the custom-made BM isolating device. A. Perforation of a 0.5-ml tube with a 21 G needle for BM isolation. B. Bones cleaned from muscles and placed in a 0.5-ml tube. C. Assembly for centrifugation. D. Isolated BM in a 1.5-ml collection tube after centrifugation.Weigh the collection tube again, record the difference in weight to define the BM weight and place on ice.
Resuspend BM in 10 ml PBS, filter through a 70-µm nylon mesh, and wash by centrifugation at 400 x g for 7 min.
Aspirate the supernatant, resuspend the cell pellet in 10 ml PBS, and repeat the filtration and washing step once more.
Weigh the spleen, prepare a single-cell suspension by passing through a syringe plunger in a 6-well plate and filtering through a nylon mesh, and centrifuge at 400 × g for 7 min.
To remove the erythrocytes, resuspend the PB and Spl pellets in 5 ml or 1 ml room-temperature RBC lysis buffer and incubate for 17 min or 1 min, respectively.
Stop the lysis procedure by adding 10 ml PBS, filter the suspension through a nylon mesh, and centrifuge at 400 × g for 7 min.
Repeat the washing-filtration-centrifugation step once more.
Resuspend the Spl, BM, and PB in PBS (10 ml, 10 ml, or 5 ml, respectively) and count the cells in an automated cell counter to define the absolute cell number.
Resuspend the cells in FCS-EDTA-PBS buffer and place ≤1 × 106 cells per sample in a 96-well V-bottomed plate for staining and subsequent flow cytometry analysis.
Adoptive transfer of monocytes into IMQ-treated mice and monocyte differentiation analysis
Notes:
Experimental procedures can only be performed after approval by the local Animal Care Committee.
Perform experiments with 8-12-week-old mice. Use B6.SJL-PtprcaPepcb/BoyJ (CD45.1+) mice as recipients (Jackson Laboratory).
Collect no more than 2 × 106 sorted monocytes in 2 ml collection buffer and carefully mix the collection tube every 20-30 min to avoid excess stress to the cells. This step enhances cell viability and ensures a high yield of donor cells for analysis after adoptive transfer.
For successful flow cytometry analysis of transferred cells, it is important to deplete recipient cells from recipient Spl and BM but not from peripheral blood.
It is critical to remove erythrocytes using density centrifugation with Histopaque 1083 and not RBC lysis for successful analysis of transferred donor cells.
Cell enrichment on magnetic columns must be performed with ice-cold FCS-EDTA-PBS buffer. Avoid excess air bubble and foam formation.
Induction of systemic inflammation using IMQ
Depilate and treat recipient CD45.1+ mice on days 1-4 with IMQ as described in A and Figure 1.
Isolation and adoptive transfer of inflammatory monocytes on day 2
Euthanize 4-5 donor CX3CR1-GFP+ CD45.2+ mice and isolate the bones (hips (Ilium), femoral (Femur), tibial (Tibia), and brachial (Humerus) bones from both sides, 8 bones/mouse total, Figure 3). Clean the muscles from the bones using scissors and a paper towel. Collect the cleaned bones in PBS and keep on ice.
Dip the bones in 70% EtOH for 3-5 s and dry on EtOH-sterilized dry paper towel. Cut one end of the bone and put 4 bones/tube in 0.5-ml Eppendorf tubes with a hole in the bottom created using a 21 G needle (Figure 4).
Close the tube and place in another 1.5-ml collection tube. Centrifuge this device in a table-top centrifuge with a fixed-angle rotor at 5,000 × g for 20 s.
Collect the bone marrow, resuspend in 50 ml PBS, filter through a 100-µm sterile cell strainer, and centrifuge in a swinging-bucket rotor at 400 × g for 7-8 min to collect the pellet.
Resuspend the cell pellet from 4-5 mice in 6-8 ml PBS, overlay 3-4 ml suspension on 3 ml Histopaque 1083 in 15-ml tubes, and centrifuge in a swinging-bucket rotor at 420 × g for 20 min without the brake.
Collect the interphase (appr. 5 ml) containing the cells in a 15-ml tube, fill the tube with PBS, and wash by centrifugation at 400 × g for 7 min.
Resuspend the pellet in 600 µl FCS-EDTA-PBS and add 3 µl Fc block (TruStain FcX). Incubate the tube at 4°C for 12-15 min.
Without washing, add PE-labeled anti-mouse CD3- (1.5 µl), CD19- (1.5 µl), B220- (0.75 µl), Ly6G- (0.75 µl), Ter119- (0.75 µl), CD90.2- (0.75 µl), NK1.1- (3 µl), CD49b- (3 µl); anti-mouse CD11b-Pacific Blue (3 µl), and anti-mouse Ly6C-PE-Cy7 (0.43 µl) antibodies (Table 1) to the 600 µl cell suspension and incubate for an additional 15 min.
Wash the cells twice in 10 ml buffer, filter through a 40-µm cell strainer, resuspend in 2 ml FCS-EDTA-PBS buffer, and sort Lin-CD11b+CX3CR1-GFPloLy6Chi cells. Collect ~2 × 106 sorted cells in 2 ml ice-cold 20% FCS-PBS and mix the collection tube every 20-30 min during sorting.
Immediately after sorting, wash the sorted cells twice with 10 ml PBS each, resuspend in PBS, and intravenously inject 0.7 × 106-2 × 106 cells per mouse in 100 µl volume into the tail vein using a 1-ml syringe and 27 G needle. To dilate the tail vein and facilitate injection, prewarm the tail in warm water.
Isolation and analysis of donor-derived CD45.2+ monocytes from CD45.1+ recipients 3 days after adoptive transfer.
Euthanize recipient mice in deep narcosis, collect blood in 200 µl Heparin-PBS, collect the spleen and 8 bones (hips, femoral, tibial, and brachial bones).
Prepare a single-cell suspension from Spl by passing through a 5-ml syringe into a 6-well plate, resuspending in 4 ml PBS, and filtering through a 70-µm nylon mesh.
Isolate BM as described in Figure 4, resuspend in 4 ml PBS, and filter through a 70-µm nylon mesh.
Dilute the blood (PB) with PBS to 7 ml.
Load Spl, BM, and PB separately onto 2.5 ml Histopaque 1083 in 15-ml tubes and centrifuge in a swinging-bucket rotor at 420 × g for 20 min without the brake.
Collect the interphase (appr. 5ml) containing the cells in a clean 15-ml tube, fill up the tube with PBS, and wash once by centrifugation at 400 × g for 7 min.
Resuspend the cell pellet in 100 µl (BM, Spl) or 50 µl (PB) FCS-PBS-EDTA buffer supplemented with Fc block (1:200 dilution) and incubate at 4°C for 15 min. Without washing the Fc block, add biotin-labeled antibodies consisting of anti-mouse CD45.1, CD3, CD19, B220, Ly6G, and Ter119 to the final dilution indicated in Table 2, but do not add the biotinylated anti-mouse NK1.1 at this stage.
Quickly vortex the tube and incubate at 4°C for 15 min.
Fill the tubes with 5 ml FCS-EDTA-PBS buffer and centrifuge at 400 × g for 8 min.
Wash PB once more with 5 ml FCS-EDTA-PBS buffer and transfer the pellet to a 96-well V-bottomed plate for further staining.
Resuspend the Spl and BM pellets in 240 µl FCS-EDTA-PBS buffer, add 60 µl anti-biotin microbeads, and incubate at 4°C for 15 min. Fill the tubes again with FCS-EDTA-PBS buffer to wash the excess beads and centrifuge at 400 × g for 7-8 min.
Place the LS columns in the Quadro MACS separation unit and equilibrate with ice-cold FCS-EDTA-PBS buffer (1 ml buffer per column).
Resuspend the Spl and BM pellets in 2 ml FCS-EDTA-PBS buffer and load onto the pre-equilibrated LS columns.
Wash the columns twice with 2 ml FCS-EDTA-PBS buffer, collect the flowthrough, pellet by centrifugation at 400 × g for 7 min, transfer to a 96-well V-bottomed plate, and stain in 30 µl volume with the appropriate antibody cocktail (Table 2).
Resuspend the cells after staining and wash in FCS-EDTA-PBS buffer. Add propidium iodide (PI 1 mg/ml stock in PBS, 1:12,000 final dilution) prior to acquisition and acquire flow cytometry data for further analysis.
Data analysis
Analysis of IMQ-induced systemic inflammation
Measure the mouse weight, ear thickness, blood count, coagulation, inflammatory cytokine levels, liver and kidney function, and myeloid cell composition of the BM, PB, and Spl, and plot the data as described in Figures 5A-5J in this manuscript and in Figure 3E, Figure 4G, Figure 3 – figure supplement 1A, Figure 4 – figure supplement 1C in Gamrekelashvili et al. (2020).
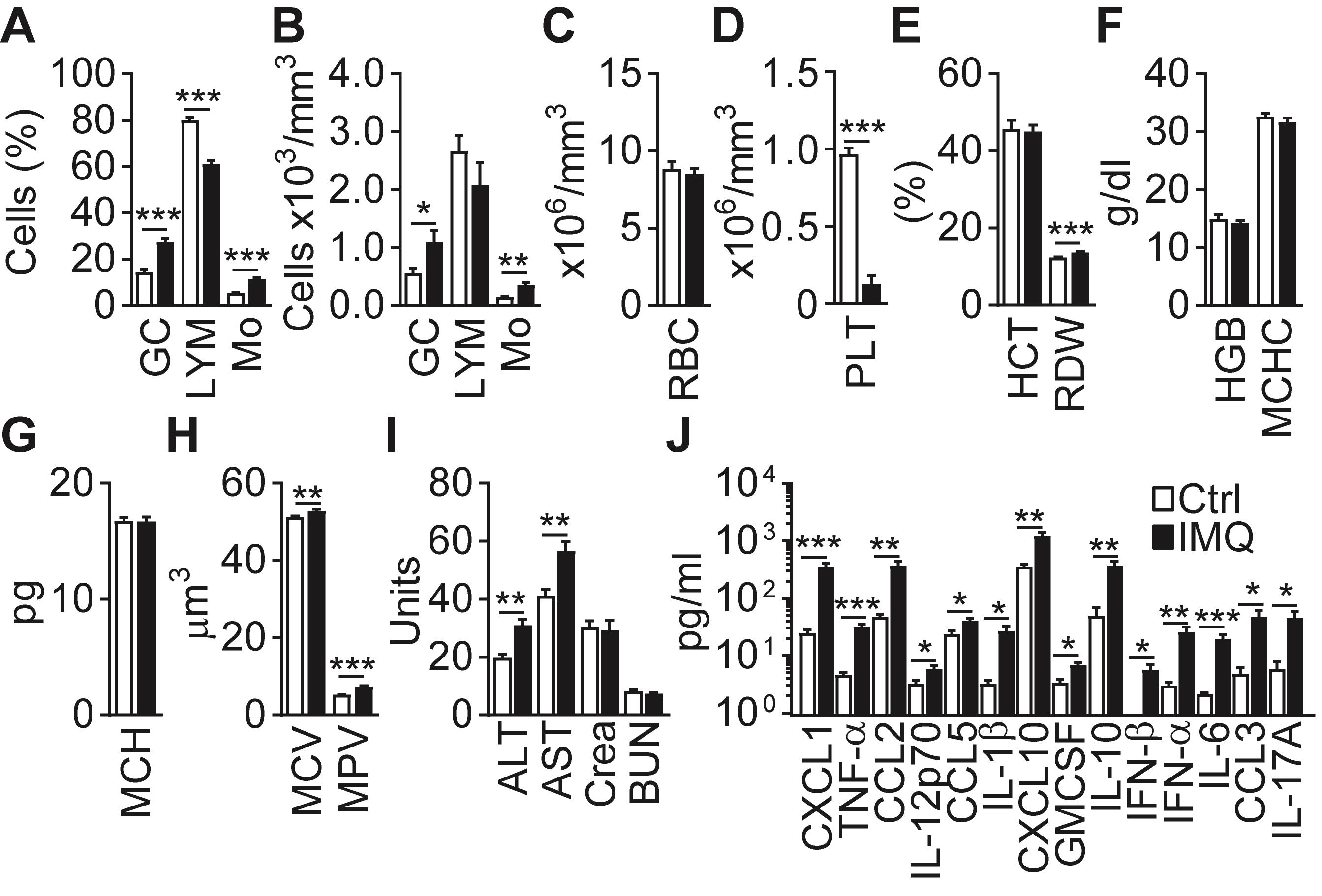
Figure 5. IMQ induces systemic inflammation. (A-H) Blood parameters in Ctrl (Vaseline) or IMQ-treated mice (GC – granulocytes, LYM – lymphocytes, Mo – monocytes, RBC – red blood cells, PLT – platelets, HCT – hematocrit, RDW – red blood cell distribution width, HGB – Hemoglobin, MCHC – mean corpuscular hemoglobin concentration, MCH – mean corpuscular hemoglobin, MCV – Mean corpuscular volume, MPV – mean platelet volume). (I) ALT/AST, creatinine (Crea) and blood urea nitrogen (BUN) in IMQ-treated mice. (J) Cytokine and chemokine concentrations in Ctrl or IMQ-treated mouse sera. (A-J) Data are pooled from 3-4 experiments, n = 4-9. *P < 0.05, **P < 0.01, ***P < 0.001; two-tailed unpaired Student’s t-test.Analyze the flow cytometry data using the Flowjo software. Calculate the relative frequency of myeloid cell subsets [for a definition of the subsets, see Figure 3E, Figure 3 – figure supplement 2, Supplementary File 1 in Gamrekelashvili et al. (2020)] in live cells after the exclusion of doublets, export the numerical data to Excel, and calculate the absolute cell numbers per mg BM, mg Spl, or µl blood.
Transfer the data to GraphPad Prism, plot the values as the mean and standard error of the mean, and calculate the statistical significance using an unpaired two-tailed Student’s t-test. Define P < 0.05 as statistically significant and indicate on the graphs.
Analysis of adoptively transferred monocytes using flow cytometry
Analyze the flow cytometry data using the Flowjo software. Calculate the relative frequency of myeloid cell subsets in live CD45.2+CD11b+CX3CR1-GFP+ donor cells after the exclusion of doublets and CD45.1+ recipient cells, export the numerical data into GraphPad Prism, and plot the values as the mean and standard error of the mean, as shown in Figures 5F and 5G in Gamrekelashvili et al. (2020). Calculate the statistical significance using an unpaired two-tailed Student’s t-test. Define P < 0.05 as statistically significant and indicate on the graphs.
Create representative flow cytometry plots, as shown in Figure 6, depicting the gating strategy, surface phenotype, and frequency of donor-derived cells. Initially, define the cells according to size and granularity (FSC-A vs. SSC-A plot), exclude doublets (SSC-W vs. SSC-A plot) and dead (PI+) cells (FSC-A vs. PI plot), and gate donor cells (CD45.2+GFP+). As a next step, define monocytes (CD11b+GFP+ population) in donor cells and create subsequent plots showing the expression of Ly6C, F4/80, CD11c, CD43, and I-A/I-E as markers of monocyte subsets.
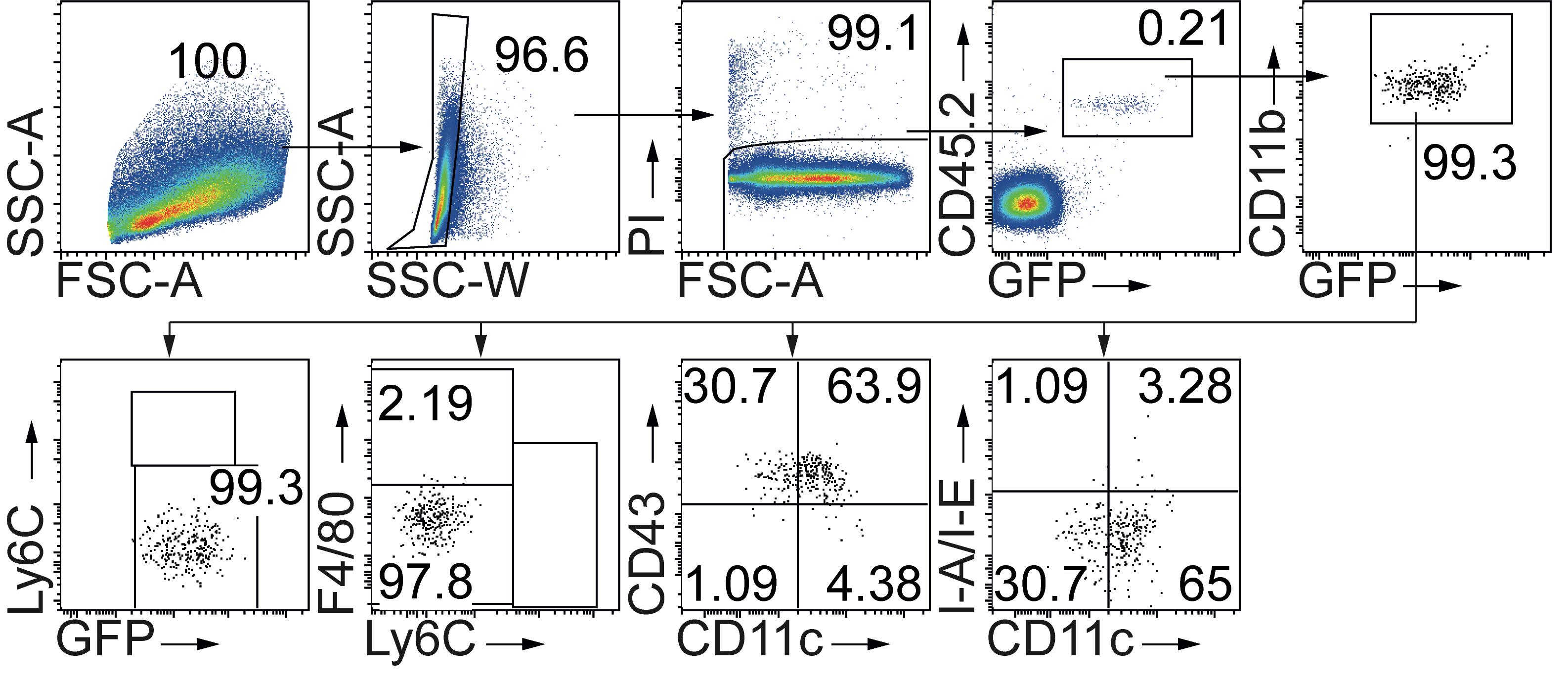
Figure 6. Adoptively transferred classic monocytes differentiate into patrolling monocytes. Flow cytometry plots showing the gating strategy and analysis of donor cells 3 days after adoptive transfer of CD45.2+ classic monocytes in the spleen of IMQ-treated CD45.1+ recipients. Donor-derived monocytes (CD45.2+CD11b+CX3CR1-GFP+Ly6ChiF4/80lo/-CD11c-CD43lo/-I-A/I-Elo/- cells) no longer show a classic monocyte phenotype in recipient mice but instead show a phenotype consistent with patrolling monocytes (CD45.2+CD11b+CX3CR1-GFP+Ly6Clo/-F4/80loCD11cloCD43hiI-A/I-Elo/-).
Recipes
FCS-EDTA-PBS buffer
FCS 20 ml (2% final)
EDTA (0.5 M stock in PBS, pH 7.2) 4 ml (2 mM final)
PBS 1,000 ml
Sorted cell-collection buffer (20% FCS-PBS buffer)
FCS 10 ml
PBS 40 ml
Filter through a 0.22-µm filter and keep sterile
Heparin-PBS (250 U/ml) for collection of peripheral blood
Heparin stock 5,000 U/ml 50 µl
PBS 950 µl
Take 100 µl heparin-PBS for ≤ 500 µl blood.
RBC lysis buffer
RBC lysis buffer (10×) 1 ml
Deionized water 9 ml
Acknowledgments
We thank the Central Animal Facility and the Research Core Facility Cell Sorting of Hannover Medical School for their support. We thank Stefan Sablotny and Herle Chlebusch for their excellent technical help. This work was funded by grants from Deutsche Forschungsgemeinschaft to JG (GA 2443/2-1) and FPL (Li948-7/1). This protocol describes experiments published in Gamrekelashvili et al. (2020).
Competing interests
The authors declare no competing financial or non-financial interests.
Ethics
All animal experiments described in this protocol were approved by the local Animal Welfare Board (LAVES, Lower Saxony, Animal Studies Committee, TVA 16/2251, 18/2777, 2018/221).
References
- Akilesh, H. M., Buechler, M. B., Duggan, J. M., Hahn, W. O., Matta, B., Sun, X., Gessay, G., Whalen, E., Mason, M., Presnell, S. R., Elkon, K. B., Lacy-Hulbert, A., Barnes, B. J., Pepper, M. and Hamerman, J. A. (2019). Chronic TLR7 and TLR9 signaling drives anemia via differentiation of specialized hemophagocytes. Science 363(6423): eaao5213-5213.
- Arazi, A., Rao, D. A., Berthier, C. C., Davidson, A., Liu, Y., Hoover, P. J., Chicoine, A., Eisenhaure, T. M., Jonsson, A. H., Li, S., Lieb, D. J., Zhang, F., Slowikowski, K., Browne, E. P., Noma, A., Sutherby, D., Steelman, S., Smilek, D. E., Tosta, P., Apruzzese, W., Massarotti, E., Dall'Era, M., Park, M., Kamen, D. L., Furie, R. A., Payan-Schober, F., Pendergraft, W. F., 3rd, McInnis, E. A., Buyon, J. P., Petri, M. A., Putterman, C., Kalunian, K. C., Woodle, E. S., Lederer, J. A., Hildeman, D. A., Nusbaum, C., Raychaudhuri, S., Kretzler, M., Anolik, J. H., Brenner, M. B., Wofsy, D., Hacohen, N., Diamond, B. and Accelerating Medicines Partnership in, S. L. E. n. (2019). The immune cell landscape in kidneys of patients with lupus nephritis. Nat Immunol 20(7): 902-914.
- Bonnardel, J. and Guilliams, M. (2018). Developmental control of macrophage function. Curr Opin Immunol 50: 64-74.
- Carlin, L. M., Stamatiades, E. G., Auffray, C., Hanna, R. N., Glover, L., Vizcay-Barrena, G., Hedrick, C. C., Cook, H. T., Diebold, S. and Geissmann, F. (2013). Nr4a1-dependent Ly6Clow monocytes monitor endothelial cells and orchestrate their disposal. Cell 153(2): 362-375.
- Chakarov, S., Lim, H. Y., Tan, L., Lim, S. Y., See, P., Lum, J., Zhang, X. M., Foo, S., Nakamizo, S., Duan, K., Kong, W. T., Gentek, R., Balachander, A., Carbajo, D., Bleriot, C., Malleret, B., Tam, J. K. C., Baig, S., Shabeer, M., Toh, S. E. S., Schlitzer, A., Larbi, A., Marichal, T., Malissen, B., Chen, J., Poidinger, M., Kabashima, K., Bajenoff, M., Ng, L. G., Angeli, V. and Ginhoux, F. (2019). Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science 363(6432): eaau0964.
- Cros, J., Cagnard, N., Woollard, K., Patey, N., Zhang, S. Y., Senechal, B., Puel, A., Biswas, S. K., Moshous, D., Picard, C., Jais, J. P., D'Cruz, D., Casanova, J. L., Trouillet, C. and Geissmann, F. (2010). Human CD14dim monocytes patrol and sense nucleic acids and viruses via TLR7 and TLR8 receptors. Immunity 33(3): 375-386.
- Diebold, S. S., Kaisho, T., Hemmi, H., Akira, S. and Reis e Sousa, C. (2004). Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303(5663): 1529-1531.
- Gamrekelashvili, J., Giagnorio, R., Jussofie, J., Soehnlein, O., Duchene, J., Briseno, C. G., Ramasamy, S. K., Krishnasamy, K., Limbourg, A., Kapanadze, T., Ishifune, C., Hinkel, R., Radtke, F., Strobl, L. J., Zimber-Strobl, U., Napp, L. C., Bauersachs, J., Haller, H., Yasutomo, K., Kupatt, C., Murphy, K. M., Adams, R. H., Weber, C. and Limbourg, F. P. (2016). Regulation of monocyte cell fate by blood vessels mediated by Notch signalling. Nat Commun 7: 12597.
- Gamrekelashvili, J., Kapanadze, T., Sablotny, S., Ratiu, C., Dastagir, K., Lochner, M., Karbach, S., Wenzel, P., Sitnow, A., Fleig, S., Sparwasser, T., Kalinke, U., Holzmann, B., Haller, H. and Limbourg, F. P. (2020). Notch and TLR signaling coordinate monocyte cell fate and inflammation. Elife 9: e57007.
- Hettinger, J., Richards, D. M., Hansson, J., Barra, M. M., Joschko, A. C., Krijgsveld, J. and Feuerer, M. (2013). Origin of monocytes and macrophages in a committed progenitor. Nat Immunol 14(8): 821-830.
- Iwasaki, A. and Pillai, P. S. (2014). Innate immunity to influenza virus infection. Nat Rev Immunol 14(5): 315-328.
- Jung, S., Aliberti, J., Graemmel, P., Sunshine, M. J., Kreutzberg, G. W., Sher, A. and Littman, D. R. (2000). Analysis of fractalkine receptor CX3CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol 20(11): 4106-4114.
- Kaminski, M. M., Ohnemus, A., Cornitescu, M. and Staeheli, P. (2012). Plasmacytoid dendritic cells and Toll-like receptor 7-dependent signalling promote efficient protection of mice against highly virulent influenza A virus. J Gen Virol 93(Pt 3): 555-559.
- Santiago-Raber, M. L., Baudino, L., Alvarez, M., van Rooijen, N., Nimmerjahn, F. and Izui, S. (2011). TLR7/9-mediated monocytosis and maturation of Gr-1hi inflammatory monocytes towards Gr-1lo resting monocytes implicated in murine lupus. J Autoimmun 37(3): 171-179.
- van der Fits, L., Mourits, S., Voerman, J. S., Kant, M., Boon, L., Laman, J. D., Cornelissen, F., Mus, A. M., Florencia, E., Prens, E. P. and Lubberts, E. (2009). Imiquimod-induced psoriasis-like skin inflammation in mice is mediated via the IL-23/IL-17 axis. J Immunol 182(9): 5836-5845.
Article Information
Copyright
Gamrekelashvili et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Gamrekelashvili, J., Haller, H. and Limbourg, F. P. (2021). Analysis of Monocyte Cell Fate by Adoptive Transfer in a Murine Model of TLR7-induced Systemic Inflammation. Bio-protocol 11(9): e4007. DOI: 10.21769/BioProtoc.4007.
- Gamrekelashvili, J., Kapanadze, T., Sablotny, S., Ratiu, C., Dastagir, K., Lochner, M., Karbach, S., Wenzel, P., Sitnow, A., Fleig, S., Sparwasser, T., Kalinke, U., Holzmann, B., Haller, H. and Limbourg, F. P. (2020). Notch and TLR signaling coordinate monocyte cell fate and inflammation. Elife 9: e57007.
Category
Immunology > Animal model > Mouse
Developmental Biology > Cell growth and fate
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








