- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
COVID-19 Sample Pooling: From RNA Extraction to Quantitative Real-time RT-PCR
(*contributed equally to this work) Published: Vol 11, Iss 9, May 5, 2021 DOI: 10.21769/BioProtoc.4005 Views: 9057
Reviewed by: Alessandro DidonnaVaibhav B. ShahRan Chen

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Binding Affinity Measurements Between DNA Aptamers and their Virus Targets Using ELONA and MST
Gregory T. Pawel [...] Ana Sol Peinetti
Nov 5, 2022 3381 Views

Development of a Rapid Epstein–Barr Virus Detection System Based on Recombinase Polymerase Amplification and a Lateral Flow Assay
Yidan Sun [...] Chao Shen
Dec 5, 2024 1619 Views

Novel Workflows for Separate Isolation of Pathogen RNA or DNA From Wastewater: Detection by Innovative and Conventional qPCR
Kristina M. Babler [...] Ayaaz Amirali
Feb 20, 2025 3197 Views
Abstract
The COVID-19 pandemic requires mass screening to identify those infected for isolation and quarantine. Individually screening large populations for the novel pathogen, Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), is costly and requires a lot of resources. Sample pooling methods improve the efficiency of mass screening and consume less reagents by increasing the capacity of testing and reducing the number of experiments performed, and are therefore especially suitable for under-developed countries with limited resources. Here, we propose a simple, reliable pooling strategy for COVID-19 testing using clinical nasopharyngeal (NP) and/or oropharyngeal (OP) swabs. The strategy includes the pooling of 10 NP/OP swabs for extraction and subsequent testing via quantitative real-time reverse transcription polymerase chain reaction (RT-qPCR), and may also be applied to the screening of other pathogens.
Keywords: COVID-19Background
The coronavirus disease (COVID-19) pandemic has resulted in significant socioeconomic and public health burden in affected countries worldwide. The unprecedented spread of the disease has placed healthcare systems under considerable pressure, particularly in efforts to detect cases as efficiently as possible, contain the disease, and treat cases in a timely manner. Disease surveillance and the confirmation of positive COVID-19 cases has also mostly relied upon quantitative real-time reverse transcription polymerase chain reaction (RT-qPCR) of nasopharyngeal/oropharyngeal (NP/OP) swab specimens for the detection of the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) following WHO guidelines (Corman et al., 2020). However, the RT-qPCR testing capacity can be highly variable, particularly in rural areas, where primary healthcare facilities may not have trained personnel or the necessary equipment and facilities to carry out confirmatory testing. Hence, samples are sent to centralized facilities, which in turn are met with an overwhelmingly high demand for testing. As COVID-19 cases continue to rise, these laboratories have begun to face a backlog of pending tests and rising turnaround times that affect the clinical utility of results and timely surveillance of populations at the greatest risk.
In efforts to increase the cost-effectiveness and capacity of COVID-19 screening, we developed a pooling strategy during nucleic acid extraction without a reduction in the sensitivity of RT-qPCR (Lim et al., 2020). The pooling of clinical samples has previously been employed for the screening of the human immunodeficiency virus (HIV), hepatitits C virus (HCV) (Hourfar et al., 2008), influenza viruses (Pilcher et al., 2005; Sullivan et al., 2011), and during the COVID-19 pandemic, SARS-CoV-2 (Lim et al., 2020). This method has been demonstrated to be cost-effective by allowing an increase in the viral detection capacity for mass screening while maintaining testing accuracy (Abdalhamid et al., 2020). Pooled testing for RT-qPCR has been frequently utilized (Hogan et al., 2020; Lohse et al., 2020), unlike the strategy described in this paper, where we pool samples during nucleic acid extraction prior to performing RT-qPCR. This method has been evaluated as an effective approach in recent studies (Garg et al., 2020; Wacharapluesadee et al., 2020), where in a similar fashion, NP/OP swab samples were pooled in Viral Transport Medium (VTM) for extraction. Findings by Yelin et al., (2020) suggest that even in pools of up to 32 samples, one positive sample could be detected.
Pooling during the extraction stage provides additional reductions in testing costs and workload. Using the strategy described in this protocol leads to a 9-fold reduction in the number of nucleic acid extractions and RT-qPCR reactions required (Lim et al., 2020). For laboratories conducting screening for SARS-CoV-2, the pooled testing strategy used in this protocol does not introduce any significant changes to the current testing workflow and may be incorporated into routine procedures. We note that the optimal pool size will be dependent upon, among others, the prevalence of COVID-19, the objective of the testing strategy, and the resources available to the testing facility; therefore, it is recommended that laboratories account for these differences when adopting this strategy.
Thus, in this paper, we aim to provide a step-by-step protocol that details the pooling strategy for the screening of SARS-CoV-2. The strategy involves the pooling of 10 NP/OP swabs for extraction and subsequent testing by RT-qPCR. Implementing such a pooling strategy can minimize the costs incurred and the reagents utilized, while simultaneously reducing turnaround times for the rapid identification and isolation of positive COVID-19 cases. This protocol may also be applied to other pathogens that are screened using similar techniques, and would be particularly useful for the community screening of large groups in resource-poor settings and/or where the prevalence of infection is low.
Materials and Reagents
Personal protective equipment (PPE) appropriate for working with SARS-CoV-2
0.2 ml low-profile PCR tube strips with caps (Bio-Rad, catalog numbers: TLS0851 and TCS0903)
1.5 ml microcentrifuge tubes (Axygen, catalog number: MCT-150-C-S)
0.5-10 µl, 0.5-50 µl, 1-200 µl, 100-1,000 µl filtered pipette tips (Axygen, catalog numbers: TF-400-R-S, TF-50-R-S , TF-200-R-S, TF-1005-WB-R-S)
Geneaid Viral Nucleic Acid Extraction Kit (Geneaid Biotech, catalog number: VR300)
Qiagen DNeasy Blood and Tissue Kit (Qiagen, catalog number: 69506)
SuperScriptTM III One-Step RT-PCR System with PlatinumTM Taq DNA Polymerase (Invitrogen, catalog number: 12574-026) (aliquot and store at -20°C)
DNase- and RNase-free water (Integrated DNA Technologies, catalog number: 11-05-01-04) (aliquot into clean 1.5-ml microcentrifuge tubes)
Primers for SARS-CoV-2 (Integrated DNA Technologies) (Table 1)
Table 1. Primers and probes targerting RNA-dependent RNA polymerase (RdRP), SARS-CoV-2 detectiona
Catalog number Oligonucleotide Sequenceb CV001 RdRp_SARSr-F 5’-GTGARATGGTCATGTGTGGCGG-3’ CV002 RdRp_SARSr-R 5’-CARATGTTAAASACACTATTAGCATA-3’ CV101 RdRP_SARSr-P1 5’-FAM-CCAGGTGGWACRTCATCMGGTGATGC-BBQ-3’ CV102 RdRp_SARSr-P2 5’-FAM-CAGGTGGAACCTCATCAGGAGATGC-BBQ-3’ a Oligonucleotide sequences were adapted from the WHO-Charité protocol (Corman et al., 2020).
b W is A/T; R is G/A; M is A/C; S is G/C. FAM: 6-carboxyfluorescein; BBQ: blackberry quencher.
Primer mix (see Recipes)
Equipment
1 µl-10 µl, 10 µl-100 µl, 20 µl-200 µl, 100 µl-1,000 µl micropipettors (Eppendorf, catalog numbers: 3123000020, 3123000047, 3123000055, 3123000063)
CFX96 RealTime System (Bio-Rad, model: C1000 touch)
Thermo Aluminium Bath (Fine PCR, model: ALB128) to be used with the Qiagen DNeasy Blood and Tissue Kit
Centrifuge 5425 (Eppenddorf, catalog number: 5405000417)
Procedure
All procedures in (A) sample pooling, (B) preparation of positive sample-pool, (C) ribonucleic acid (RNA) extraction, and (D) quantitative reverse transcription polymerase chain reaction (RT-qPCR) were carried out separately inside different biosafety cabinets. All clinical waste and consumables were autoclaved prior to being sent for incineration.
Sample pooling (see Figure 1)
We chose 10 sample-pools since the positive detection rate in our laboratory is less than 2%. You may optimize the sample-pool size according to the positive detection rate in your laboratory (Abdalhamid et al., 2020).
Pipette 60 µl each of the 10 clinical NP/OP swabs into a single 1.5-ml microcentrifuge tube. The total volume is 600 µl. The original volume of the specimens ranged from 1 ml to 3 ml.
Aliquot 300 µl pooled swabs to each of two clean 1.5-ml microcentrifuge tubes and label with the same sample ID.
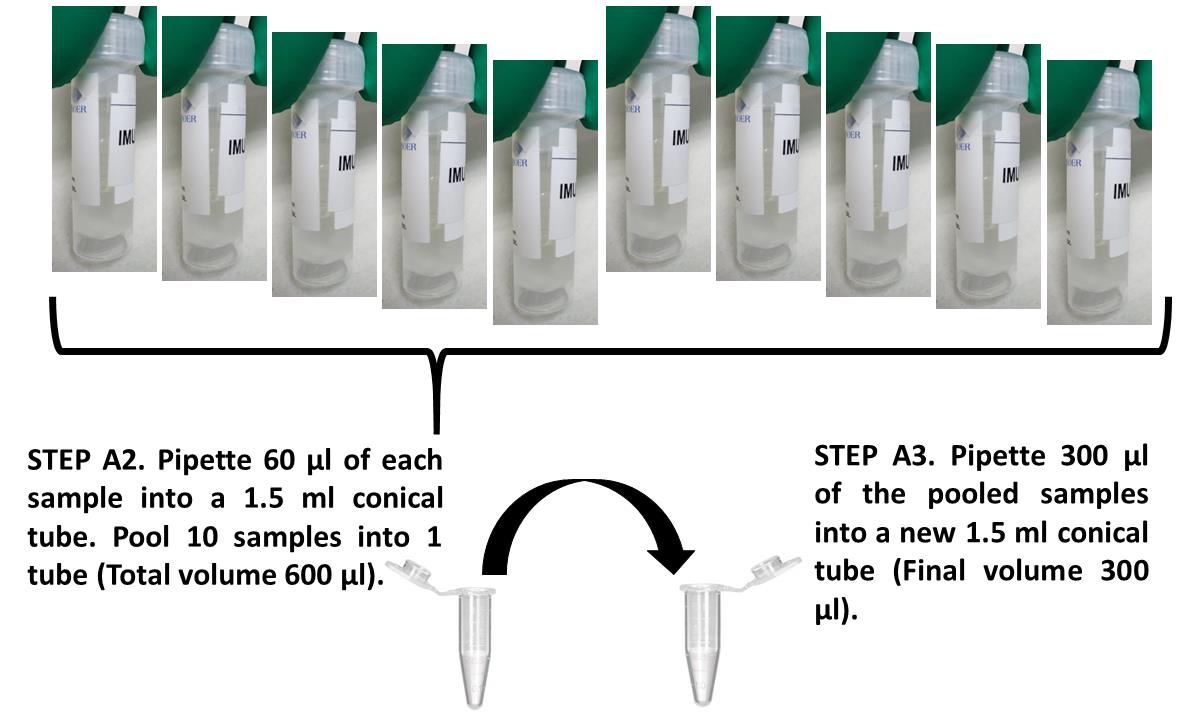
Figure 1. Pooling of 10 individual samples
Preparation of positive sample-pool
Identify a positive NP/OP swab that has a CT value ≥15.00 but ≤36.00 (refer to Table S2 in Lim et al., 2020).
Aliquot the identified positive NP/OP swab and store at -20°C.
Prepare the positive sample-pool according to Steps A2-A3 by mixing 1 positive with 9 unknown samples.
RNA extraction
Choose either of the kits below for RNA extraction
Geneaid Viral Nucleic Acid Extraction Kit
Add 400 µl VB Lysis Buffer to each of the 1.5-ml microcentrifuge tubes. Shake the tube virgorously for 10 s.
Incubate at room temperature for 10 min.
Add 450 µl AD Buffer (make sure that the ethanol has been added) to each of the 1.5-ml microcentrifuge tubes. Shake the tube vigorously to mix.
Transfer 700 µl lysate mixture to the VB column in a 2-ml collection tube. Centrifuge at 14,000 × g for 30 s. Discard the flowthrough and place the VB Column back into the 2-ml collection tube. Repeat Step (iv) until all the mixture (two 1.5-ml microcentrifuge tubes prepared in Step A3) has completely flowed through the VB column.
Add 400 µl W1 Buffer to the VB column and centrifuge at 14,000 × g for 30 s. Discard the flowthrough and place the VB column back into the 2-ml collection tube.
Add 600 µl Wash Buffer (make sure that the ethanol has been added) to the VB column. Centrifuge at 14,000 × g for 30 s. Discard the flowthrough and place the VB column back into the 2-ml collection tube.
Centrifuge at 14,000 × g for 3 min to dry the column matrix.
Place the dried VB column in a clean 1.5-ml microcentrifuge tube. Add 35 µl DNAse RNase-free water to the center of the VB column matrix and incubate for 3 min at room temperature (15-25°C).
Centrifuge at ≥6,000 × g for 1 min to elute the purified nucleic acid.
Qiagen DNeasy Blood and Tissue Kit
Pipette 20 µl Proteinase K into each 1.5-ml microcentrifuge tube.
Add 200 µl Buffer AL and mix thoroughly by shaking vigorously.
Incubate at 56°C for 10 min.
Add 200 µl ethanol (96-100%) and mix thoroughly by shaking vigorously.
Pipette 700 µl mixture into a DNeasy Mini spin column placed in a 2-ml collection tube. Centrifuge at ≥6,000 × g for 1 min. Repeat Step (v) until all the mixture (two 1.5-ml microcentrifuge tubes prepared in Step A3) has completely flowed through the spin column. Discard the flowthrough and collection tube.
Place the spin column into a clean 2-ml collection tube.
Add 500 µl Buffer AW1 and centrifuge for 1 min at ≥6,000 × g.
Discard the flowthrough and collection tube.
Place the spin column in a clean 2-ml collection tube, add 500 µl Buffer AW2, and centrifuge for 3 min at 14,000 × g.
Discard the flowthrough and collection tube.
Transfer the spin column into a clean 1.5-ml microcentrifuge tube.
Elute the RNA by adding 35 µl DNAse- and RNAse-free water to the center of the spin column membrane. Incubate for 3 min at room temperature (15-25°C).
Centrifuge for 1 min at ≥6,000 × g to elute the purified nucleic acid.
RT-qPCR
Prepare the RdRP-Primer/Probe mixture (10 µM) (see Recipe).
Prepare 3 tests: (1) positive sample-pool; (2) non-template control (NTC); and (3) unknown sample-pool.
Prepare the RT-qPCR mixture using the SuperScriptTM III One-Step RT-PCR System with PlatinumTM Taq DNA Polymerase as described in Table 2.
Table 2. RT-qPCR mixture preparationReagent Final concentration Volume (µl) for one reaction 2× reaction buffer 1× 10.0 SuperScriptTM III RT/PlatinumTM Taq Mix - 0.5 RdRP-Primer/Probe mixture (10 µM) 2 µM 1.0 Purified nucleic acid (10 sample pool)a - 8.5 Total volume 20.0 a Replace purified nucleic acid with DNAse- and RNAse-free water for NTC.
Run the RT-qPCR cycles as described below:
55°C for 15 min
95°C for 2 min
95°C for 15 s and 56°C for 30 s for 50 cycles
Data analysis
Intepret and record the results using the criteria below (see Notes for explanation):
If the CT value is ≤14.00, consider it as contamination. Repeat the RT-qPCR of the pooled purified nucleic acid. Upon detection of the same CT value in the repeated RT-qPCR, deconvolute the positive pool(s) and repeat the nucleic acid extraction and RT-qPCR on individual samples from the positive pool(s). Test results are confirmed by a conventional RT-qPCR clinical test on a single sample.
If the CT value is >14.01 but ≤45, record the CT value and consider positive detection of SARS-CoV-2 in the pooled samples. Deconvolute the positive pool(s) and repeat the nucleic acid extraction and RT-qPCR on individual samples from the positive pool(s). Test results are confirmed by a conventional RT-qPCR clinical test on a single sample.
If the CT value is ≥45.01 or no CT value is detected, record the pooled samples as negative. Each single sample is reported as negative in this pool.
An example of the analysis is shown in Tables 3, 4, and 5 (extracted Dataset S1 from Lim et al., 2020). Here, we briefly describe the data shown in Tables 3-5.
On 6th April 2020, the total number of specimens to be extracted was 101 (including an internal positive control). We performed sample pooling according to Steps A1-A3. Groups A-I were pools of 10, while Groups J and K were pools of 5 (total volume 300 µl) and 6 (total volume 360 µl), respectively. The RNA was extracted from the pooled samples as per Steps C1a.i-ix. The RT-qPCR was performed as per Steps D1-D4. Results of pooled testing are indicated in Table 3. SARS-CoV-2 was detected in Groups I and J. Individual specimens from both groups were individually extracted again according to the manufacturer’s protocol and RT-qPCR was performed as per Steps D1-D4. The results of individual testing are stated in Tables 4 and 5 for Groups I and J, respectively.
Table 3. April 7th, 2020. IMU COVID-19 pooled testing results
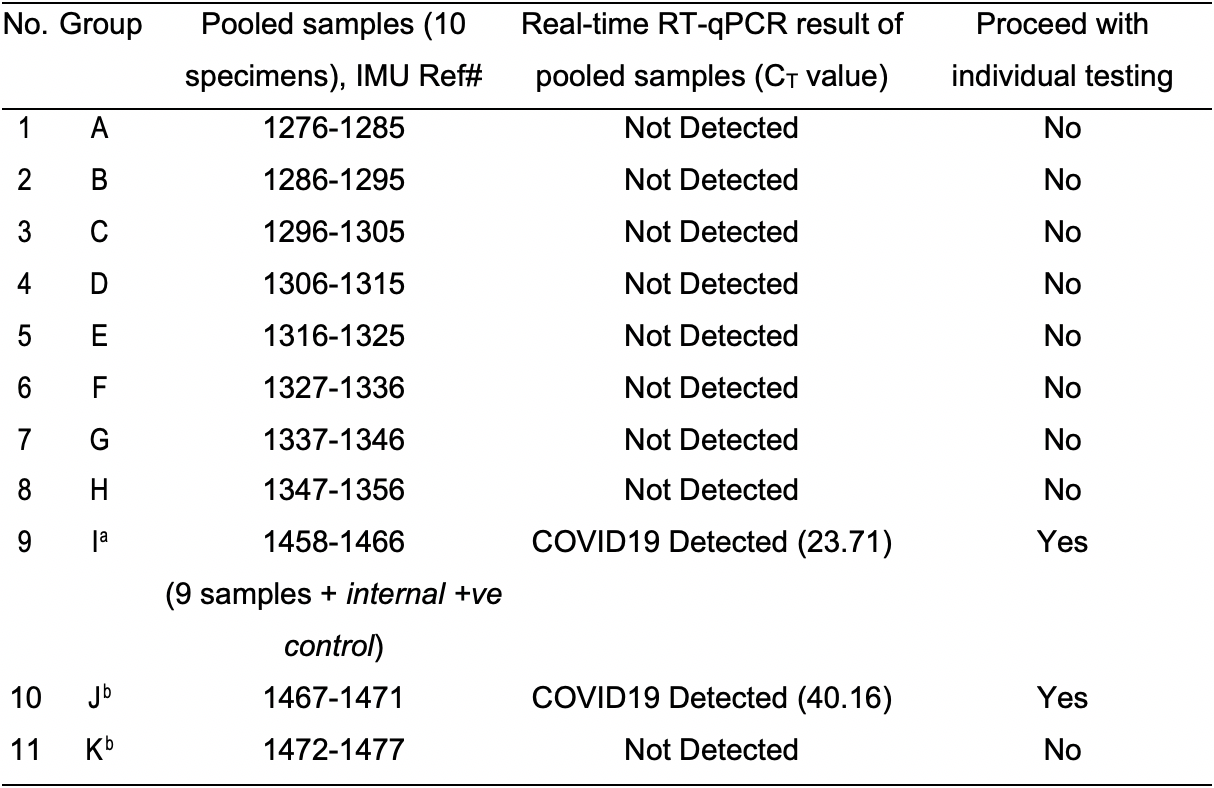
aGroup I was prepared as per step B3, spiked with one internal positive control (9 samples + 1 internal +ve control).
bGroups J and K were pools of 5 (total volume 300 µl) and 6 (total volume 360 µl), respectively. Aliquots of 150 µl (Group J) and 180 µl (Group K) were pipetted into two 1.5-ml microcentrifuge tubes as per step A3.
Table 4. Individual testing RT-qPCR results for pooled Group IGroup I IMU Ref# CT value Pooled testing (1st qRT-PCR) Individual (2nd qRT-PCR) 1 1458
COVID19 Detected (23.71)Not Detected 2 1459 Not Detected 3 1460 Not Detected 4 1461 Not Detected 5 1462 Not Detected 6 1463 Not Detected 7 1464 Not Detected 8 1465 Not Detected 9 1466 Not Detected 10 Internal +ve control COVID19 Detected (23.23)
Table 5. Individual testing RT-qPCR results for pooled Group JGroup J IMU Ref# CT value Pooled testing (1st RT-qPCR) Individual (2nd RT-qPCR) 1 1467
COVID19 Detected (40.16)Not Detected 2 1468 Not Detected 3 1469 Not Detected 4 1470 Not Detected 5 1471 Not Detected
Notes
CT value: We chose the CT value cut-off based on our preliminary data obtained by optimizing the pooling. We noticed that a weak positive CT value of 38 may increase to 40 after pooling; thus, we recommend a CT value cut-off of 45 to safeguard the accuracy and precision of the detection method. Any pooled samples considered to be positive, with CT values detected below 45, will need to be individually tested for accurate reporting. Please see the example provided in Table 3.
Recipes
Primer mix
Prepare each one of the primers and probes at 100 µM
Primer/probe mix (100 µl):
RdRp_SARSr-F 10 µl (10 µM final concentration)
RdRp_SARSr-R 10 µl (10 µM final concentration)
RdRP_SARSr-P1 10 µl (10 µM final concentration)
RdRp_SARSr-P2 10 µl (10 µM final concentration)
DNase- and RNase-free water 60 µl
Aliquot and store at -20°C
Acknowledgments
This study was supported by a grant from the International Medical University (IMU), Malaysia (IMU 477-2020). We thank the National Public Health Laboratory (NPHL), Ministry of Health Malaysia for their generosity and support in sharing the necessary reagents for laboratory testing. This protocol was derived from our previous work (Lim et al., 2020).
Competing interests
The author(s) declare no potential conflicts of interest with respect to the research, authorship, and/or publication of this article. While performing this research, no company had invested in the research and no commercialization was intended.
Ethics
This study obtained ethical clearance from the International Medical University Joint Research and Ethical Committee (EC/IRB Ref. No. 4.41/JCM-196/2020). All clinical specimens used in this study were fully anonymized and deidentified by assigning new laboratory reference numbers before access was given to the researchers. The researchers were blinded to patient information.
References
- Abdalhamid, B., Bilder, C. R., McCutchen, E. L., Hinrichs, S. H., Koepsell, S. A. and Iwen, P. C. (2020). Assessment of Specimen Pooling to Conserve SARS CoV-2 Testing Resources. Am J Clin Pathol 153(6): 715-718.
- Corman, V. M., Landt, O., Kaiser, M., Molenkamp, R., Meijer, A., Chu, D. K., Bleicker, T., Brunink, S., Schneider, J., Schmidt, M. L., Mulders, D. G., Haagmans, B. L., van der Veer, B., van den Brink, S., Wijsman, L., Goderski, G., Romette, J. L., Ellis, J., Zambon, M., Peiris, M., Goossens, H., Reusken, C., Koopmans, M. P. and Drosten, C. (2020). Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25(3): 2000045.
- Garg, J., Singh, V., Pandey, P., Verma, A., Sen, M., Das, A. and Agarwal, J. (2021). Evaluation of sample pooling for diagnosis of COVID-19 by real time-PCR: A resource-saving combat strategy. J Med Virol 93(3): 1526-1531.
- Hogan, C. A., Sahoo, M. K. and Pinsky, B. A. (2020). Sample pooling as a strategy to detect community transmission of SARS-CoV-2. JAMA 323(19): 1967-1969.
- Hourfar, M. K., Jork, C., Schottstedt, V., Weber-Schehl, M., Brixner, V., Busch, M. P., Geusendam, G., Gubbe, K., Mahnhardt, C., Mayr-Wohlfart, U., Pichl, L., Roth, W. K., Schmidt, M., Seifried, E., Wright, D. J. and German Red Cross, N. A. T. S. G. (2008). Experience of German Red Cross blood donor services with nucleic acid testing: results of screening more than 30 million blood donations for human immunodeficiency virus-1, hepatitis C virus, and hepatitis B virus. Transfusion 48(8): 1558-1566.
- Lim, K. L., Johari, N. A., Wong, S. T., Khaw, L. T., Tan, B. K., Chan, K. K., Wong, S. F., Chan, W. L. E., Ramzi, N. H., Lim, P. K. C., Hakim, S. L. and Voon, K. (2020). A novel strategy for community screening of SARS-CoV-2 (COVID-19): Sample pooling method. PLoS One 15(8): e0238417.
- Lohse, S., Pfuhl, T., Berko-Gottel, B., Rissland, J., Geissler, T., Gartner, B., Becker, S. L., Schneitler, S. and Smola, S. (2020). Pooling of samples for testing for SARS-CoV-2 in asymptomatic people. Lancet Infect Dis 20(11): 1231-1232.
- Pilcher, C. D., Fiscus, S. A., Nguyen, T. Q., Foust, E., Wolf, L., Williams, D., Ashby, R., O'Dowd, J. O., McPherson, J. T., Stalzer, B., Hightow, L., Miller, W. C., Eron, J. J., Jr., Cohen, M. S. and Leone, P. A. (2005). Detection of acute infections during HIV testing in North Carolina. N Engl J Med 352(18): 1873-1883.
- Sullivan, T. J., Patel, P., Hutchinson, A., Ethridge, S. F. and Parker, M. M. (2011). Evaluation of pooling strategies for acute HIV-1 infection screening using nucleic acid amplification testing. J Clin Microbiol 49(10): 3667-3668.
- Wacharapluesadee, S., Kaewpom, T., Ampoot, W., Ghai, S., Khamhang, W., Worachotsueptrakun, K., Wanthong, P., Nopvichai, C., Supharatpariyakorn, T., Putcharoen, O., Paitoonpong, L., Suwanpimolkul, G., Jantarabenjakul, W., Hemachudha, P., Krichphiphat, A., Buathong, R., Plipat, T. and Hemachudha, T. (2020). Evaluating the efficiency of specimen pooling for PCR-based detection of COVID-19. J Med Virol 92(10): 2193-2199.
- Yelin, I., Aharony, N., Tamar, E. S., Argoetti, A., Messer, E., Berenbaum, D., Shafran, E., Kuzli, A., Gandali, N., Shkedi, O., Hashimshony, T., Mandel-Gutfreund, Y., Halberthal, M., Geffen, Y., Szwarcwort-Cohen, M. and Kishony, R. (2020). Evaluation of COVID-19 RT-qPCR Test in Multi sample Pools. Clin Infect Dis 71(16): 2073-2078.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Voon, K., Johari, N. A., Lim, K. L., Wong, S. T., Khaw, L. T., Wong, S. F., Chan, E. W. L., Chan, K. K., Tan, B. K., Ramzi, N. H., Lim, P. K. C. and Sulaiman, L. H. (2021). COVID-19 Sample Pooling: From RNA Extraction to Quantitative Real-time RT-PCR. Bio-protocol 11(9): e4005. DOI: 10.21769/BioProtoc.4005.
Category
Microbiology > Pathogen detection > PCR
Biological Sciences > Microbiology
Biochemistry > Virus
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








