- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Analysis of TORC1-body Formation in Budding Yeast
(*contributed equally to this work) Published: Vol 11, Iss 7, Apr 5, 2021 DOI: 10.21769/BioProtoc.3975 Views: 4483
Reviewed by: Juan Facundo Rodriguez AyalaJohn P PhelanAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

TUNEL Labeling to Detect Double-stranded DNA Breaks in Caenorhabditis elegans Gonads
Peter A. Kropp [...] Andy Golden
Mar 20, 2022 2611 Views

Assessment of Chemosensory Response to Volatile Compounds in Healthy, Aged, and Neurodegenerative Caenorhabditis elegans Models
Cira Crespo and Roberto Grau
May 5, 2023 1516 Views

A Reproducible Method to Evaluate Sublethal Acoustic Stress in Aquatic Invertebrates Using Oxidative Biomarkers
Francesca Maria Mitton [...] María Paz Sal Moyano
Jan 20, 2026 158 Views
Abstract
The Target of Rapamycin kinase Complex I (TORC1) is the master regulator of cell growth and metabolism in eukaryotes. In the presence of pro-growth hormones and abundant nutrients, TORC1 is active and drives protein, lipid, and nucleotide synthesis by phosphorylating a wide range of proteins. In contrast, when nitrogen and/or glucose levels fall, TORC1 is inhibited, causing the cell to switch from anabolic to catabolic metabolism, and eventually enter a quiescent state. In the budding yeast Saccharomyces cerevisiae, TORC1 inhibition triggers the movement of TORC1 from its position around the vacuole to a single focus/body on the edge of the vacuolar membrane. This relocalization depends on the activity of numerous key TORC1 regulators and thus analysis of TORC1 localization can be used to follow signaling through the TORC1 pathway. Here we provide a detailed protocol for measuring TORC1 (specifically, Kog1-YFP) relocalization/signaling using fluorescence microscopy. Emphasis is placed on procedures that ensure: (1) TORC1-bodies are identified (and counted) correctly despite their relatively low fluorescence and the accumulation of autofluorescent foci during glucose and nitrogen starvation; (2) Cells are kept in log-phase growth at the start of each experiment so that the dynamics of TORC1-body formation are monitored correctly; (3) The appropriate fluorescent tags are used to avoid examining mislocalized TORC1.
Keywords: TORC1Background
The Target of Rapamycin kinase Complex I (TORC1) is the central hub in the cell growth control network of eukaryotes (Loewith and Hall, 2011; Gonzalez and Hall, 2017; Liu and Sabatini, 2020). In the presence of pro-growth hormones and abundant nutrients, TORC1 is active and phosphorylates a wide array of proteins to drive protein and ribosome synthesis, activate lipid and nucleotide synthesis, tune nitrogen and amino acid metabolism/transport, and repress autophagy (Kamada et al., 2000; Huber et al., 2009; Hsu et al., 2011; Kim et al., 2011; Loewith and Hall, 2011; Peterson et al., 2011; Robitaille et al., 2013; Ben-Sahra et al., 2016; Gonzalez and Hall, 2017; Ben-Sahra and Manning, 2017; Liu and Sabatini, 2020). In contrast, when cells are exposed to stress or starvation conditions, TORC1 is inactivated to limit cell growth and redirect available resources to the appropriate stress or starvation response (Barbet et al., 1996; Duvel et al., 2010).
Over the last 15 years, detailed analysis of TORC1 signaling has shed light on the protein network that transmits stress and starvation signals to TORC1, but many questions remain about how TORC1 is regulated in the wide range of stimuli that influence cell growth and survival (Loewith and Hall, 2011; Gonzalez and Hall, 2017; Liu and Sabatini, 2020). In the model organism Saccharomyces cerevisiae (where TORC1 was first discovered) (Loewith and Hall, 2011; Gonzalez and Hall, 2017), progress mapping the TORC1 regulatory circuit has been hindered by the absence of a rapid and scalable TORC1 signaling assay. Here we describe a protocol for monitoring the movement of TORC1 (made up of the TOR kinase Tor1, the essential regulatory protein Kog1, and two accessory proteins, Lst8 and Tco89) into a focus/body during glucose and/or nitrogen starvation. Since TORC1 relocalization is controlled by all known TORC1 regulators in yeast (Hughes Hallett et al., 2015; Sullivan et al., 2019), the assay can be used to (1) measure changes in TORC1 signaling at the single cell level and (2) rapidly test the impact that novel genes, mutations and/or drugs have on TORC1 activity.
Materials and Reagents
8-well micro-slide (Ibidi, catalog number: 80826 )
YEPD plates (VWR, catalog number: 10128-392 )
28 ml test tube (VWR, catalog number: 47729-583 )
Wooden applicator sticks (Key scientific, catalog number: CA958701 )
BD Difco Yeast Nitrogen Base without Amino Acids (Fisher Scientific, BD Biosciences, catalog number: BD 291920 )
BD Difco Yeast Nitrogen Base without Amino Acids and Ammonium Sulfate (Fisher Scientific, BD Biosciences, catalog number: DF0335-15-9 )
Drop-out Mix Complete w/o Yeast Nitrogen Base (US Biological, catalog number: D9515 )
Dextrose Anhydrous Crystalline Granules (Fisher Scientific, Fisher BioReagents, catalog number: BP350-1 )
Ammonium sulfate (Fisher Scientific, catalog number: BP212-212 )
Glycerol (Sigma-Aldrich, catalog number: G7893 )
Concanavalin A (Fisher Scientific, MP Biomedicals, catalog number: ICN15071001 )
SD (see Recipes)
S-glucose (see Recipes)
S-nitrogen (see Recipes)
2 mg/ml Concanavalin A solution (see Recipes)
Equipment
Drum style test-tube rotator (New Brunswick, TC-7 Roller Drum ) at 30 °C
150 ml Erlenmeyer flasks (Fisher Scientific, catalog number: 50-172-0176 )
Platform Shaker (New Brunswick Innova 2300) at 30 °C
Genesys 30 UV/Vis Spectrophotometer (ThermoFisher, catalog number: 840-297300 )
Nikon Eclipse Ti-E Microscope, 100× objective, Photometrics Prime 95-B Camera (or equivalent)
Software
NIS-Elements (Nikon, https://www.microscope.healthcare.nikon.com/products/software/nis-elements)
Fiji (free image analysis software that includes ImageJ and several useful plugins; https://imagej.net/Fiji); refer to Schindelin et al., (2012).
Procedure
This protocol accompanies Hughes Hallett et al. (2015) and Sullivan et al. (2019).
We generally follow movement of TORC1 on the vacuolar membrane in a standard W303 lab strain (trp1-1;can1-100;leu2-3,112;his3-11,15;ura3;GAL+;ADE+) carrying Kog1 with a yellow fluorescent protein tag at its native locus (Kog1-ECitrine, or Kog1-YFP for short). Tags on other TORC1 subunits (particularly Tco89) can also be used, but Kog1-YFP gives the strongest signal. It is worth noting fluorescent tags on Tor1 disrupt TORC1 localization and activity.
Prepare an 8-well glass-bottom chamber slide by adding 200 μl of 2 mg/ml Concanavalin A (ConA) solution to each well (ensuring the bottom of each slide is completely coated). Incubate the slide for 10 min at 25 °C, and then remove the solution and allow the wells to air-dry overnight (in the dark) with the lid in place. The ConA treatment ensures that the yeast adheres to the coverslip at the bottom of each chamber, making medium swaps and high-quality imaging possible.
Patch out the strains that are going to be examined onto fresh YEPD plates, starting from glycerol stocks (yeast in 15% glycerol and YEPD, stored at -80 °C) using sterile applicator sticks, and then incubate the plates at 30 °C overnight (or up to three days).
Transfer approximately 5 μl of each strain/patch into a separate 28 ml tube containing 5 ml of SD medium (again using sterile applicator sticks).
Grow the 5 ml starter cultures at 30 °C in a drum rotator (rotating at approximately 40 rpm) until they reach mid-log phase (OD600 between 0.5 and 0.7) – this usually takes 5 h.
Transfer approximately 4 ml of each starter culture into a separate 150 ml Erlenmeyer flask containing 20 ml of SD medium, so that the final OD600 is 0.1.
Grow the 20 ml cultures at 30 °C, shaking at 200 rpm, until they reach an OD600 ~0.4.
While the cultures grow, heat the microscope chamber, 15 ml of fresh SD medium, and 15 ml of fresh starvation medium (e.g., S-glucose or S-nitrogen) to 30 °C.
Pipet 300 μl of each culture into the wells of a ConA-treated chamber slide and then allow the cells to settle for 5 min at 30 °C.
Gently aspirate the medium from each well and discard.
Add 300 μl fresh SD medium at 30 °C.
Capture log-growth (time 0) images in each well using a 100× objective and YFP excitation/emission filter (λEX 510/25; λEM 540/21) collecting a stack of sixteen images, each separated by 0.4 µm in the z-plane, with a 200 ms exposure per plane*. In general you need to capture images in 2-3 fields to ensure you have data for enough cells (>100) to get an accurate TORC1-body count. Once the fluorescence imaging is complete, capture a Differential Interference Contrast (DIC) or Brightfield reference image.
*Note: The imaging is difficult since there are only ~100 TORC1 molecules per cell. First, you must have a high-quality microscope and a sensitive camera to see Kog1-YFP – particularly when the TORC1 molecules are spread across the vacuole. We currently use a Nikon Ti-E inverted microscope with a Photometrics Prime 95-B camera. Second, you can only image a given field once before significant photobleaching occurs. Third, you must take z-stacks (3D images) that span the entire depth of the cell to identify all of the TORC1 foci since the bodies form on the edge of the vacuole and are thus often at the top or bottom of the cell.
Remove the chamber slide from the microscope, aspirate and discard medium from each well.
Wash each well three times with synthetic medium missing glucose (S-glucose), or synthetic medium missing nitrogen (S-nitrogen), at 30 °C, using 350 μl, 400 μl, and then 450 μl of medium, by gently pipetting against the same corner of each well and then aspirating and discarding each wash except the last (which is left in the well during imaging).
Start a timer after the first wash and capture z-stacks at each time-point in all wells as described in Step 11 (we typically take pictures every 10 min for 1 h), keeping the slide at 30 °C during the entire experiment.
In each experiment, a wild-type (or relevant mutant strain) missing a fluorescent (YFP) tag should also be imaged as a control. We have found that starvation (particularly glucose starvation) can trigger the formation of autofluorescent puncta. The intensity and number of puncta increase over time, and their appearance is highly dependent on the batch of medium being used. We discard data from experiments/time-points where a significant number of autofluorescent foci form in the control strain and rerun the experiment in a fresh batch of medium.
Data analysis
Compress the z-stack for each time-point and strain (composed of all 16 planes) into a maximum intensity projection, using Fiji or other software. Identify in focus cells using the DIC image, and then count the fraction of (in focus) cells that contain a TORC1 (Kog1-YFP) body. A mother cell with an attached bud should be counted as two individual cells due to the observation that a newly formed daughter cell can contain its own TORC1-body. To establish statistical significance, experiments must be completed in triplicate on three different days, and the average fraction of cells with a body at each time-point, and the corresponding standard deviation, were calculated.
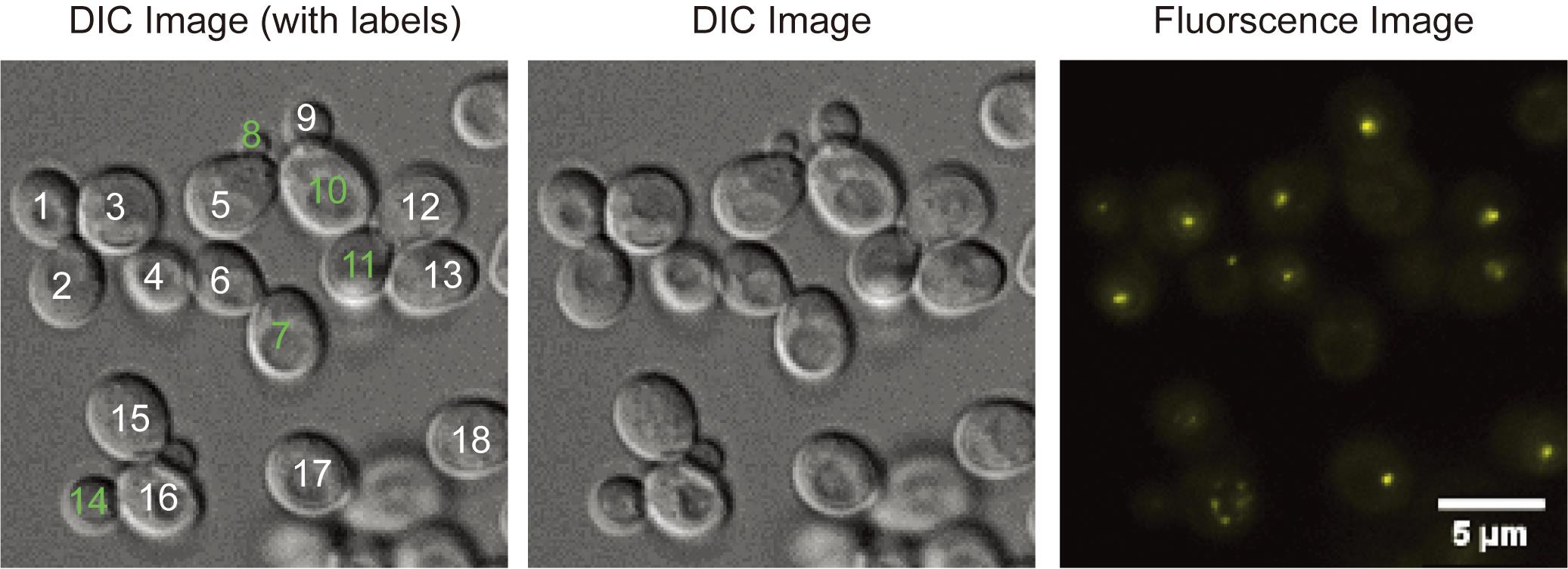
Figure 1. Kog1-YFP (TORC1) foci in starvation conditions. Cells that are in the focal plane and used in the analysis are numbered and colored white if they contain a TORC1 body.
In Figure 1 there are 18 in-focus cells, including buds. The fluorescence (maximum projection) image shows the presence of 13 cells containing TORC1-bodies, and thus 72% of cells in this field contain a TORC1-body. For other sample images see Hughes Hallett et al. (2015) and Sullivan et al. (2019).
Recipes
Medium:
SD
200 ml of 5× amino acids stock (US Biological)
100 ml of 10× YNB stock (BD Biosciences)
50 ml of 40% glucose stock (Fisher Scientific)
650 ml ddH2O
S-glucose
200 ml of 5× amino acids stock (US Biological)
100 ml of 10× YNB stock (BD Biosciences)
700 ml ddH2O
S-nitrogen
100 ml of 10× YNB stock w/o ammonium sulfate (BD Biosciences)
50 ml of 40% glucose stock (Fisher Scientific)
850 ml ddH2O
2 mg/ml Concanavalin A (ConA)
Dissolve 20 mg of ConA (MP Biomedicals) into 10 ml ddH2O and aliquot into 1 ml tubes and stored at -20 °C
Acknowledgments
This work was supported by the National Institutes of Health (NIH) grants (R01GM097329 and T32GM008659). This protocol is based on our previous work, described in Hughes Hallett et al. (2015) and Sullivan et al. (2019).
Competing interests
No competing interests.
References
- Barbet, N. C., Schneider, U., Helliwell, S. B., Stansfield, I., Tuite, M. F. and Hall, M. N. (1996). TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell 7(1): 25-42.
- Ben-Sahra, I. and Manning, B. D. (2017). mTORC1 signaling and the metabolic control of cell growth. Curr Opin Cell Biol 45: 72-82.
- Ben-Sahra, I., Hoxhaj, G., Ricoult, S. J. H., Asara, J. M. and Manning, B. D. (2016). mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science 351(6274): 728-733.
- Duvel, K., Yecies, J. L., Menon, S., Raman, P., Lipovsky, A. I., Souza, A. L., Triantafellow, E., Ma, Q., Gorski, R., Cleaver, S., Vander Heiden, M. G., MacKeigan, J. P., Finan, P. M., Clish, C. B., Murphy, L. O. and Manning, B. D. (2010). Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell 39(2): 171-183.
- Gonzalez, A. and Hall, M. N. (2017). Nutrient sensing and TOR signaling in yeast and mammals. EMBO J 36(4): 397-408.
- Hsu, P. P., Kang, S. A., Rameseder, J., Zhang, Y., Ottina, K. A., Lim, D., Peterson, T. R., Choi, Y., Gray, N. S., Yaffe, M. B., Marto, J. A. and Sabatini, D. M. (2011). The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332(6035): 1317-1322.
- Huber, A., Bodenmiller, B., Uotila, A., Stahl, M., Wanka, S., Gerrits, B., Aebersold, R. and Loewith, R. (2009). Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev 23(16): 1929-1943.
- Hughes Hallett, J. E., Luo, X. and Capaldi, A. P. (2015). Snf1/AMPK promotes the formation of Kog1/Raptor-bodies to increase the activation threshold of TORC1 in budding yeast. Elife 4: e09181.
- Kamada, Y., Funakoshi, T., Shintani, T., Nagano, K., Ohsumi, M. and Ohsumi, Y. (2000). Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol 150(6): 1507-1513.
- Kim, J., Kundu, M., Viollet, B. and Guan, K. L. (2011). AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 13(2): 132-141.
- Liu, G. Y. and Sabatini, D. M. (2020). mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol 21: 183-203.
- Loewith, R. and Hall, M. N. (2011). Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189(4): 1177-1201.
- Peterson, T. R., Sengupta, S. S., Harris, T. E., Carmack, A. E., Kang, S. A., Balderas, E., Guertin, D. A., Madden, K. L., Carpenter, A. E., Finck, B. N. and Sabatini, D. M. (2011). mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 146(3): 408-420.
- Robitaille, A. M., Christen, S., Shimobayashi, M., Cornu, M., Fava, L. L., Moes, S., Prescianotto-Baschong, C., Sauer, U., Jenoe, P. and Hall, M. N. (2013). Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science 339(6125): 1320-1323.
- Schindelin, J., Arganda-Carreras, I. and Frise, E. et al. (2012). Fiji: an open-source platform for biological-image analysis.Nature Methods 9(7): 676-682.
- Sullivan, A., Wallace, R. L., Wellington, R., Luo, X. and Capaldi, A. P. (2019). Multilayered regulation of TORC1-body formation in budding yeast. Mol Biol Cell 30(3): 400-410.
Article Information
Copyright
Wallace et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Wallace, R. L., Lu, E., Sullivan, A., Hughes Hallett, J. E. and Capaldi, A. P. (2021). Analysis of TORC1-body Formation in Budding Yeast. Bio-protocol 11(7): e3975. DOI: 10.21769/BioProtoc.3975.
- Hughes Hallett, J. E., Luo, X. and Capaldi, A. P. (2015). Snf1/AMPK promotes the formation of Kog1/Raptor-bodies to increase the activation threshold of TORC1 in budding yeast. Elife 4: e09181.
Category
Developmental Biology > Cell signaling > Stress response
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








