- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Atomic Force Microscopy to Characterize Ginger Lipid-Derived Nanoparticles (GLDNP)
Published: Vol 11, Iss 7, Apr 5, 2021 DOI: 10.21769/BioProtoc.3969 Views: 5974
Reviewed by: RAMESH KUDIRABrahma MuluguAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Fluorescent Biosensor Imaging of Nitrate in Arabidopsis thaliana
Yen-Ning Chen and Cheng-Hsun Ho
Aug 20, 2023 3635 Views

Vegetative Propagation of Cannabis sativa and Resin Obtained From its Female Inflorescences
Sebastián D´Ippolito [...] Silvana L. Colman
Feb 20, 2025 1735 Views

A New Approach to Detect and Semi-quantify All Molecular Species and Classes of Anionic Phospholipids Simultaneously in Plant Samples
Manon Genva [...] Laetitia Fouillen
Apr 20, 2025 1718 Views
Abstract
We have demonstrated that a specific population of ginger-derived nanoparticles (GDNP-2) could effectively target the colon, reduce colitis, and alleviate colitis-associated colon cancer. Naturally occurring GDNP-2 contains complex bioactive components, including lipids, proteins, miRNAs, and ginger secondary metabolites (gingerols and shogaols). To construct a nanocarrier that is more clearly defined than GDNP-2, we isolated lipids from GDNP-2 and demonstrated that they could self-assemble into ginger lipid-derived nanoparticles (GLDNP) in an aqueous solution. GLDNP can be used as a nanocarrier to deliver drug candidates such as 6-shogaol or its metabolites (M2 and M13) to the colon. To characterize the nanostructure of GLDNP, our lab extensively used atomic force microscopy (AFM) technique as a tool for visualizing the morphology of the drug-loaded GLDNP. Herein, we provide a detailed protocol for demonstrating such a process.
Keywords: Atomic force microscopyBackground
Developing new drug-based therapeutic approaches against Intestinal Bowel Disease (IBD) must overcome numerous challenges, including potential off-target effects, large-scale production costs, and the need to ensure tissue-specific delivery, systemic safety, and low toxicity. Our group and others have recently demonstrated that artificially synthesized nanoparticles could target low doses of drugs (e.g., siRNAs, proteins, or peptides) to colonic tissues or colonic immune cells, such as macrophages (Ulbrich and Lamprecht, 2010; Chen et al., 2017). However, these synthetic NPs to date have two major limitations: i) each constituent of the synthesized nanoparticle must be examined for potential in vivo toxicity before clinical application; and ii) the production scale is limited. The use of nanoparticles derived from natural sources may overcome these limitations. In this context, we reported that a special population of ginger-derived nanoparticles (GDNP-2) could reduce colitis and colitis-associated colon cancer (Zhang et al., 2016). Naturally occurring GDNP-2 is also safer and cheaper than synthetic NPs. We further identified 6-shogaol as a major candidate that may account for the anti-inflammatory and anti-cancer activities of GDNP-2 (Yang et al., 2020). To construct a nanocarrier that is more clearly defined than GDNP-2, we characterized ginger lipid-derived nanoparticles (GLDNP) and demonstrated that they could be used as a natural carrier to deliver natural anti-inflammatory drug candidates such as 6-shogaol, M2, or M13 to the colon and reduced colitis in mice (Yang et al., 2020).
Atomic force microscopy (AFM) is a versatile and powerful technique to characterize the morphology of nanoscale and submicron structured materials. It has been widely used to visualize different types of NPs, including metal-, inorganic- (non-metallic), and organic-NPs. AFM has the advantages of simplicity in sample preparation and no need for electric conducting treatment (Morris et al., 2010). In previous studies, our group and others have extensively used AFM to analyze the morphology and structure of GLDNP (Zhang et al., 2017; Wang et al., 2019; Sung et al., 2020; Yang et al., 2020). However, no attempt has been taken to document the procedure of sample preparation and AFM parameter setting. In the following protocol, we will use the GLDNP as the specimen to demonstrate the process of obtaining AFM pictures for nanostructure characterization.
Materials and Reagents
Pipette tips 0.1-10 μl, 1-200 μl and 100-1,000 μl (Sorenson Bioscience, catalog numbers: 70600 , 70520 , 70540 )
Powder-free gloves (Denville Scientific, catalog number: G4162 )
50 ml conical tubes (Denville Scientific, catalog number: C1062-P )
Phosphate-buffered Saline (PBS) (Corning, catalog number: 21-040-CV )
Methanol (Sigma-Aldrich, catalog number: 34860-1L-R )
Potassium chloride (KCl, Millipore, catalog number: 7447-40-7 )
Dichloromethane (Sigma-Aldrich, catalog number: 650463-1L )
Deionized-distilled water (ddH2O)
Mica sheet (Electron Microscopy Sciences, catalog number: 71855-15 )
1 M KCl solution (see Recipes)
Equipment
Pipettes 0.5-10 μl, 10-100 μl and 100-1,000 μl (Eppendorf, model: Research® Plus , Variable Adjustable Volume Pipettes)
Milli-Q advantage A10 water purification system (Millipore-Sigma, catalog number: C10117 )
Glass separatory funnel (Southern Labware, model: 3964-3)
Centrifuge (Thermo Fisher Scientific, model: Sorvalis ST16R )
Vortexer (Scientific Industries, model: 200-SI0236 )
Rotary evaporator (Buchi, model: R-210 )
Vacuum pump (Buchi, model: V-700 )
Vacuum controller (Buchi, model: V-800 )
Heating bath (Buchi, model: B-491 )
Evaporating flask (Buchi, catalog number: Z402982 )
Notebook computer (ThinkPad, model: T570 )
CoreAFM controller (Nanosurf, model: CoreAFM controller)
CoreAFM system (Nanosurf, model: CoreAFM system)
Isostage 300 controller (Nanosurf, model: Isostage 300 controller)
CoreAFM tool set (Nanosurf, model: CoreAFM tool set)
Software
CoreAFM control software (Version 3.10.0, https://www.nanosurf.net/en/software/coreafm)
Procedure
Preparation of GLDNP
Please refer to our published bio-protocols Sung et al. (2019 and 2020).
Note: Stored GLDNP (in 1× PBS) can be enriched after ultracentrifugation (30,000 × g, 4 °C, and 45 min) and remove the supernatant.
Setting up the AFM (see Note 1)
Assemble the Nanosurf CoreAFM system according to the steps in the operating instructions of CoreAFM. The AFM equipment after assembly is shown in Figure 1.
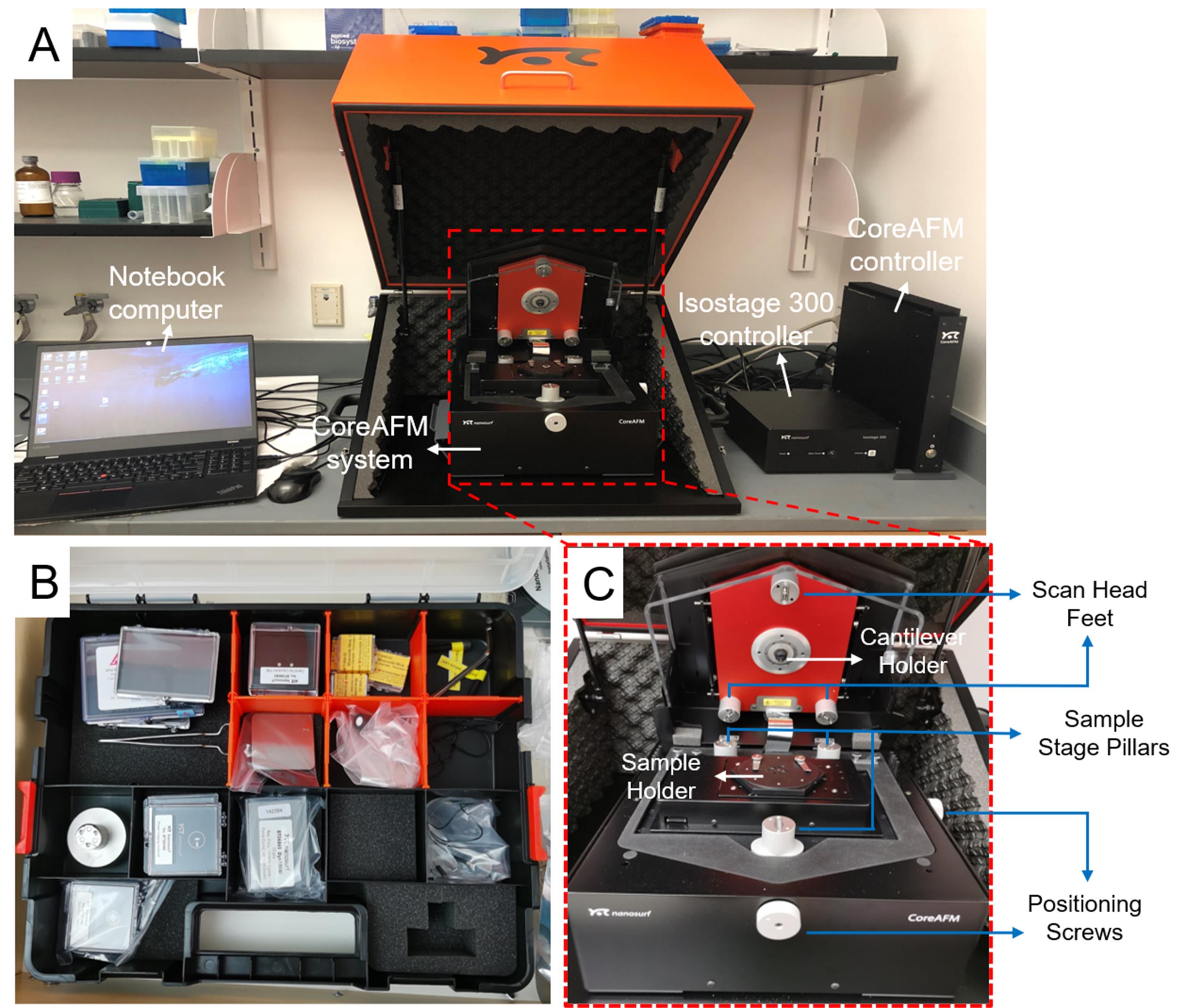
Figure 1. Assembly of the Nanosurf CoreAFM system. A. The assembled Nanosurf CoreAFM system, which contains CoreAFM system, Isostage 300 controller, CoreAFM controller, and a laptop, according to the instructions. B. The CoreAFM toolset (see Note 2). C. Enlarged photo of CoreAFM in its opened configuration.Switch on the Nanosurf CoreAFM system (see Note 3).
Insert a Nanosurf Dyn190AI dynamic mode cantilever in the cantilever holder, ensuring that the tip is not damaged and attaching the cantilever holder to the magnets at the bottom of the CoreAFM scan head (see Note 3).
Loading GLDNP samples for AFM imaging
Dilute GLDNP solution in ddH2O to 1 mg/ml.
Deposit 2 μl of nanoparticle sample to mica sheet (Figure 2A).
Note: As a drug delivery vehicle, the precise dose-response is one of the factors that must be considered. For different samples, the solution is usually diluted from high concentration to low concentration, while the volume of the solution is continuously reduced. According to the imaging effect of AFM, the most suitable solution concentration and volume are finally selected. The concentration of GLDNP samples used for imaging in this procedure should be > 0.01 μg/ml.
Dry the sample at room temperature (RT) for 2 h.
Gently rinse the mica sheet with 5 μl of distilled water three times (Figure 2B).
Note: First, drop distilled water gently into the middle of the mica sheet using the pipette, and then use filter paper to absorb water from the edge of the mica sheet. Since GLDNP samples are stored in PBS, the salt particles from PBS will affect the imaging effect of AFM, so the purpose of gently rinsing the mica sheet is to remove the salt in GLDNP samples.
Dry the sample at RT for 2 h again.
Leave the mica sheet for 30 min at RT until the sample becomes flat.
Note: To judge whether a sample is dried and becomes flat, we can place the mica sheet in a vertical position, and if we observe no sign of flow from the sample spot, it generally means that the sample is flat and dried. The drying and flatness of the sample on the mica sheet will affect the imaging effect of AFM.
Fix the mica sheet on the sample magnet in the center of the standard sample holder (Figure 2C).
Close the CoreAFM scan head lid, and the three scan head feet will align with the corresponding sample stage pillars.
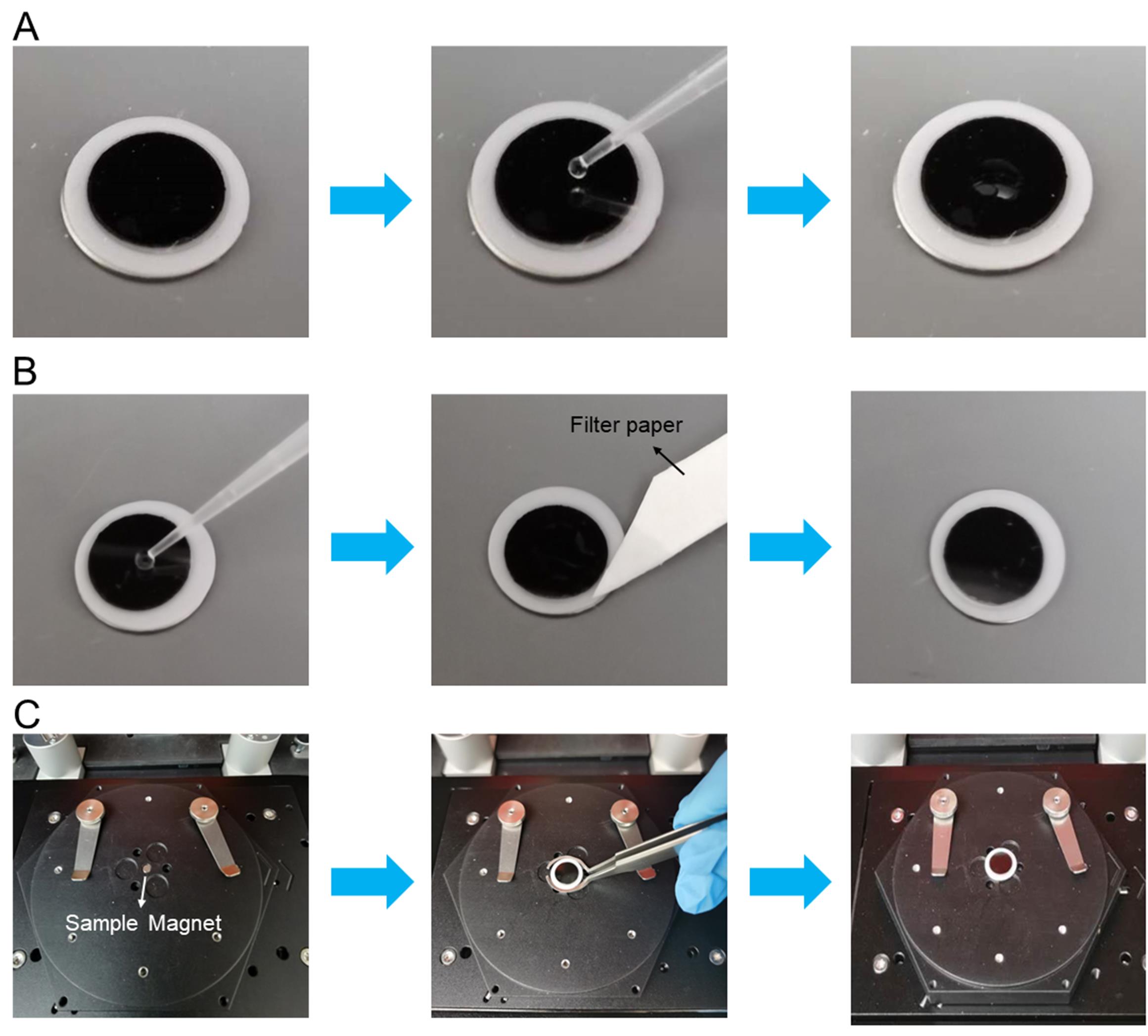
Figure 2. Loading the GNDLN sample to the mica sheet and fixing it on the sample magnet. A. The process of depositing 2 μl of nanoparticle sample to a mica sheet. B. The process of gently rinsing the mica sheet with distilled water and adsorbing water from the edge of the mica sheet with filter paper. C. The process of fixing the mica sheet on the sample magnet of the standard sample holder (see Note 3).
AFM imaging of the GLDNP samples
Checking the laser position and quality (see Note 3).
Attach the CoreAFM system to the CoreAFM controller, and start the equipment and software.
In the CoreAFM control software, open the Laser Alignment dialog (Figure 3A) and switch the CoreAFM to the top view camera (Figure 3B) using the Camera selector (Figure 3C).
Note: The Laser Alignment dialog displays the current position of the AFM laser spot on the detector and the used laser power. The dialog is opened by clicking the “Laser Align” button in the Preparation group of the Acquisition tab.
Use the laser alignment tool that came with the CoreAFM system to turn the screws in a clockwise or counterclockwise direction (Figure 3C). An optimally aligned laser would result in Figure 3A.
Note: CoreAFM system comes with 4 holes in the scan head’s top cover that provide access to the laser and detector alignment screws that change different aspects of the laser beam’s optical pathway (Figure 3C). Open the scan head lid to an angle of approximately 30°, and you should now see a red laser spot somewhere on the sample holder or sample holder platform (Figure 3C).
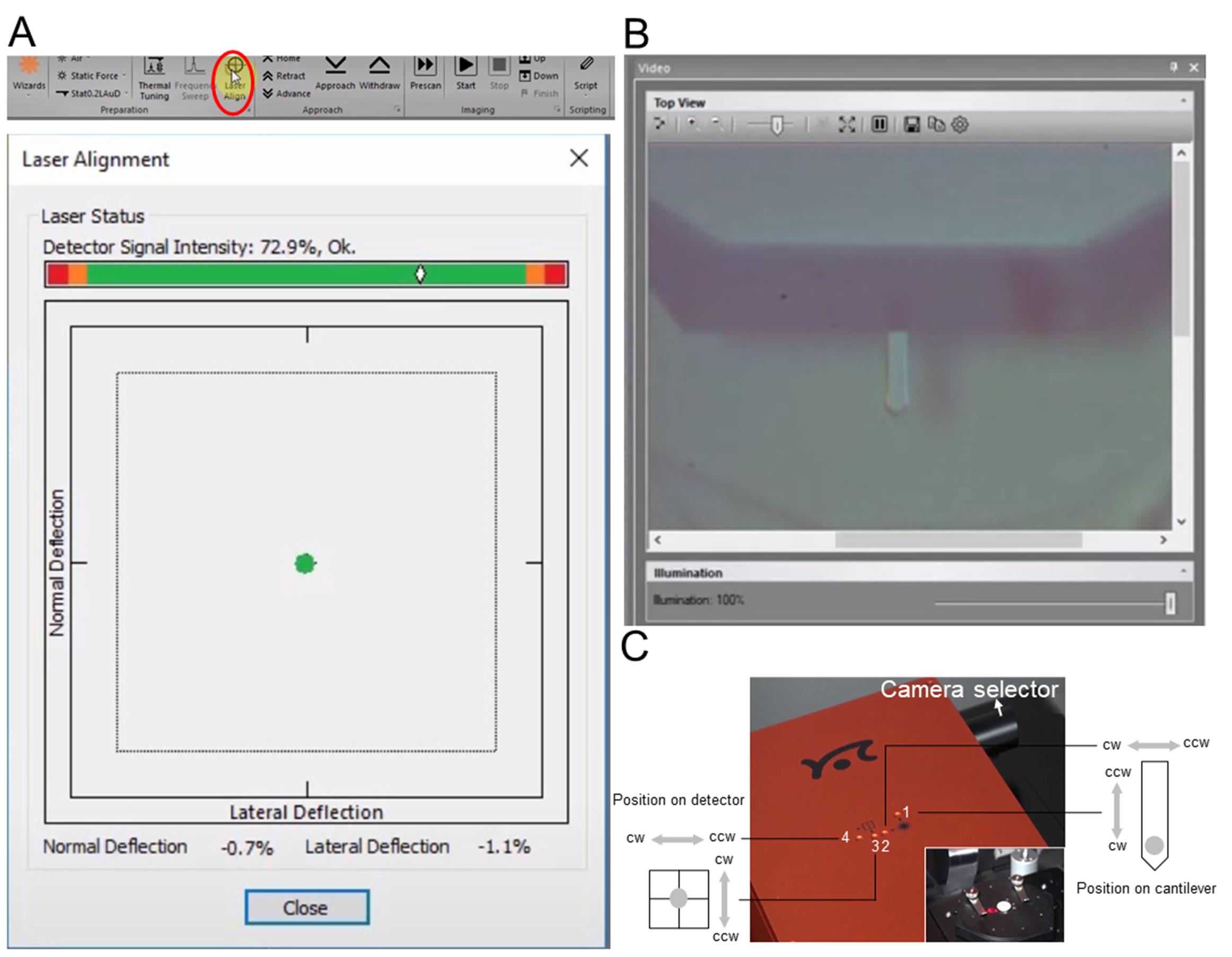
Figure 3. Checking the laser position and quality. A. The menu of CoreAFM control software application and the proofread completed Laser Alignment dialog. This graphical area shows where the deflected laser beam hits the photodiode detector. A green spot anywhere within the area enclosed by the dotted square means that the cantilever deflection detection system (consisting of laser, cantilever, and detector) is properly aligned. If the laser spot falls outside this area, it will become red, meaning that the alignment does not allow proper measurements to take place. B. Top view image of a cantilever and its chip structure. C. Physical photos of the CoreAFM scan head and the sample holder.Configuring measurement parameters of the CoreAFM (see Note 3).
In the menu of CoreAFM control software, click the “Air” to select the measurement environment from the Measurement environment drop-down menu (Figure 4A).
Click the “Static Force” to select the desired operating mode from the Operating mode drop-down menu (Figure 4B).
Click the “ACL-A” to select the desired cantilever type from the Cantilever selector drop-down menu (Figure 4C).
Click the “Frequency Sweep” button that opens the Vibration frequency search dialog. Click the “Find Vibration Frequency” button in this dialog (Figure 4D). Leave the dialog by clicking “OK”.
Note: After the automatic frequency search is finished, you should see a clean resonance curve and a marker showing the vibration frequency.
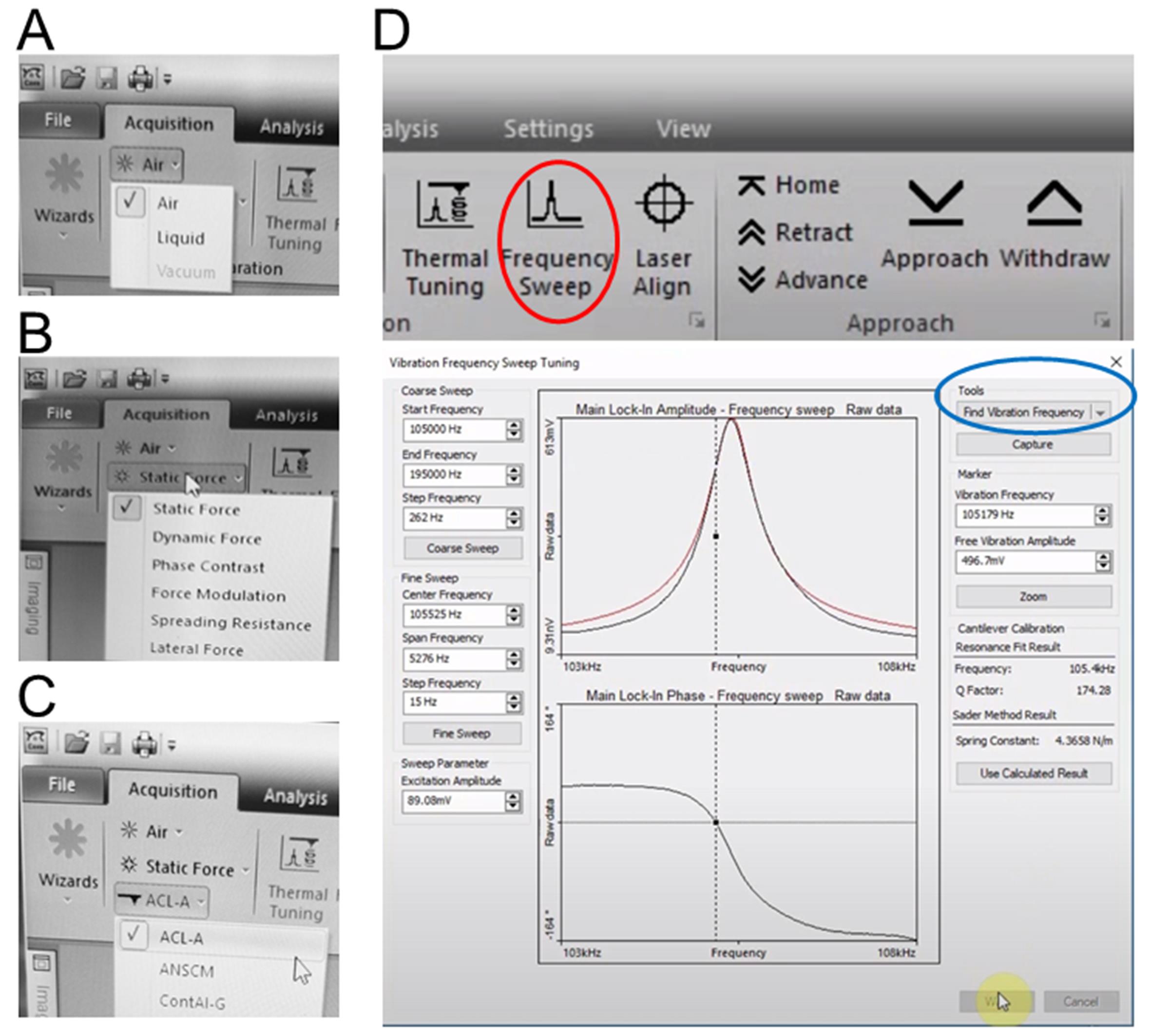
Figure 4. Configuring measurement parameters of the CoreAFM. A-C. Set the Measurement environment to “Air” (A), set Operating mode to “Static Force” (B) and select “ACL-A” as cantilever in the Cantilever selector (C). D. The menu of CoreAFM control software application and the proofread completed Vibration frequency search dialog.Approaching the GLDNP samples (see Note 3).
Observe the distance between tip and sample in the side view (Figure 5A).
While observing the tip-sample distance, click and hold the “Advance” button in the Approach group of the Acquisition tab until the tip is close enough to the sample (Figure 5B).
Click the “Approach” button in the Approach group of the Acquisition tab (Figure 5B). Click the “OK” button (Figure 5C).
Note: In this last step, this sample automatically approaches the tip until a given setpoint is reached.
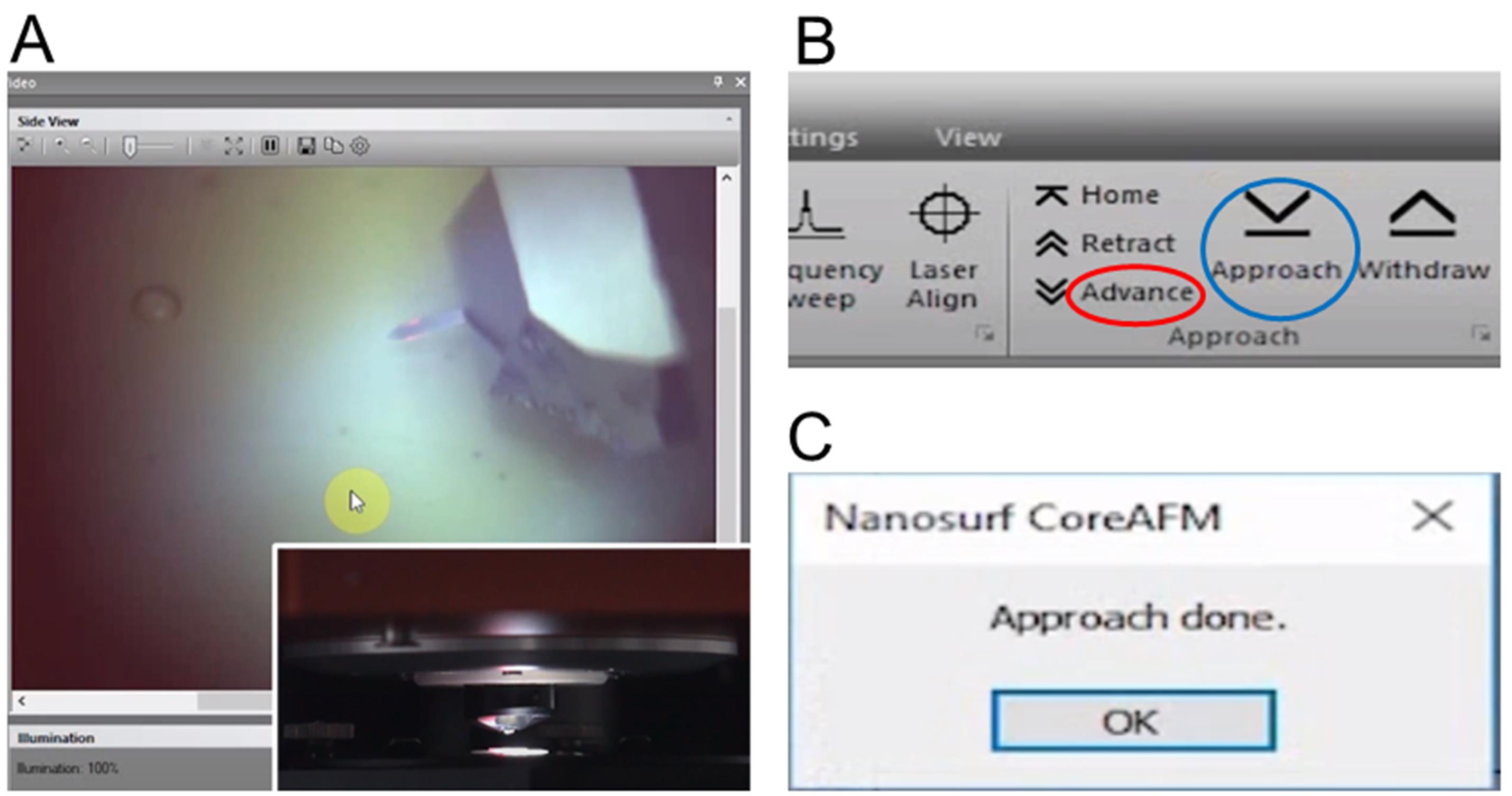
Figure 5. Approaching the GNDLN samples. A. A side view image of a cantilever. B. The “Advance” and “Approach” buttons in the Approach group of the Acquisition tab. C. When the Setpoint is reached, the “Approach done” message is displayed.Starting a measurement (see Note 3).
Select the initial scan parameters in the Imaging dialog as follows (Figure 6A):
Image Scan Size = 10.00 μm
Time/Line = 0.78 s
Points/Line = 256
Note: The other values on the master panel are the values automatically filled in by the instrument upon the finishing of probe tuning; thus, no changes of these values are needed.
Click the “Start” button in the Imaging group of the Acquisition tab and start scanning for 1 min to stabilize the cantilever and adjust the instrument to surrounding environmental vibrations (Figure 6B).
Note: Manually lower the “Setpoint” value until surface features start to appear on the height image. Imaging optimization can be achieved by adjusting “Setpoint”, “P-Gain”, and “I-Gain” values (see Note 4). Finally, the “Image Scan Size” can be decreased to 5.00 μm or 4.00 μm for adjusting the scan size of the image, and the “Points/Line” can be increased to 512 or even 1,024 (see Note 5) for more pixels in each image thus enhance the image quality.
Once satisfied with the image quality, click the “Start” button and restart the scan with optimized parameters. After scanning, click the “Stop” button in the Imaging group of the Acquisition tab (Figure 6B).
Note: Adjust the positioning screws of the CoreAFM system (see Figure 1C) so that the cantilever can be positioned in different locations of the mica sheet surface for each sample. Then click the “Prescan” button in the Imaging group of the Acquisition tab (Figure 6B). When the desired position is located by pre-scanning, click the “Start” button for scanning.
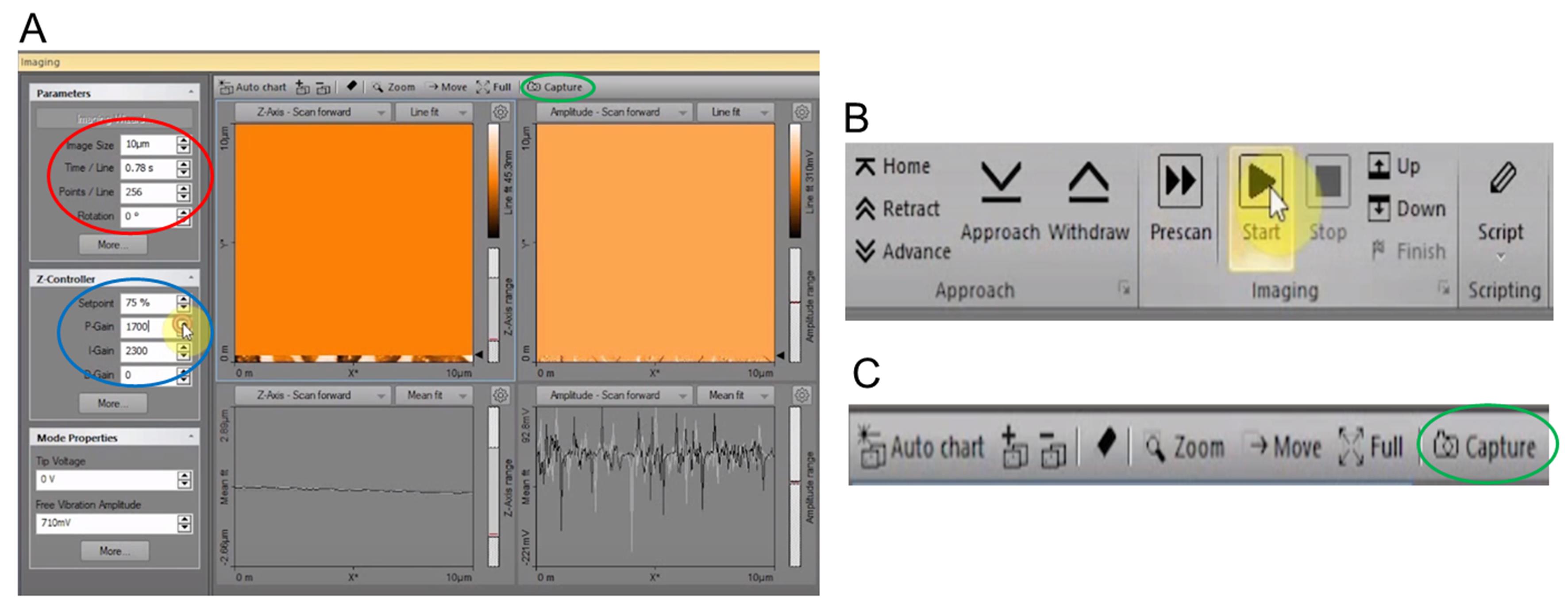
Figure 6. Starting and storing the measurement of the GLDNP samples. A. Select the initial scan parameters and adjust the “Setpoint”, “P-Gain”, and “I-Gain” values to enhance the image quality in the Imaging dialog. B. The “Prescan”, “Start” and “Stop” buttons in the Imaging group of the Acquisition tab. C. The “Capture” button in the Imaging toolbar.Storing the measurement and working with documents.
By default, each completed measurement is automatically stored (temporarily) on your computer so that it can be used later. You can also take snapshots of measurements still in progress by clicking the “Capture” button (Figure 6C).
The captured document will remain open in the document space of the CoreAFM control software (Figure 7A). Adding or removing a chart, or setting chart parameters is all performed in the Chart Properties dialog (Figure 7B).
Click the “Add Chart” button (“+” in Figure 7B) to create a copy of the currently selected or active chart and add it to the active window in the last position. Click the “Remove Chart” button (“-” in Figure 7B) to remove the currently active chart.
Selects the chart type (Line graph, Color map, 3D view, XY Line Graph, or Force Curve graph) to be used for the display of the measurement data from the “Type” of Chart Parameters (Figure 7B). Figure 7A shows three different chart types of the GLDNP samples.
Use the Chart Properties dialog to set the various parameters of the corresponding chart type of the GLDNP samples.
After setting, click "Close" button (Figure 7B) to close the Chart Properties dialog and click the “Save icon” (Figure 7A) to save the documents.
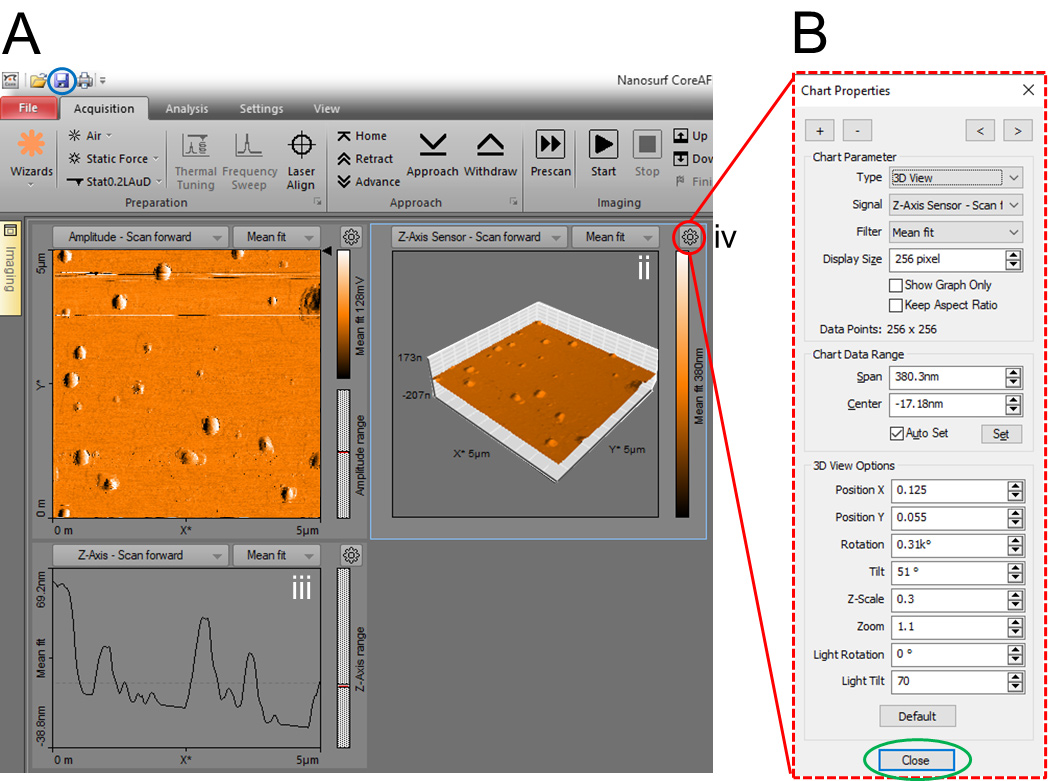
Figure 7. Different chart types of the GLDNP samples and the Chart Properties dialog. A. (i) Color map, (ii) 3D view and (iii) Line graph of the GLDNP samples, and (iv) the “Chart Properties” button. B. The Charts Properties dialog is used to set all chart properties that influence data display by the respective chart.
Data analysis
We used the above process to perform AFM imaging of the GLDNP samples (Figures 8A-8C).
Select the final scan parameters in the Imaging dialog for the GLDNP as follows:
Image Scan Size = 5.00 μm
Time/Line = 0.78 s
Points/Line = 256
In Figures 8A-8C, AFM images showed that GLDNPs are nano-sized particles and have a spherical shape and presented a size-homogenized appearance.
We prepared the synthetic material Poly(lactic-co-glycolic Acid) (PLGA)-based nanoparticles, then used the above process and set the same parameters to obtain the AFM image of PLGA samples (Figures 8D-8F), and compared it with GLDNP. Through the comparison of the images, we can observe that PLGA nanoparticles are also spherical with a size-homogenized appearance, and their particle size is slightly smaller than GLDNP. PLGA nanoparticles show a certain degree of aggregation on mica sheet compared to GLDNP.
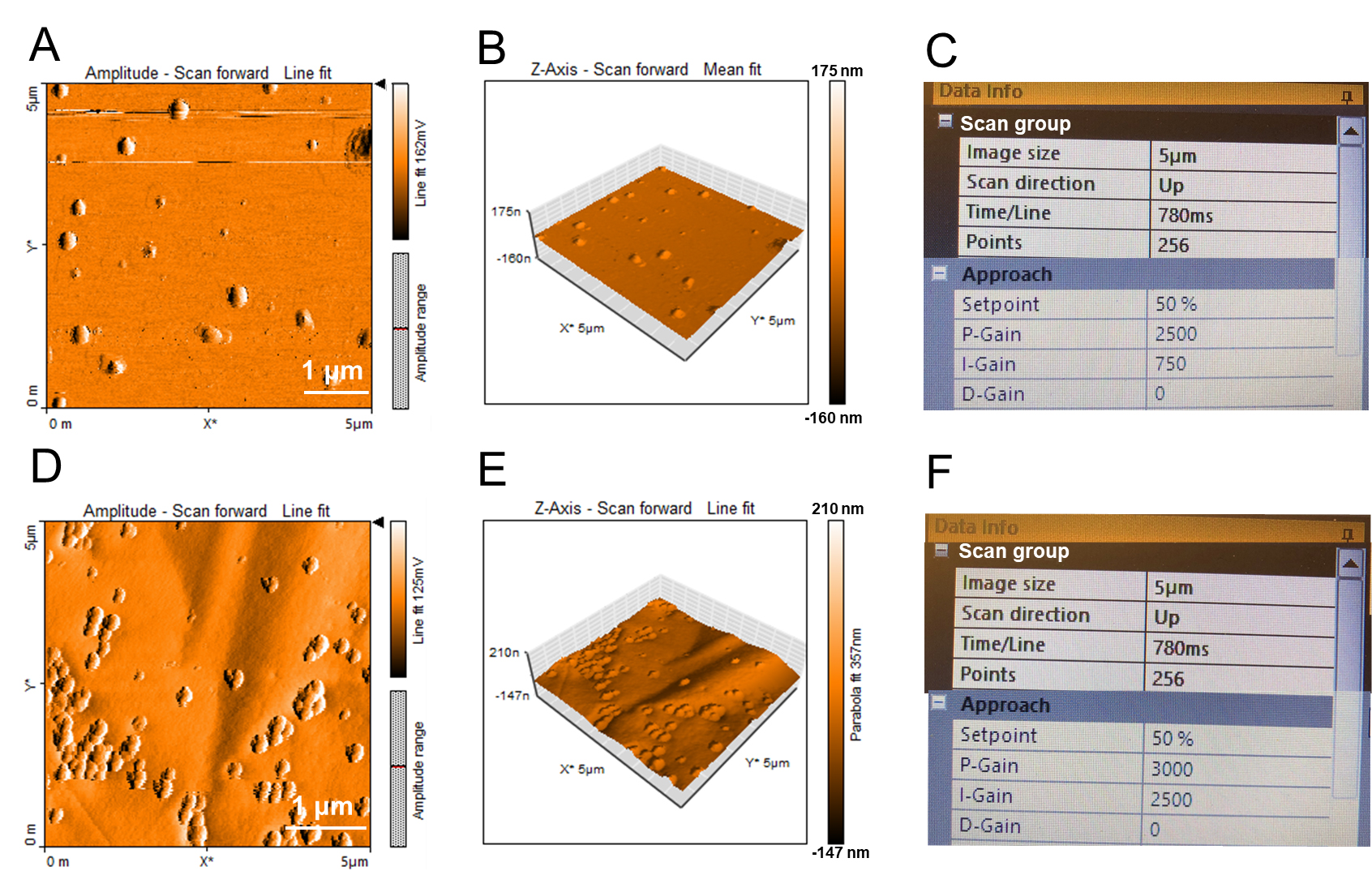
Figure 8. AFM characterization of the GLDNP samples and the poly lactic-co-glycolic acid (PLGA) samples (as control). A-C. The Color map (A), 3D View (B), and the data information of image scan (C) for AFM characterization of GLDNP. D-F. The Color map (D), 3D View (E), and the data information of image scan (F) for AFM characterization of PLGA.
Notes
All procedures here are described for a Nanosurf CoreAFM system. For different models and brands of AFM, the protocol will need to be adapted according to the manufacturer’s instructions.
The content of the toolset depends on the options included in the user's order. It minimally contains the following items:
Ground cable
Cantilever tweezers: (103A C.A.)
Cantilever exchange tool
Laser alignment tool: Allen key 1.5-mm (ballpoint hex) with a screwdriver handle
Allen key 5-mm: used for the CoreAFM transportation locks
Standard sample holder
Cantilever holder liquid-air flat
Prepare and operate the CoreAFM system and use this system to measure the brief program. Video is available at the product official website (https://www.nanosurf.net/en/products/coreafm-the-essence-of-atomic-force-microscopy). It will take about 4 h 30 min to prepare nanoparticles-loaded mica sheet for AFM imaging, and 15-20 min for AFM debugging and photography. Therefore, AFM imaging excluding data analysis can be completed within 5 h. In particular, it should be noted that the main limitation of the above procedure is that AFM imaging can only be done on dry samples in an indoor air environment, and AFM imaging of prepared samples (loaded on mica sheet) usually needs to be completed within a limited time (no more than 6 h). In addition, the minimum resolvable particle size of the sample is about 10 nm by the CoreAFM system.
Use a low “P-Gain” and “I-Gain” in the beginning to protect the cantilever. These values can be elevated in a later scan for better image quality.
“Points/Line” determines the number of pixel points on each line and the number of lines scanned in the image. In most cases, this value should be kept the same.
Recipes
1 M KCl solution
Dissolve 0.745 g potassium chloride (KCl) in 10 ml of ddH2O and mix well
Acknowledgments
This work is supported by the National Institute of Diabetes and Digestive and Kidney Diseases (RO1-DK-116306 and RO1-DK-107739 to D.M.), the Department of Veterans Affairs (Merit Award BX002526 to D.M.), and the Crohn’s and Colitis Foundation of America. D.L. is a recipient of the Research Fellowship Award from the Crohn’s and Colitis Foundation of America (Award Number #689659). D.M. is a recipient of the Senior Research Career Scientist Award from the Department of Veterans Affairs (BX004476). This protocol is based on our previously published study (Yang et al., 2020).
Competing interests
The authors declare no conflicts of interest within the work.
References
- Chen, Q., Xiao, B. and Merlin, D. (2017). Nanotherapeutics for the treatment of inflammatory bowel disease. Expert Rev Gastroenterol Hepatol 11(6): 495-497.
- Morris, V. J., Kirby, A. R. and Gunning, A. P. (2010). Atomic Force Microscopy for Biologists. 2nd edition. Imperial College Press, ISBN-10: 184816467X.
- Sung, J., Yang, C., Viennois, E., Zhang, M. and Merlin, D. (2019). Isolation, Purification, and Characterization of Ginger-derived Nanoparticles (GDNPs) from Ginger, Rhizome of Zingiber officinale. Bio-protocol 9(19): e3390.
- Sung, J., Yang, C., Collins, J. F. and Merlin, D. (2020). Preparation and Characterization of Ginger Lipid-derived Nanoparticles for Colon-targeted siRNA Delivery. Bio-protocol 10(14): e3685.
- Ulbrich, W. and Lamprecht, A. (2010). Targeted drug-delivery approaches by nanoparticulate carriers in the therapy of inflammatory diseases. J R Soc Interface 7 Suppl 1: S55-66.
- Wang, X., Zhang, M., Flores, S. R., Woloshun, R. R., Yang, C., Yin, L., Xiang, P., Xu, X., Garrick, M. D., Vidyasagar, S., Merlin, D. and Collins, J. F. (2019). Oral gavage of ginger nanoparticle-derived lipid vectors carrying Dmt1 siRNA blunts iron loading in murine hereditary hemochromatosis. Mol Ther 27(3): 493-506.
- Yang, C., Zhang, M., Lama, S., Wang, L. and Merlin, D. (2020). Natural-lipid nanoparticle-based therapeutic approach to deliver 6-shogaol and its metabolites M2 and M13 to the colon to treat ulcerative colitis. J Control Release 323: 293-310.
- Zhang, M., Viennois, E., Prasad, M., Zhang, Y., Wang, L., Zhang, Z., Han, M. K., Xiao, B., Xu, C., Srinivasan, S. and Merlin, D. (2016). Edible ginger-derived nanoparticles: A novel therapeutic approach for the prevention and treatment of inflammatory bowel disease and colitis-associated cancer. Biomaterials 101: 321-340.
- Zhang, M., Wang, X., Han, M. K., Collins, J. F. and Merlin, D. (2017). Oral administration of ginger-derived nanolipids loaded with siRNA as a novel approach for efficient siRNA drug delivery to treat ulcerative colitis. Nanomedicine (Lond) 12(16): 1927-1943.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Long, D., Yang, C., Sung, J. and Merlin, D. (2021). Atomic Force Microscopy to Characterize Ginger Lipid-Derived Nanoparticles (GLDNP). Bio-protocol 11(7): e3969. DOI: 10.21769/BioProtoc.3969.
Category
Cancer Biology > Cancer biochemistry > Drug resistance
Plant Science > Plant biochemistry > Metabolite
Biochemistry > Other compound > Edible nanoparticles
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








