- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Isolation of GFP-expressing Malarial Hypnozoites by Flow Cytometry Cell Sorting
Published: Vol 11, Iss 9, May 5, 2021 DOI: 10.21769/BioProtoc.4006 Views: 4646
Reviewed by: Dhiman Sankar PalShalini AgarwalAndrew Sylwester

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Thrombopoietin-independent Megakaryocyte Differentiation of Hematopoietic Progenitor Cells from Patients with Myeloproliferative Neoplasms
Chloe A. L. Thompson-Peach [...] Daniel Thomas
Jan 20, 2023 2361 Views

Isolation, Purification, and Culture of Embryonic Melanoblasts from Green Fluorescent Protein–expressing Reporter Mice
Melissa Crawford [...] Lina Dagnino
Sep 5, 2023 1984 Views

Identification and Sorting of Adipose Inflammatory and Metabolically Activated Macrophages in Diet-Induced Obesity
Dan Wu [...] Weidong Wang
Oct 20, 2025 2233 Views
Abstract
Hypnozoites are dormant liver-stage parasites unique to relapsing malarial species, including the important human pathogen Plasmodium vivax, and pose a barrier to the elimination of malaria. Little is known regarding the biology of these stages, largely due to their inaccessible location. Hypnozoites can be cultured in vitro but these cultures always consist of a mixture of hepatocytes, developing forms, and hypnozoites. Here, using a GFP-expressing line of the hypnozoite model parasite Plasmodium cynomolgi, we describe a protocol for the FACS-based isolation of malarial hypnozoites. The purified hypnozoites can be used for a range of ‘-omics’ studies to dissect the biology of this cryptic stage of the malarial life cycle.
Keywords: MalariaBackground
Many gaps in knowledge surround hypnozoite biology, which hampers drug discovery efforts. This is largely due to the inaccessible location of hypnozoites inside the liver in vivo and their presence in low numbers (in contrast to the size of the liver, which contains an estimated 3.6 × 1011 cells) (Bianconi et al., 2013.)
In vitro liver-stage cultures have been developed but the percentage of infected hepatocytes is at maximum 3% (Zeeman et al., 2014), which complicates ‘-omics’ studies aiming to characterize hypnozoites in more detail. The host cell genome (Yan et al., 2011) is 100-fold larger in size than the parasite genome (Pasini et al., 2017), indicating that hepatocyte-derived material will be a major source of contamination for ‘-omics’ studies of hypnozoites. Moreover, in vitro cultures (Sattabongkot et al., 2006; Dembele et al., 2011; Gural et al., 2018; Roth et al., 2018) contain a mixture of uninfected hepatocytes and liver-stage schizonts and hypnozoites; therefore, not only is parasite enrichment needed but also separation of hypnozoites from the developing forms.
For several malarial species, including P. yoelii, P. berghei, and P. falciparum, methods have been described to purify liver stages by FACS using fluorescent reporter lines (Natarajan et al., 2001; Tarun et al., 2006; Prudencio et al., 2008); however, these species do not develop into hypnozoites. Only a few primate malarial parasite species form hypnozoites.
P. vivax liver-stage research is dependent on patient material, which complicates experimentation; nonetheless, RNAseq of P. vivax hypnozoites has been performed (Gural et al., 2018) through selective drug treatment of liver-stage cultures, precluding a direct comparison between hypnozoites and liver-stage schizonts.
The closely phylogenetically related monkey parasite, P. cynomolgi, is more accessible to experimentation and has been used extensively as a model for P. vivax (Joyner et al., 2016; Zeeman et al., 2016; Gupta et al., 2019). In fact, hypnozoites were first identified in P. cynomolgi (Krotoski et al., 1983) and extensive drug studies involving P. cynomolgi have shown very similar responses between P. vivax and P. cynomolgi (Schmidt, 1983). While working with monkeys is ethically restricted and limited to labs that have access to non-human primates, the P. cynomolgi parasite model offers unique possibilities for screening compounds for hypnozoiticidal activity and to gain a better understanding of hypnozoite biology. A previous study used laser-capture dissection of P. cynomolgi hypnozoites for transcriptomics (Cubi et al., 2017), which enables small-scale sampling of fixed parasites. To allow sampling of larger quantities of (live) hypnozoites, here, we describe a method for the FACS sorting of hypnozoites.
P. cynomolgi is accessible to genetic engineering (Kocken et al., 1999) and this has enabled the development of fluorescent reporter lines for FACS (Voorberg-van der Wel et al., 2013; Voorberg-van der Wel, 2020), permitting the large-scale isolation of live malarial hypnozoites and liver-stage schizonts as described in this protocol (Voorberg-van der Wel et al., 2017; Bertschi et al., 2018).
Materials and Reagents
Collagen-coated 96-well plates (PerkinElmer, catalog number: 6055700)
Cellstar 50 ml polypropylene tubes (Greiner Bio-One, catalog number: 227261)
5 ml serological pipettes (for example, VWR, catalog number: 612-5523)
10 ml serological pipettes (for example, Greiner Bio-One, catalog number: 607-107)
25 ml serological pipettes (for example, VWR, catalog number: 612-5544)
1.5 ml Sarstedt tubes, sterile (Sarstedt, catalog number: 72.692.405)
5 ml round-bottomed polystyrene test tubes (Falcon, catalog number: 352052)
5 ml round-bottomed polystyrene test tubes, with cell strainer snap cap (Falcon, catalog number: 352235)
Reagent reservoir (Greiner Bio-One, catalog number: 960305)
Pipette tips 300 µl and 1,000 µl (Rainin, SR LTS filter tip 768)
Phosphate-buffered saline (PBS, 1×) (Thermo Fisher Scientific, catalog number: 14200-067)
0.25% trypsin/EDTA (Thermo Fisher Scientific, catalog number: 25200072)
TRIzol reagent (Thermo Fisher Scientific, catalog number: 15596026)
William’s E medium + Glutamax (Thermo Fisher Scientific, catalog number: 32551-087)
Human serum A+ (pooled, heat inactivated; Sanquin blood bank)
Insulin/transferrin/selenium supplement 100× (Thermo Fisher Scientific, catalog number: 41400-045)
Sodium pyruvate 100 mM (Thermo Fisher Scientific, catalog number: 11360-036)
MEM-NEAA 100× (Thermo Fisher Scientific, catalog number: 11140-035)
Pen/strep 100× (Thermo Fisher Scientific, catalog number: 15140-122)
Hydrocortisone (Sigma, catalog number: H0888)
50 mM β-mercaptoethanol (Thermo Fisher Scientific, catalog number: 31350-010)
DMSO hybrimax (Sigma, catalog number: P1860)
William’s B medium (see Recipes)
20% William’s B medium (see Recipes)
0.1 M hydrocortisone (see Recipes)
Primary cells and parasites
Primary macacque hepatocytes (freshly isolated or thawed from a cryopreserved stock; Macaca mulatta or Macaca fascicularis; source: BPRC; hepatocytes may be obtained through commercial suppliers
GFP-expressing P. cynomolgi lines as described in Voorberg-van der Wel et al. (2013 and 2020); lines can be obtained upon request from the corresponding author
Equipment
(Multichannel) pipettes (Rainin)
Pipette controller (Integra Biosciences, model: Pipetboy acu 2)
Refrigerated benchtop centrifuge (Beckman Coulter, model: Allegra X-15R)
Inverted microscope (Leica, model: DMi1)
Laminar flow biosafety cabinet IIB (Clean-Air EF/B6)
Fume hood
Humidified 5% CO2 incubator at 37°C
Water bath at 37°C
Ice machine
Vortex
High Content Imager – Operetta (PerkinElmer)
Cell sorter – FACS Aria I flow cytometer (BD Biosciences) equipped with a blue laser (20 mW)
-80°C freezer (Eppendorf)
Software
FlowJo software version 9.9 (https://www.flowjo.com/solutions/flowjo/downloads/previous-versions)
FACS Diva version 8.0.1 (https://www.bdbiosciences.com/en-us/instruments/research-instruments/research-software/flow-cytometry-acquisition/facsdiva-software)
Columbus 2.8.2 (PerkinElmer) (https://www.perkinelmer.com/nl/product/image-data-storage-and-analysis-system-columbus)
Procedure
The different steps of the protocol are depicted in Figure 1 and described in detail below.
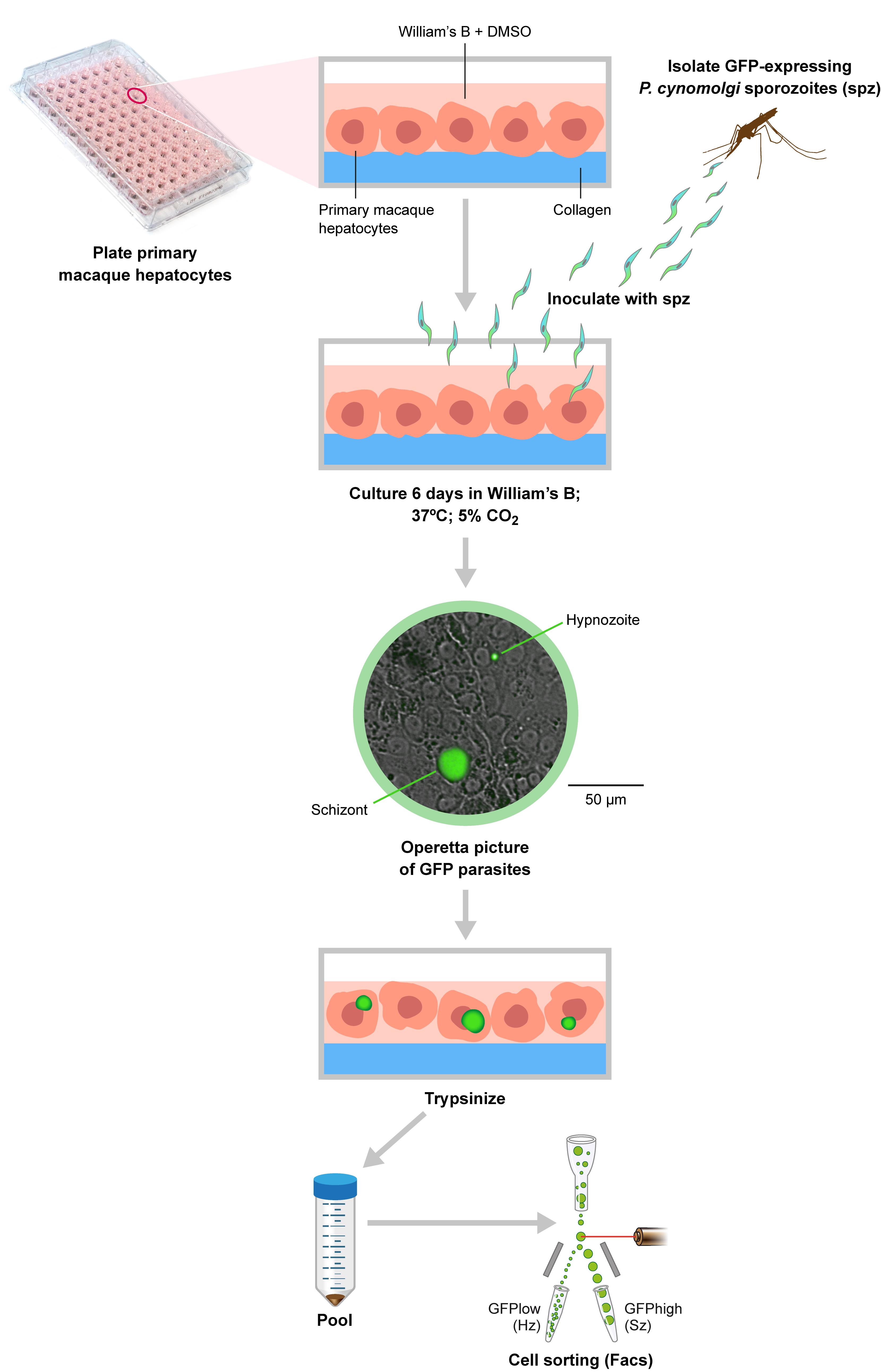
Figure 1. Overview of the procedure. Briefly, primary macaque hepatocytes are plated into collagen-coated 96-well plates, and after 2-3 days are inoculated with GFP-expressing P. cynomolgi sporozoites isolated from A. stephensi mosquitoes. At 6 days post-inoculation the culture is imaged using an Operetta high content imager to assess the presence of hypnozoites and schizonts. The cells are then detached by trypsinization and this pooled material is used for flow cytometry cell sorting of hypnozoites and schizonts.
Infection and culture of macaque primary hepatocytes with transgenic P. cynomolgi parasites
Plate 65 × 103 macaque primary hepatocytes per well (96-well plates; see Note 2) and maintain the culture at 5% CO2 and 37°C in William’s B medium (100 µl per well; see Recipe 1) supplemented with 2% DMSO. Hepatocytes can be freshly isolated or used from a cryopreserved stock. After 2-3 days, inoculate the cultures with 50 × 103 fluorescent transgenic P. cynomolgi sporozoites per well (Voorberg-van der Wel et al., 2013 ) as described in https://bio-protocol.org/e3722.
Maintain the culture in William’s B medium without DMSO (this affects parasite development) at 5% CO2 and 37°C; refresh the medium three times per week by removing as much culture medium as possible, followed by gently adding fresh William’s B medium (100 µl per well). Routinely, the culture is maintained for 6 days because at that time, hypnozoites and developing liver stages can be distinguished based on their size.
Culture alongside uninfected wells with macaque primary hepatocytes as the control (≥6 wells of a 96-well plate).
Harvesting cells
On the day of harvest, check the quality of infection (for reference) by live imaging on a fluorescence microscope/High Content Imager (Voorberg-van der Wel et al., 2013 ). Please see the image in the flowchart for an example. Further insights into liver-stage parasite development can be gained from Voorberg-van der Wel et al. (2020) .
Remove the supernatant from the 96-well plates and wash with 1× in PBS (100 µl/well).
Remove the supernatant and add 30-50 µl pre-warmed (at 37°C) 0.25% trypsin/EDTA per well; incubate for 3 min at 37°C until the cells have detached (check with an inverted microscope).
Add 100 µl William’s B medium per well to stop trypsinization.
Mix by gently pipetting up and down (using a multichannel pipette) and pool the contents of the wells (containing the detached cells and fluids) into a reagent reservoir.
Transfer the material into a 50-ml tube and spin (2 min 200 × g, RT, brake medium).
Wash twice in 20% William’s B medium (each time spin 2 min 200 × g, RT, brake medium).
Pass the sample, while gently pipetting up and down, through a cell strainer snap cap (on top of 5-ml round-bottomed polystyrene test tubes) to exclude clumps.
Keep cells on ice until sorting.
Fluorescence-activated cell sorting
Pre-cool the sample loader at 4°C and rotate at 300 rpm during cell sorting.
Set up the FACSAria according to the manufacturer’s procedures, e.g., run Cytometer Setup and Tracking beads (CS&T).
Install a 100-µm nozzle and set the sheath pressure to 20 psi (default setting).
Record 100,000 events of the uninfected hepatocyte culture as well as the transgenic liver-stage parasite culture to check the gating settings.
Insert 1.5-ml collection tubes filled with 300 µl TRIzol if the material is to be used for transcriptomics
NB: TRIzol is toxic: fill the tubes in the fume hood and dispose of the waste separately.
Sort (sort precision: 4-way purity mode) the fractions of interest (Figure 2).
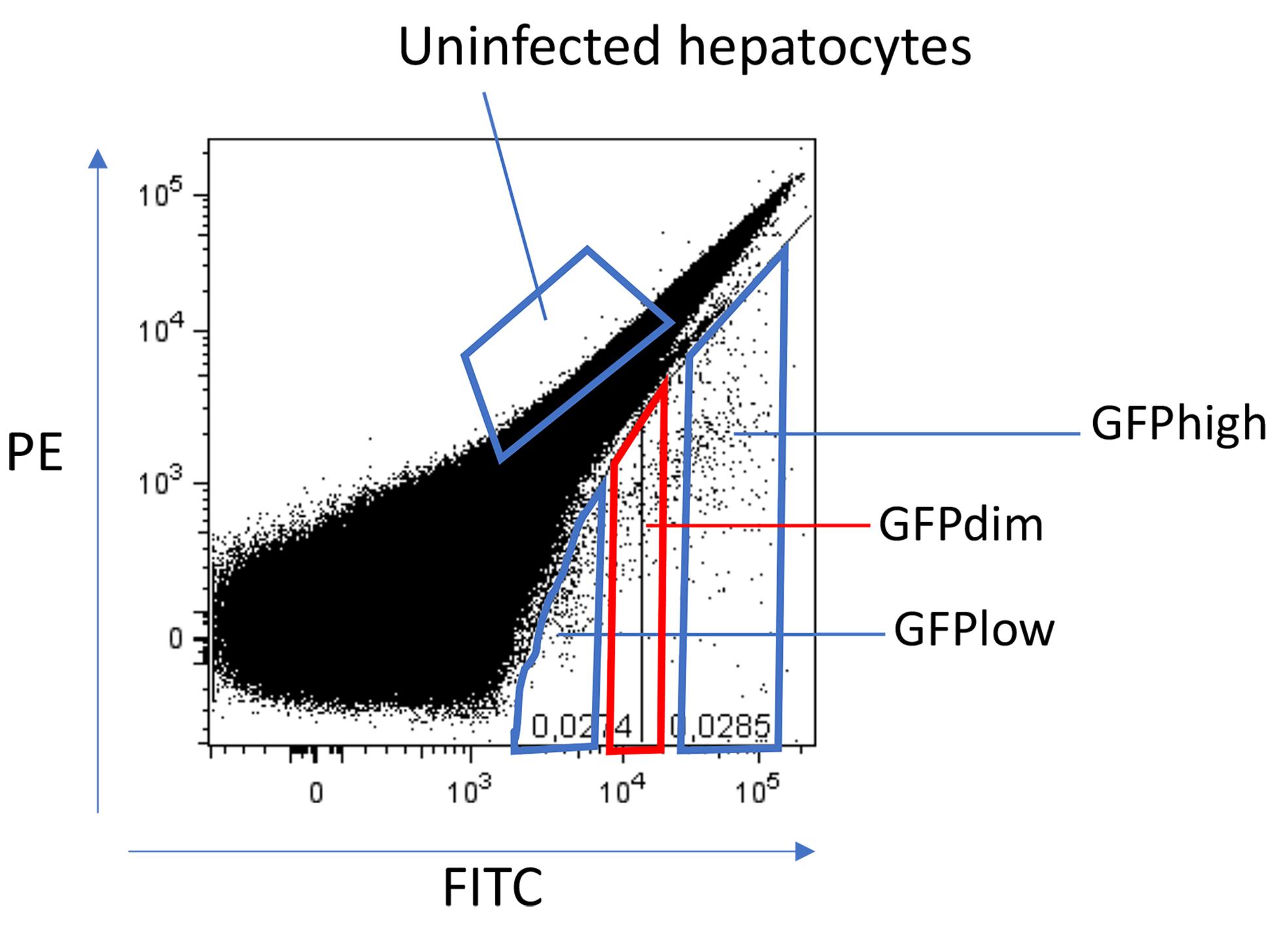
Figure 2. Gating strategy for sorting hypnozoites and schizonts on day 6. Parasite populations are gated based on FITC expression. The GFPlow gate contains hypnozoites and the GFPhigh gate contains developing forms (schizonts). The GFPdim gate contains a mixture of parasites.When approximately half of the sample has been sorted, sort a small amount of sample into 100 µl 20% William’s B (collect about 100 GFPlow and GFPhigh events each; also take a sample of the other fractions at the same time) and transfer to a 96-well plate for a quality check of the Operetta system (Voorberg-van der Wel et al., 2013 ).
When the sort has finished, vortex the TRIzol tubes for 30 s and transfer to the -80°C freezer.
Notes
Please note that working with the materials listed above potentially involves biological risks. Monkey-derived hepatocytes should be treated as biohazard material and may contain infectious agents. P. cynomolgi parasites are infectious to humans. All handling of these materials should be performed under BSL2 conditions.
While the original work was performed in 6-well plates, we have noticed that infection grades in 96-well plates are higher.
Instead of using uninfected hepatocytes as the negative control, macaque primary hepatocytes infected with P. cynomolgi wild-type parasites may be used (Voorberg-van der Wel et al., 2013 ).
Hepatocytes are highly autofluorescent; therefore, it is difficult to distinguish between uninfected cells and those containing GFPlow-expressing parasites (hypnozoites). We have empirically determined that using the PE channel in combination with the FITC channel provides an efficient method to separate the different parasitic populations.
Implementing a doublet and debris exclusion through the addition of a scatterplot of FSC versus SSC, followed by a scatterplot of FSC-A versus FSC-H, may be beneficial for removing subcellular debris and multi-cell aggregates.
Instead of harvesting all the wells, it is recommended to perform live imaging and fix a few wells for IFA (https://bio-protocol.org/e3722) to establish the infection characteristics.
Co-staining with Sytox Blue can be included as a live/dead indicator, as described by others ( Posfai et al., 2018 ). Alternatively, a surface stain found on all intact hepatocytes could potentially be used in combination with, for example, PE-Cy7.
Depending on the cell sorting apparatus, a 130-µm nozzle can be used as an alternative to a 100-µm nozzle. This may be beneficial for maintaining host cell integrity.
Recipes
William’s B medium
William’s E medium with glutamax
10% human serum (A+)
1% MEM nonessential amino acids (NEAA)
2% penicillin/streptomycin
1% insulin/transferrin/selenium
1% sodium pyruvate
50 µM β-mercaptoethanol
0.05 µM hydrocortisone
500 ml William’s E medium
50 ml Human Serum A+
5 ml 100× pen/strep (10,000 U/ml)
5 ml 100× insulin/transferin/selenium supplement
5 ml 100 mM sodium pyruvate
5 ml 100× MEM-NEAA
250 µl 0.1 M hydrocortisone
500 µl 50 mM β-mercaptoethanol
Store at 4°C and use within 1 month
20% William’s B medium
Dilute William’s B medium 1:5 with William’s E medium
0.1 M hydrocortisone
Dissolve 50 mg hydrocortisone in 1 ml DMSO
Store 250-µl aliquots at 4°C
Acknowledgments
This work was supported by a grant from the Medicines for Malaria Venture, a translational research grant (WT078285) from the Wellcome Trust, FP7 EU grants MALSIG (contract number 223044) and EVIMALAR (contract number 242095), and the Bill and Melinda Gates foundation (OPP1141292).
Ethics
Nonhuman primates were used because no other models (in vitro or in vivo) were suitable for the aims of this project. The local independent ethics committee conforming to Dutch law (BPRC Dier Experimenten Commissie, DEC) approved the research protocol (agreement number DEC# 708) prior to the beginning of the study, and the experiments were performed according to Dutch and European laws. Further details can be found in the original publication (Voorberg-van der Wel et al., 2017).
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have influenced the work reported in this paper.
References
- Bertschi, N. L., Voorberg-van der Wel, A., Zeeman, A. M., Schuierer, S., Nigsch, F., Carbone, W., Knehr, J., Gupta, D. K., Hofman, S. O., van der Werff, N., Nieuwenhuis, I., Klooster, E., Faber, B. W., Flannery, E. L., Mikolajczak, S. A., Chuenchob, V., Shrestha, B., Beibel, M., Bouwmeester, T., Kangwanrangsan, N., Sattabongkot, J., Diagana, T. T., Kocken, C. H. and Roma, G. (2018). Transcriptomic analysis reveals reduced transcriptional activity in the malaria parasite Plasmodium cynomolgi during progression into dormancy. Elife 7: e41081.
- Bianconi, E., Piovesan, A., Facchin, F., Beraudi, A., Casadei, R., Frabetti, F., Vitale, L., Pelleri, M. C., Tassani, S., Piva, F., Perez-Amodio, S., Strippoli, P. and Canaider, S. (2013). An estimation of the number of cells in the human body. Ann Hum Biol 40(6): 463-471.
- Cubi, R., Vembar, S. S., Biton, A., Franetich, J. F., Bordessoulles, M., Sossau, D., Zanghi, G., Bosson-Vanga, H., Benard, M., Moreno, A., Dereuddre-Bosquet, N., Le Grand, R., Scherf, A. and Mazier, D. (2017). Laser capture microdissection enables transcriptomic analysis of dividing and quiescent liver stages of Plasmodium relapsing species. Cell Microbiol 19(8): e12735.
- Dembele, L., Gego, A., Zeeman, A. M., Franetich, J. F., Silvie, O., Rametti, A., Le Grand, R., Dereuddre-Bosquet, N., Sauerwein, R., van Gemert, G. J., Vaillant, J. C., Thomas, A. W., Snounou, G., Kocken, C. H. and Mazier, D. (2011). Towards an in vitro model of Plasmodium hypnozoites suitable for drug discovery. PLoS One 6(3): e18162.
- Gupta, D. K., Dembele, L., Voorberg-van der Wel, A., Roma, G., Yip, A., Chuenchob, V., Kangwanrangsan, N., Ishino, T., Vaughan, A. M., Kappe, S. H., Flannery, E. L., Sattabongkot, J., Mikolajczak, S., Bifani, P., Kocken, C. H. and Diagana, T. T. (2019). The Plasmodium liver-specific protein 2 (LISP2) is an early marker of liver stage development. Elife 8: e43362.
- Gural, N., Mancio-Silva, L., Miller, A. B., Galstian, A., Butty, V. L., Levine, S. S., Patrapuvich, R., Desai, S. P., Mikolajczak, S. A., Kappe, S. H. I., Fleming, H. E., March, S., Sattabongkot, J. and Bhatia, S. N. (2018). In Vitro Culture, Drug Sensitivity, and Transcriptome of Plasmodium vivax Hypnozoites. Cell Host Microbe 23(3): 395-406 e394.
- Joyner, C., Moreno, A., Meyer, E. V., Cabrera-Mora, M., Ma, H. C., Kissinger, J. C., Barnwell, J. W. and Galinski, M. R. (2016). Plasmodium cynomolgi infections in rhesus macaques display clinical and parasitological features pertinent to modelling vivax malaria pathology and relapse infections.Malar J 15(1): 451.
- Kocken, C. H., van der Wel, A. and Thomas, A. W. (1999). Plasmodium cynomolgi: transfection of blood-stage parasites using heterologous DNA constructs. Exp Parasitol 93(1): 58-60.
- Krotoski, W. A., Garnham, P. C., Bray, R. S., Krotoski, D. M., Killick-Kendrick, R., Draper, C. C., Targett, G. A. and Guy, M. W. (1982). Observations on early and late post-sporozoite tissue stages in primate malaria. I. Discovery of a new latent form of Plasmodium cynomolgi (the hypnozoite), and failure to detect hepatic forms within the first 24 hours after infection. Am J Trop Med Hyg 31(1): 24-35.
- Natarajan, R., Thathy, V., Mota, M. M., Hafalla, J. C., Menard, R. and Vernick, K. D. (2001). Fluorescent Plasmodium berghei sporozoites and pre-erythrocytic stages: a new tool to study mosquito and mammalian host interactions with malaria parasites. Cell Microbiol 3(6): 371-379.
- Pasini, E. M., Bohme, U., Rutledge, G. G., Voorberg-Van der Wel, A., Sanders, M., Berriman, M., Kocken, C. H. and Otto, T. D. (2017). An improved Plasmodium cynomolgi genome assembly reveals an unexpected methyltransferase gene expansion. Wellcome Open Res 2: 42.
- Posfai, D., Sylvester, K., Reddy, A., Ganley, J. G., Wirth, J., Cullen, Q. E., Dave, T., Kato, N., Dave, S. S. and Derbyshire, E. R. (2018). Plasmodium parasite exploits host aquaporin-3 during liver stage malaria infection. PLoS Pathog 14(5): e1007057.
- Prudencio, M., Rodrigues, C. D., Ataide, R. and Mota, M. M. (2008). Dissecting in vitro host cell infection by Plasmodium sporozoites using flow cytometry. Cell Microbiol 10(1): 218-224.
- Roth, A., Maher, S. P., Conway, A. J., Ubalee, R., Chaumeau, V., Andolina, C., Kaba, S. A., Vantaux, A., Bakowski, M. A., Thomson-Luque, R., Adapa, S. R., Singh, N., Barnes, S. J., Cooper, C. A., Rouillier, M., McNamara, C. W., Mikolajczak, S. A., Sather, N., Witkowski, B., Campo, B., Kappe, S. H. I., Lanar, D. E., Nosten, F., Davidson, S., Jiang, R. H. Y., Kyle, D. E. and Adams, J. H. (2018). A comprehensive model for assessment of liver stage therapies targeting Plasmodium vivax and Plasmodium falciparum. Nat Commun 9(1): 1837.
- Sattabongkot, J., Yimamnuaychoke, N., Leelaudomlipi, S., Rasameesoraj, M., Jenwithisuk, R., Coleman, R. E., Udomsangpetch, R., Cui, L. and Brewer, T. G. (2006). Establishment of a human hepatocyte line that supports in vitro development of the exo-erythrocytic stages of the malaria parasites Plasmodium falciparum and P. vivax. Am J Trop Med Hyg 74(5): 708-715.
- Schmidt, L. H. (1983). Appraisals of compounds of diverse chemical classes for capacities to cure infections with sporozoites of Plasmodium cynomolgi. Am J Trop Med Hyg 32(2): 231-257.
- Tarun, A. S., Baer, K., Dumpit, R. F., Gray, S., Lejarcegui, N., Frevert, U. and Kappe, S. H. (2006). Quantitative isolation and in vivo imaging of malaria parasite liver stages. Int J Parasitol 36(12): 1283-1293.
- Voorberg-van der Wel, A. M., Zeeman, A. M., Nieuwenhuis, I. G., van der Werff, N. M., Klooster, E. J., Klop, O., Vermaat, L. C., Kumar Gupta, D., Dembele, L., Diagana, T. T. and Kocken, C. H. M. (2020). A dual fluorescent Plasmodium cynomolgi reporter line reveals in vitro malaria hypnozoite reactivation. Commun Biol 3: 7.
- Voorberg-van der Wel, A., Roma, G., Gupta, D. K., Schuierer, S., Nigsch, F., Carbone, W., Zeeman, A. M., Lee, B. H., Hofman, S. O., Faber, B. W., Knehr, J., Pasini, E., Kinzel, B., Bifani, P., Bonamy, G. M. C., Bouwmeester, T., Kocken, C. H. M. and Diagana, T. T. (2017). A comparative transcriptomic analysis of replicating and dormant liver stages of the relapsing malaria parasite Plasmodium cynomolgi.Elife 6: e29605.
- Voorberg-van der Wel, A., Zeeman, A. M., van Amsterdam, S. M., van den Berg, A., Klooster, E. J., Iwanaga, S., Janse, C. J., van Gemert, G. J., Sauerwein, R., Beenhakker, N., Koopman, G., Thomas, A. W. and Kocken, C. H. (2013). Transgenic fluorescent Plasmodium cynomolgi liver stages enable live imaging and purification of Malaria hypnozoite-forms. PLoS One 8(1): e54888.
- Yan, G., Zhang, G., Fang, X., Zhang, Y., Li, C., Ling, F., Cooper, D. N., Li, Q., Li, Y., van Gool, A. J., Du, H., Chen, J., Chen, R., Zhang, P., Huang, Z., Thompson, J. R., Meng, Y., Bai, Y., Wang, J., Zhuo, M., Wang, T., Huang, Y., Wei, L., Li, J., Wang, Z., Hu, H., Yang, P., Le, L., Stenson, P. D., Li, B., Liu, X., Ball, E. V., An, N., Huang, Q., Zhang, Y., Fan, W., Zhang, X., Li, Y., Wang, W., Katze, M. G., Su, B., Nielsen, R., Yang, H., Wang, J., Wang, X. and Wang, J. (2011). Genome sequencing and comparison of two nonhuman primate animal models, the cynomolgus and Chinese rhesus macaques. Nat Biotechnol 29(11): 1019-1023.
- Zeeman, A. M., Lakshminarayana, S. B., van der Werff, N., Klooster, E. J., Voorberg-van der Wel, A., Kondreddi, R. R., Bodenreider, C., Simon, O., Sauerwein, R., Yeung, B. K., Diagana, T. T. and Kocken, C. H. (2016). PI4 Kinase Is a Prophylactic but Not Radical Curative Target in Plasmodium vivax-Type Malaria Parasites. Antimicrob Agents Chemother 60(5): 2858-63.
- Zeeman, A. M., van Amsterdam, S. M., McNamara, C. W., Voorberg-van der Wel, A., Klooster, E. J., van den Berg, A., Remarque, E. J., Plouffe, D. M., van Gemert, G. J., Luty, A., Sauerwein, R., Gagaring, K., Borboa, R., Chen, Z., Kuhen, K., Glynne, R. J., Chatterjee, A. K., Nagle, A., Roland, J., Winzeler, E. A., Leroy, D., Campo, B., Diagana, T. T., Yeung, B. K., Thomas, A. W. and Kocken, C. H. (2014). KAI407, a potent non-8-aminoquinoline compound that kills Plasmodium cynomolgi early dormant liver stage parasites in vitro.Antimicrob Agents Chemother 58(3): 1586-95.
Article Information
Copyright
Voorberg-van der Wel et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Voorberg-van der Wel, A., Hofman, S. O. and Kocken, C. H. M. (2021). Isolation of GFP-expressing Malarial Hypnozoites by Flow Cytometry Cell Sorting. Bio-protocol 11(9): e4006. DOI: 10.21769/BioProtoc.4006.
- Voorberg-van der Wel, A., Roma, G., Gupta, D. K., Schuierer, S., Nigsch, F., Carbone, W., Zeeman, A. M., Lee, B. H., Hofman, S. O., Faber, B. W., Knehr, J., Pasini, E., Kinzel, B., Bifani, P., Bonamy, G. M. C., Bouwmeester, T., Kocken, C. H. M. and Diagana, T. T. (2017). A comparative transcriptomic analysis of replicating and dormant liver stages of the relapsing malaria parasite Plasmodium cynomolgi.Elife 6: e29605.
Category
Microbiology > Microbial cell biology > Cell isolation and culture
Microbiology > in vivo model > Protozoan
Cell Biology > Cell isolation and culture > Cell isolation > Flow cytometry
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









