- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Strategy of Isolating ‘Primed’ Tumor Initiating Cells Based on Mitochondrial Transmembrane Potential
Published: Vol 11, Iss 5, Mar 5, 2021 DOI: 10.21769/BioProtoc.3945 Views: 5434
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Thrombopoietin-independent Megakaryocyte Differentiation of Hematopoietic Progenitor Cells from Patients with Myeloproliferative Neoplasms
Chloe A. L. Thompson-Peach [...] Daniel Thomas
Jan 20, 2023 2361 Views

Isolation, Purification, and Culture of Embryonic Melanoblasts from Green Fluorescent Protein–expressing Reporter Mice
Melissa Crawford [...] Lina Dagnino
Sep 5, 2023 1984 Views

Identification and Sorting of Adipose Inflammatory and Metabolically Activated Macrophages in Diet-Induced Obesity
Dan Wu [...] Weidong Wang
Oct 20, 2025 2233 Views
Abstract
Various stem cells have been found to be dependent on mitochondrial energetics. The role of mitochondria in regulating the self-renewal of normal stem cells and stem-like tumor initiating cells (TICs) is increasingly being appreciated. We proposed that TIC populations have a sub population of cells that are “primed” by mitochondria for self-renewal. Using ovarian cancer model, we have developed a protocol to identify and isolate these “primed” cells using Fluorescence-Assisted Cell Sorting (FACS). We combined live cell stains for a functional marker of TICs and for mitochondrial transmembrane potential to enrich TICs with higher mitochondrial potential that form in vitro spheroids 10-fold more than the other TICs with lower mitochondrial potential. This protocol can be directly used or modified to be used in various cell types. Thus, this protocol is anticipated to be invaluable for the basic understanding of mitochondrial and energetic heterogeneity within stem cell population, and may also prove valuable in translational studies in regenerative medicine and cancer biology.
Keywords: Tumor initiating cellsBackground
Cellular heterogeneity in tumors poses tough challenges for cancer therapy (Magee et al., 2012). Certain tumors are arranged hierarchically with the chemotherapy resistant tumor initiating cells (also called cancer stem cells) residing at the bottom of the cellular hierarchy as they maintain the tumor through self-renewal and differentiation (Magee et al., 2012). Plasticity of the tumor initiating cells (TICs) makes them difficult to identify, isolate and study. Heterogeneity and plasticity of energetics of TICs is only beginning to be appreciated and is far from clear (Martinez-Outschoorn et al., 2017) (Intlekofer and Finley, 2019). Moreover, energetics status of TICs appears to be tumor specific, and certain TICs are dependent on mitochondrial energetics and can be eliminated by mitochondrial inhibitors (Pasto et al., 2014; Sancho et al., 2015; Viale et al., 2015; Martinez-Outschoorn et al., 2017; Spurlock et al., 2019).
Mitochondrial energetics is maintained by complex biochemical regulation of multiple bioenergetic parameters, including mitochondrial transmembrane potential maintained across the mitochondrial inner membrane (ΔΨ), which is consumed to generate ATP, heat, or reactive oxygen species. Higher mitochondrial transmembrane potential is expected to maintain higher ATP synthesis depending on various other parameters like substrate availability, etc. Previously, stem cells isolated based on mitochondrial transmembrane potential have been found to differ in their stem cell properties (Schieke et al., 2008; Sukumar et al., 2016). Recently, we developed a flowcytometric protocol to simultaneously sort cells for ΔΨ, using the red fluorescent potentiometric dye TMRE, and the activity of a functional TIC marker aldehyde dehydrogenase (ALDH), using the AldeFluor reagent (Figures 1-2) (Spurlock et al., 2019). Using such a flow cytometric sorting strategy, we were able to identify a subpopulation of human ovarian TICs with 10 fold higher self-renewing and proliferating capacity, as determined by in vitro limiting dilution assay. Thus, our protocol enables teasing out mitochondrial heterogeneity in stem cell populations.
The role of mitochondrial energetics in maintenance of normal stem cells and TICs is more appreciated as our understanding of stem cell energetics increases (Viale et al., 2015; Chandel et al., 2016; Intlekofer and Finley, 2019). Adult stem cells are majorly quiescent and considered to be highly glycolytic and release from quiescence involves metabolic/energetic shifts that remain poorly understood (Margineantu et al., 2002; Ito and Suda, 2014; Folmes and Terzic, 2016). We proposed that a subpopulation of ovarian TICs with particularly elevated mitochondrial transmembrane potential, associated with other specific mitochondrial properties, are primed for greater self-renewal and proliferation ability (Spurlock et al., 2019). Similar mitochondria based priming has been observed in hematopoietic lineage and has been linked to exit from quiescence and entry into the cell cycle for self-renewal and differentiation (Liang et al., 2020). Our protocol specifically isolates these mitochondria primed stem cells (mpSCs) by combining a TIC marker (Aldh activity) with ΔΨ. Aldh activity is a TIC marker for several tumor types, including ovarian, breast, cervical, lung, liver, prostate, bone marrow, skin, colon, and pancreatic (Marcato et al., 2011; Toledo-Guzmán, et al., 2019; Vassalli, 2019). We developed this protocol in human ovarian cancer cell lines; however, we have also performed these assays in human ovarian cancer cells derived from a mouse xenograft model as well as patient ascites. The sorted cells can be studied for stem cell or other properties with relevant methods. We have studied mitochondrial structural and functional properties of the sorted cells using our novel assay mito-SinCe2 (Spurlock et al., 2019) (example in Figure 3). This protocol is a simple and valuable tool for the isolation and study of normal and neoplastic stem cells from cell lines as well as primary cell populations. Thus, this protocol will allow a basic understanding of mitochondrial and energetic heterogeneity within stem cell population, and may also prove valuable in translational studies in regenerative medicine and cancer biology.
Materials and Reagents
1.5 ml microfuge tubes (Eppendorf, catalog number: 0 22363204 )
15 ml centrifuge tubes (Corning, catalog number: 430790 )
5 ml flow cytometry tubes (Falcon, catalog number: 352003 )
96-well Ultra-Low Attachment Plates (Corning, catalog number: 3474 )
8-well Lab-Tek II Chambered Coverglass (Nunc, catalog number: 155409 )
Appropriate cell culture materials and reagents for cells in use
DPBS (Sigma, catalog number: 59331C-1000ML )
Trypsin 0.05% (GE Healthcare Life Sciences, catalog number: SH30236.02 or similar)
AldeFluor Kit (StemCell Technologies, catalog number: 01700 )
For lineages where ALDH activity does not mark stemness, our protocol can be modified for use with any lineage by including lineage specific culture conditions and live cell markers (CD133 etc.), ensuring minimal or no overlap with TMRE fluorescence.
Tetramethylrhodamine, Ethyl Ester, Perchlorate (TMRE) (Invitrogen, catalog number: T669 )
RPMI medium for the A2780 ovarian cancer cell line in use (Corning, catalog number: 15-040-CM ) or appropriate base media
N1 supplement (Sigma, catalog number: N6530-5ML )
Insulin (Sigma, catalog number: 10516-5ML )
Sodium pyruvate (GE Healthcare Life Sciences, catalog number: SH30239.01 )
L-glutamine (GE Healthcare Life Sciences, catalog number: SH30034.01 )
Penicillin-Streptomycin Solution (GE Healthcare Life Sciences, catalog number: SV30010 )
FGF (Sigma, catalog number: F0291-25ug )
EGF (Sigma, catalog number: E9644-0.2ug )
TIC media (see Recipes)
Equipment
BD FACSAria II or similar with cytometer setup and tracking (CS&T) beads, accudrop calibration beads, 100 μm FACS cell nozzle and a minimum of 488 nm and 561 nm laser and with emission filters of 525/50 BP for AldeFlour and 576/26 BP for TMRE
Centrifuges for 1.5 ml microfuge tubes and 15 ml tubes
Standard biosafety hood with laminar flow
Standard incubator for cell culture maintained at 37 °C and 5% CO2 with humidification
Light microscope with 4× and 10× objectives
Any confocal microscope with 555 nm laser (e.g., Zeiss LSM700 )
Software
BD FACSDiva (Version 8.0.1) (with automatic compensation calculation feature) or similar for flow cytometric sorting and analyses
Procedure
See Figure 1.
Note: Given the protocol pertains to live cells, carefully consider the time taken for each procedure for maximizing the rigor and reproducibility of the procedure. Do not halt the protocol at any step and implement efficient planning for minimizing time taken for each procedure. The time taken for each procedure in our hands is as follows:
Sample preparation for FACS: 2 h
FACS to isolate four populations: 2 h per sample tube.
Spheroid formation and ELDA: 15 d for the cell lineage used here. This time may vary with cell lineage.
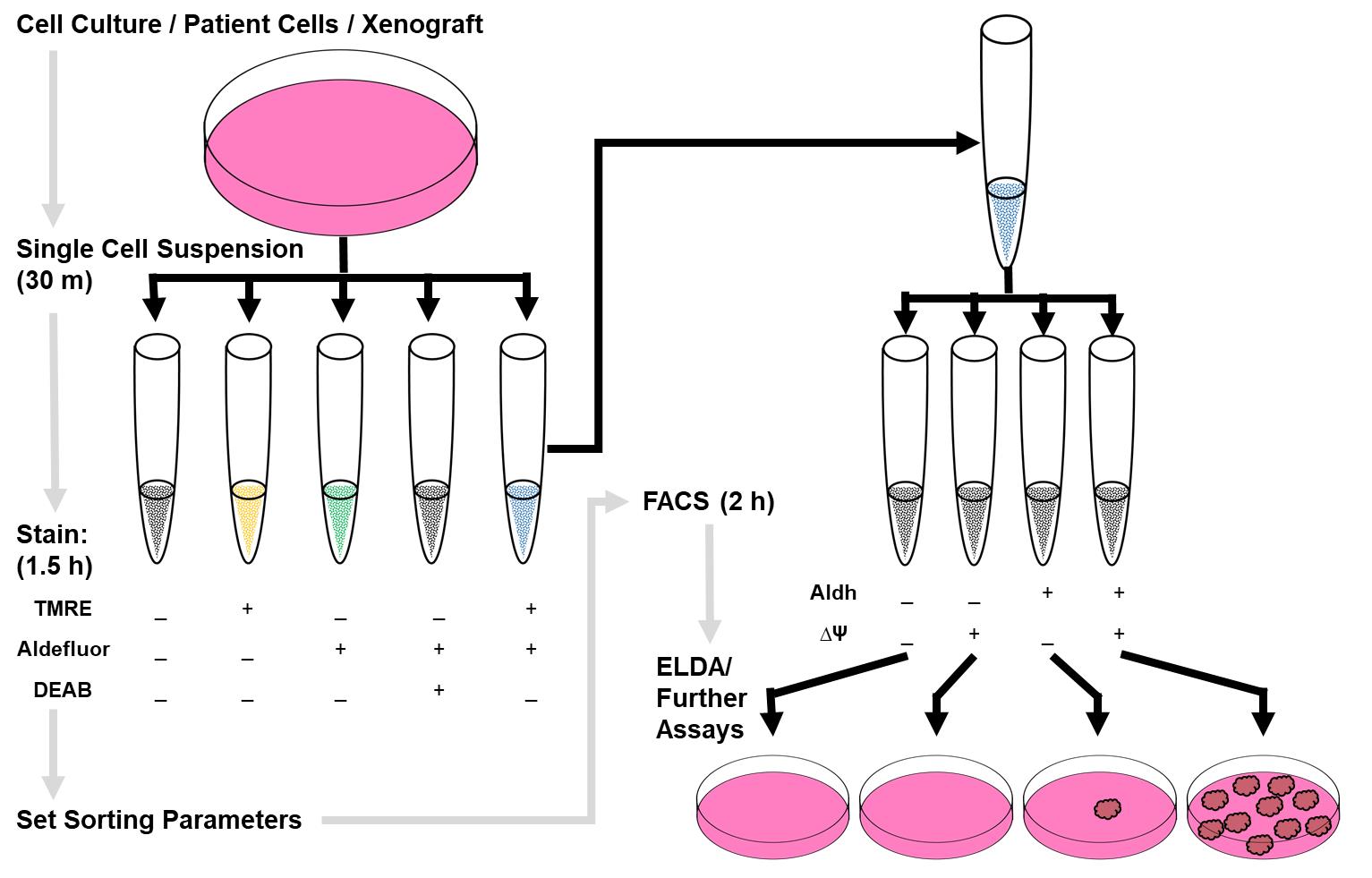
Figure 1. Graphical representation of work flow. Prepare control and sample tubes by bringing cells to single cell suspension and staining as described. Use control tubes to set parameters on the sorter and then sort four populations. The double positive population is enriched for mpSCs.
Sample preparation for Fluorescence-Activated Cell Sorting (FACS)
Preparations for Staining:
Reconstitute, aliquot and store reagents of the AldeFluor kit and TMRE according to manufacturer guidelines. Check manufacturer protocol for chemistry of the AldeFluor reagent.
Culture cells normally until ready to begin protocol. Two days prior to sorting, seed cells as typical to ~40% confluence in normal growth media, ensuring sorted populations will have enough cells required for subsequent experiments as determined from a mock experiment with 10 million cells.
Note: Although we developed the protocol with ovarian cancer cells, it can be used for any other cells with appropriate culture conditions and stem cell medium (in subsequent steps). Moreover, the staining and sorting protocol is also amenable to be used for cells directly isolated from any tissue or tumor xenografts. The sorted cells can be injected directly to animals for analyses of stem cell properties employing in vivo limiting dilution assay or other specific assays.
Warm aliquots of trypsin, DPBS, culture media, and AldeFluor buffer (from the AldeFluor kit) to 37 °C at least three hours prior to staining.
Trypsinize cells and transfer them to a 15 ml conical tube. Deactivate trypsin by adding an equal amount of culture media to the cell suspension. Make a homogenous cell suspension by pipetting thoroughly to ensure single cell suspension.
Note: As written, this protocol is for cell lines. To modify for primary cells and tissues, bring cells to a homogenous single cell suspension using appropriate techniques and begin from Step A1e.
Count cells from the homogenous cell suspension (in Step A1d) using preferred method.
Wash cells by first centrifuging cell suspension at 300 × g for 10 min at Room Temperature (RT), and removing the supernatant and resuspending cells in 3 ml warm DPBS. Finally, centrifuge cells at 300 × g for 10 min at RT.
Staining with AldeFluor and TMRE:
Aspirate the supernatant from the cell pellet (obtained in Step A1f) and resuspend cells in warm AldeFluor buffer to the concentration of 2 × 106 cells/ml, pipetting thoroughly to ensure single cell suspension.
Label 3, 1.5 ml microfuge tubes as “Unstained”, “AldeFluor”, and “TMRE” to serve as compensation controls to correct for overlap in the emission spectra of the AldeFluor reagent and TMRE. Label a fourth 1.5 ml microfuge tube as “DEAB” that, along with the “TMRE” tube, will serve as biological and gating controls for the respective AldeFluor and TMRE staining. Finally, label one 15 ml conical tube as “AldeFluor+TMRE” for the experimental sample to be sorted.
Distribute 1 × 106 cells (0.5 ml) each from cells suspension in Step A2a in Unstained, AldeFluor, TMRE and DEAB tubes. Transfer the rest of the cells to the AldeFluor+TMRE tube.
Follow manufacturer’s protocol to stain cells with AldeFlour reagent:
- Add 5 μl reagent per 2 × 106 cells in the AldeFluor, DEAB and AldeFluor+TMRE tubes.
- Inhibit Aldh activity in the DEAB tube using 5 μl per 1 × 106 cells.
Incubate all five tubes at 37 °C and 5% CO2 for 1 h (or as optimized from manufacturer’s protocol). Tap tubes to resuspend cells every 15 min.
Centrifuge cells in all 5 tubes at 300 × g for 10 min at RT.
While the previous step is ongoing, prepare 25 nM solution of TMRE in warm AldeFluor buffer.
Stain cells in the TMRE and AldeFluor+TMRE tubes with TMRE by aspirating the supernatant and resuspending in the 25 nM TMRE solution to 2 × 106 cells/ml.
Resuspend cells in the Unstained, AldeFluor, and DEAB tubes in warm AldeFluor Buffer to 2 × 106 cells/ml.
Incubate tubes at 37 °C and 5% CO2 for 15 min.
Centrifuge all five tubes at 300 × g for 10 min at RT.
Label five 5 ml flow cytometry tubes as in Step A2b.
Transfer appropriately stained cells to equivalently labeled flow cytometry tubes by aspirating the supernatant and resuspending cells in AldeFluor buffer to a concentration of 5 × 106 cells/ml. Ensure no bubbles are introduced to the tubes.
This TMRE staining protocol effectively stains energetically active mitochondria in any given cell. However, TMRE staining of mitochondria should be confirmed by adding stained cells to a live cell chambered coverglass, allowing them to settle for 5 min, and looking at them under a confocal microscope (Figure 2A). Staining with MitoTracker Green may be necessary to ensure the staining is mitochondrial. Protocol described in (Mitra and Lippincott-Schwartz, 2010). If TMRE does not incorporate to mitochondria, TMRE staining should be re-optimized by varying first TMRE concentration and incubation time in a given cell density.
Flow cytometry data acquisitions for sorting (as optimized using the BD FacsAria II)
Note: The following precautions are necessary:
Data acquisition and sorting should be accomplished within 2 h or less of transferring sample to 5 ml flow cytometry tubes. Longer time taken may cause loss of signal. For large numbers of cells, seed multiple plates so that the first set of cells can be sorted within two hours. Process the next set while the first is sorting and so on.
Optimal sorter performance should be ensured by running cytometer setup and tracking (CS&T) beads. Drop delay should be calculated using accudrop calibration beads.
Data acquisition can be performed using software such as BD FACSDiva (Version 8.0.1) or similar on the sorter.
Attach the 100 μm nozzle to sort at 20 psi pressure. Maintain 4 °C for both the sample and collection tubes through the sort. Select the following parameters: Forward Scatter (FSC in linear scale) and Side Scatter (SSC in linear scale), channels for AldeFluor (Ex: 488 nm, Em filter: 525/50 BP; log scale) and TMRE (Ex: 561 nm, Em filter: 576/26 BP, log scale).
Adjust parameters using the compensation controls as follows:
Using the unstained sample do the following adjustments:
Adjust PMT voltages of FSC and SSC to display and distinguish the target population of interest from cell debris.
Gate out the cell debris using the parameters FSC Area (FSC-A) and SSC Area (SSC-A) (Figure 2B).
Gate out cell doublets using FSC-Height (FSC-H), FSC-A, SSC Width (SSC-W) and FSC-Height (SSC-H) in two subsequent gates (Figures 2C, 2D).
Adjust PMT voltages of Unstained sample to have the auto fluorescence signal peak below the second decade of the log scale for both TMRE (Figure 2E, left) and AldeFluor (Figure 2E, right).
Using the TMRE stained sample, adjust PMT voltage for TMRE detection to ensure the signal is within the dynamic range (Figure 2F).
Using the AldeFluor stained sample, adjust PMT voltage for AldeFluor detection to ensure the signal is within the dynamic range (Figure 2G).
Once the voltages are finalized, record data with 20,000 events from the Unstained, AldeFluor and TMRE tubes, and use the automatic compensation calculation feature to calculate the compensation of fluorescence signal and apply to the experiment.
Note: Compensation and biological controls may not be necessary for every sort, after reproducibility of the sort has been established. However, re-introduce controls if the staining has been modified or if dealing with samples with large inherent variations like patient samples. If dealing with patient samples, extra steps have to be taken for gating out dead cells.
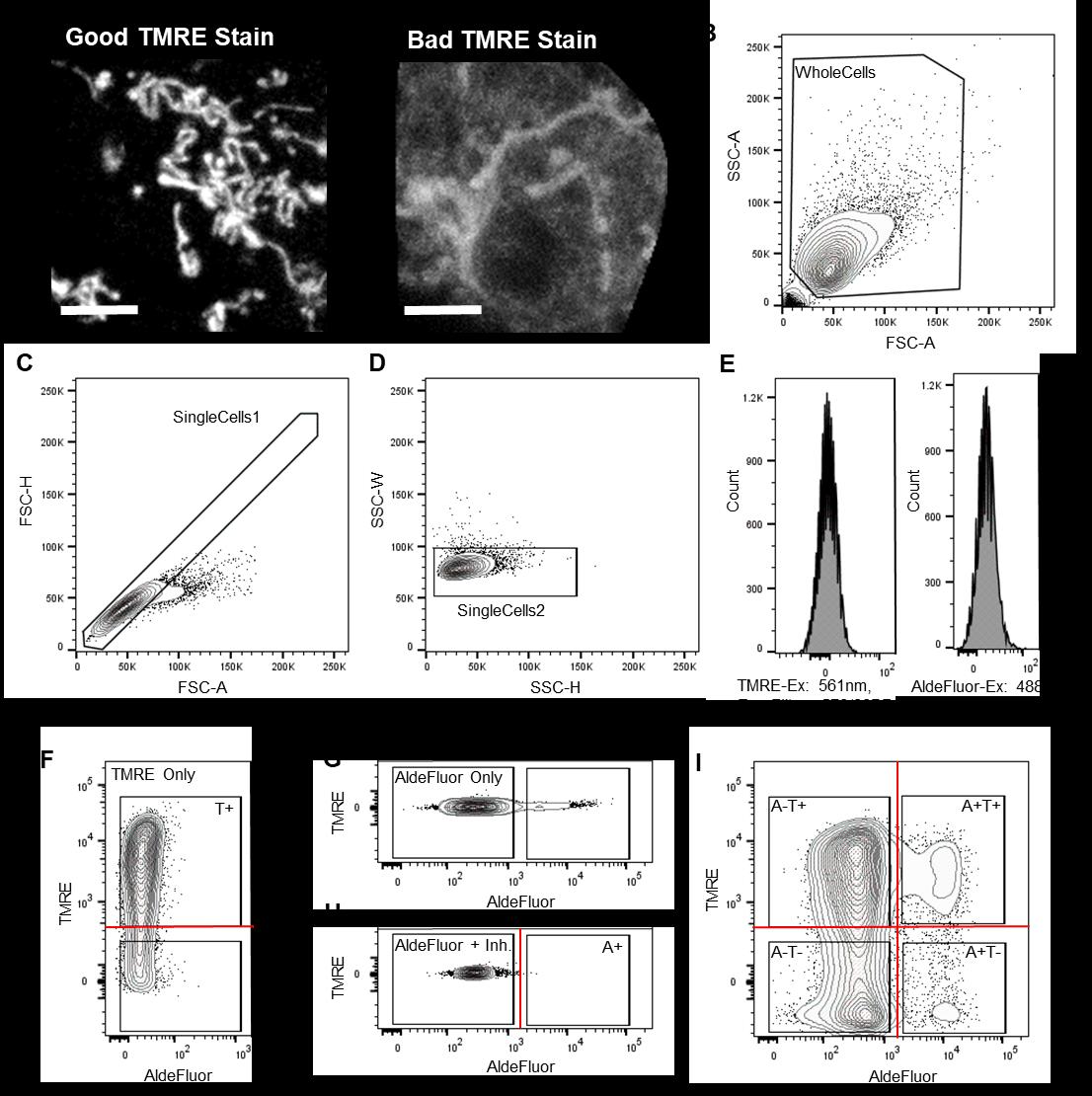
Figure 2. FACS strategy for enriching and depleting mitochondria primed TICs. A. Micrographs of iPSCs stained with TMRE as examples of good (left) and bad (right) incorporation of the dye into mitochondria. Scale bars represent 5 µm. B. Contour plot showing gating of whole cells using FSC-A and SSC-A, respectively, for the epithelial ovarian cancer cell line A2780-CP. C. Contour plot showing first gating of single cells using FSC-H and FSC-A for A2780-CP. D. Contour plot showing second gating of single cells using SSC-H and SSC-W for A2780-CP. E. Histograms showing auto-fluorescence of A2780-CP cells using TMRE parameters (left) and AldeFluor parameters (right) set within second decades of fluorescence intensity. F. Contour plot showing TMRE staining of A2780-CP cells within the dynamic range of the instrument. G. Contour plot showing AldeFluor staining of A2780-CP cells within the dynamic range of the instrument. H. Contour plot showing AldeFluor staining after inhibiting Aldh activity using DEAB. This plot marks the boundary of AldeFluor– cells. I. Contour plot showing final sort gating of four populations of A2780-CP: AldeFluor+ TMRE+, AldeFluor+ TMRE–, AldeFluor– TMRE+, AldeFluor– TMRE–.
FACS to isolate four populations: AldeFluor+ TMRE+, AldeFluor+ TMRE–, AldeFluor– TMRE+, AldeFluor– TMRE–
After executing Step B2c, use biological/gating controls to set the sorting gates as follows:
Record 20,000 events using DEAB tube and TMRE tube.
Set thresholds for AldeFluor+ (Figure 2H, red line) and TMRE+ (Figure 2F, red line).
Record 20,000 events using the AldeFluor+TMRE stained sample.
Draw sorting gates around distinct populations within each quartile as shown in Figure 1H (black rectangles) for sorting the following cell populations: AldeFluor+ TMRE+, AldeFluor+ TMRE–, AldeFluor– TMRE+, AldeFluor– TMRE–.
Note: Gating out debris was sufficient to exclude dead cells in our cell lines, where viability after acquiring a single cell suspension typically exceeded 90%. Fresh tissues and cells of other lineages may require a viability stain to exclude dead cells. Stains must be compatible with TMRE fluorescence (e.g., gating live cells by DAPI exclusion).
Set the flow cytometer to record 100,000 events from the gated pre-sort sample. Use the BD FACSDiva (Version 8.0.1) software to determine the percentage of cells in each population. The AldeFluor+TMRE+ population is enriched for mitochondria primed TICs.
For collection of population to be sorted, add 0.5 ml of either sterile culture media or appropriate stem cell media to four 5 ml flow cytometry collection tubes and label the tubes “AldeFluor+ TMRE+”, “AldeFluor+ TMRE–”, “AldeFluor– TMRE+” and “AldeFluor– TMRE–”. Place the collection tubes in the position in the FACSAria corresponding to the sorting gate (Figure 2I) for the population labeled on the tube.
Notes:
The TIC medium or stem cell medium composition may differ between lineages. Therefore, use the appropriate stem cell medium for the spheroid assay.
If the FACS instrument is in a different location than that of the sample preparation, transfer cells, collection tubes, extra flow cytometry tubes, and aliquots of AldeFluor Buffer and cell culture media at RT in a sterile container protected from exposure to light.
Distribution of TMRE fluorescence in the example cells provided here is bimodal, lending easily to separate positive and negative populations. Should the distribution be not bimodal, take the bottom 10% of the normal distribution for the negative population and the top 10% for the positive.
Use 100 μm nozzle to sort at 20 psi pressure. Maintain 4 °C for both the sample and collection tubes through the sort. Maintain the flow rate to have a threshold rate of 9,000 to 10,000 events per second. Vortex the sample at medium speed before loading to the sample port. Maintain sample agitation at 200 rpm through the sort.
Allow the sort to continue until the targeted cell number is reached for each population, replacing collection tubes as needed.
Maintain sorted cells at 4 °C until needed for subsequent experiments.
Before seeding or harvesting sorted cells, transfer cells to labeled 15 ml conical tubes and centrifuge at 300 × g for 10 min at 4 °C.
Aspirate supernatant and resuspend cells in DPBS or desired cell culture media. Centrifuge at 300 × g for 10 min at 4 °C and aspirate the supernatant.
Use these cell pellets or cell suspension for further experiments, as appropriate.
Extreme Limiting Dilution Analysis (ELDA) (Hu and Smyth, 2009) to quantitate TIC frequency from spheroids formed by the sorted population.
Note: Although this can be optional, we recommend performing this assay when using the sorting protocol for the first time in any new lineage or cell type. This is because the sorted population primed for maximal self-renewal and proliferation, as quantified by this assay, may or may not be similar to the ovarian cancer system shown here. This in vitro assay can also be replaced by the in vivo limiting dilution assay using xenografts.
Spheroids are formed as follows:
Based on the number of cells sorted, resuspend each population in stem cell media to a concentration of 104 cells/ml (final volume of at least 3 ml).
Serially dilute a portion of the above suspension to 103, 102 and 10 cells/ml.
Note: Each dilution of 10 fold can be achieved by adding 300 μl of each successive cell suspension to 2.7 ml stem cell media in 15 ml conical tubes. We always seeded cells using serial dilution based on counting live cells by Trypan Blue exclusion after the wash steps. However, seeding can also be accomplished by sorting the desired number of cells directly into each well of the 96-well Ultra-Low Attachment plate.
Distribute each cell suspension among at least 24 wells in a 96-well Ultra-Low Attachment plate (100 μl /well). The more total wells used for each cell concentration, the higher the statistical power of the assay. Leave plates in a cell culture incubator. In the example experiment in Figure 3, we used 4 plates, one for each population. In each plate, we seeded 24 wells with 1 cell/well, 24 wells with 10 cells/well, 24 wells with 100 cells/well, and 24 wells with 103 cells/well.
Every second day, examine each well using the 10× objective of a light microscope for the formation of dense spheroids with tight edges (Figure 3A).
Spike cells every second day with 10% of the initial volume of stem cell media until conspicuous tumorspheres with tight edges are detected in the positive control group (Figure 3A); in the example provided Aldh+ (A+) groups are positive control while Aldh- (A-) groups are negative control (Figure 3).
On the final day, note the number of wells that contain spheroids for each concentration for each population.
Note: Self-renewing stem cells will maintain the abundance of the stem cell marker, while differentiation will reduce it. Self-renewal in the spheroids can be confirmed by quantifying the abundance of the cells positive for the stem cell marker in the spheroids, and comparing it to that of the original population. This is important to rule out mitochondria driven priming of stem cell differentiation (Chandel et al., 2016).
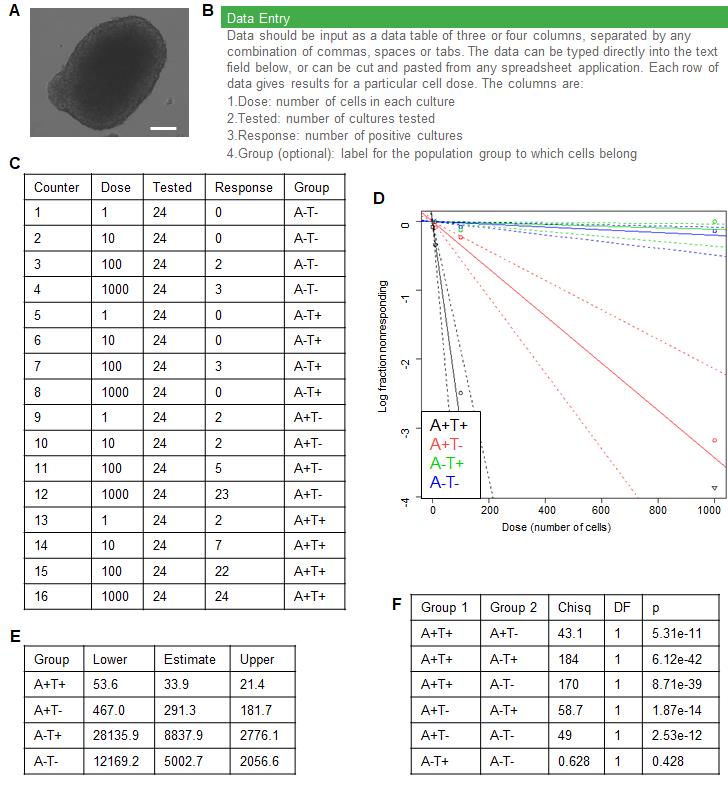
Figure 3. Extreme Limiting Dilution Analysis of Four Sorted Populations. A. Example of an advanced spheroid 21 days after seeding acquired using transmitted light on an EVOS fluorescence microscope. Scale bar represents 200 μm. B. Screen capture of the instructions from the ELDA webtool. C. Table showing example data from an ELDA conducted on FACS sorted A2780-CP cells. D. Output of the online ELDA tool plotting input data along with a linear fit used to extrapolate the frequency of sphere-forming cells in each population. E. Table showing estimate and 95% confidence interval of frequency of sphere-forming cells for each population in the A2780-CP cell line. F. Table showing pairwise hypothesis testing of differences in frequencies of sphere-forming cells in each population as output by the online ELDA tool.Data is analyzed to obtain TIC frequency as follows:
To access the ELDA statistics web tool, open url: http://bioinf.wehi.edu.au/software/elda/.
Enter the population, cells/well, total number of wells, and number of wells containing spheroids into the online ELDA statistics tool (Figures 3B, 3C).
Report the sphere forming capacity and p-value for each population for each day (Figures 3D-3F).
The AldeFluor+ TMRE+ (A+T+) population with maximum frequency of self-renewing and proliferating TICs is deemed as the mitochondria primed TIC population. The A- population has a markedly lower frequency of self-renewing and proliferating TICs, recapitulating the in vivo limiting dilution assay (see Spurlock et al., 2019 for more details).
Example applications of the method to study mitochondrial morphology by confocal microscopy in ovarian TICs:
Note: We have applied the protocol to characterize specific mitochondrial properties (by confocal microscopy), cell cycle markers (by immunoblotting), and drug sensitivity (by crystal violet staining after drug incubation) within the 4 sorted populations.
- Coat 8-well live cell chamber with Geltrex or lineage-appropriate matrix.
- Resuspend cell pellets from Step C9 half in normal culture media and half in stem cell media.
- Seed cells in live cell chamber to 80% confluence and allow to recover overnight.
- Stain cells with MitoTracker Far Red to measure mitochondrial morphology and MitoSox to measure mitochondrial oxidation according to manufacturer’s protocols.
- Acquire images according to the MitoGraph section of our protocol (Spurlock et al., 2019 and 2020).
- Figures 4A, 4B show the comparisons among the four populations allowed to recover in normal culture media using our metrics for Fission and Fusion-1. Figure 4C shows the relative mitochondrial oxidation of the four populations.
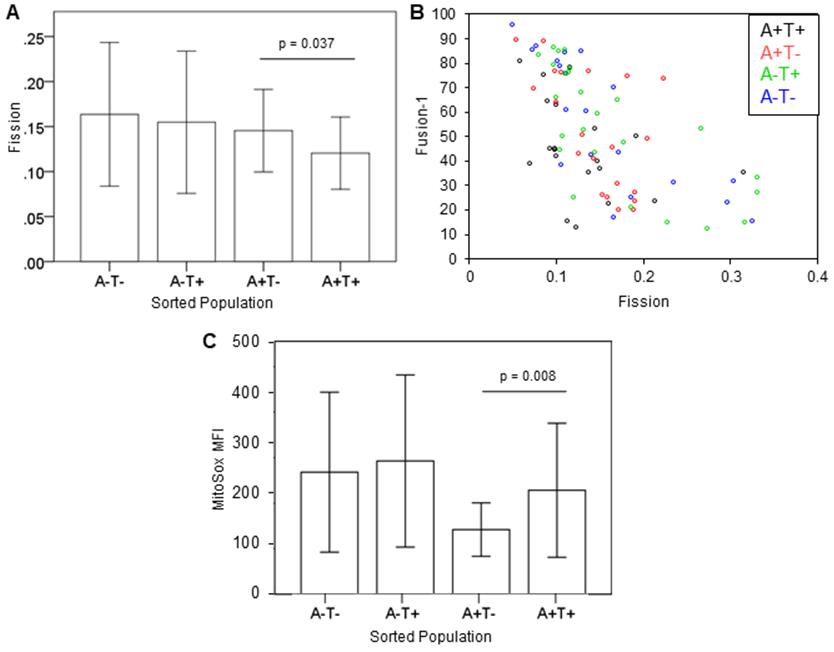
Figure 4. Application of Method to Study Mitochondrial Morphology of Ovarian TICs. A. Bar graph showing the mean Fission metric of cells from the 4 sorted population. N=18-25. Error Bar shows ± 1SD. P-value is from Student’s t-test. B. Dot plot showing an inverse relationship between Fission and Fusion metrics of each population. C. Bar graph showing the mean MitoSox fluorescence intensity of cells from each population. N = 19-22. Error Bar shows ± 1SD. P-value is from Student’s t-test.
Recipes
TIC media
Serum-free media base (RPMI or DME/F12)
1× N1 supplement
10 μg/ml insulin
20 ng/ml EGF
10 ng/ml bFGF
1% sodium pyruvate
1% L-glutamine
1% pen-strep
Discard unused media after 1 month and make fresh
Acknowledgments
This work was supported by the National Institutes of Health (NIH) [R33ES025662] to B.S. and K.M. This protocol was originally reported in “New quantitative approach reveals heterogeneity in mitochondrial structure-function relations in tumor-initiating cells” published in 2019 in the Journal of Cell Science (Spurlock et al., 2019).
Competing interests
The authors have no competing interests.
References
- Chandel, N. S., Jasper, H., Ho, T. T. and Passegue, E. (2016). Metabolic regulation of stem cell function in tissue homeostasis and organismal ageing. Nat Cell Biol 18(8): 823-832.
- Folmes, C. D. and Terzic, A. (2016). Energy metabolism in the acquisition and maintenance of stemness.Semin Cell Dev Biol 52: 68-75.
- Hu, Y. and Smyth, G. K. (2009). ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods 347(1-2): 70-78.
- Intlekofer, A. M. and Finley, L. W. S. (2019). Metabolic signatures of cancer cells and stem cells. Nat Metab 1(2): 177-188.
- Ito, K. and Suda, T. (2014). Metabolic requirements for the maintenance of self-renewing stem cells.Nat Rev Mol Cell Biol 15(4): 243-256.
- Liang, R., Arif, T., Kalmykova, S., Kasianov, A., Lin, M., Menon, V., Qiu, J., Bernitz, J. M., Moore, K., Lin, F., Benson, D. L., Tzavaras, N., Mahajan, M., Papatsenko, D. and Ghaffari, S. (2020). Restraining lysosomal activity preserves hematopoietic stem cell quiescence and potency. Cell Stem Cell 26(3): 359-376 e357.
- Magee, J. A., Piskounova, E. and Morrison, S. J. (2012). Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell 21(3): 283-296.
- Marcato, P., Dean, C. A., Giacomantonio, C. A. and Lee, P. W. (2011). Aldehyde dehydrogenase: its role as a cancer stem cell marker comes down to the specific isoform. Cell Cycle 10(9): 1378-1384.
- Margineantu, D. H., Gregory Cox, W., Sundell, L., Sherwood, S. W., Beechem, J. M. and Capaldi, R. A. (2002). Cell cycle dependent morphology changes and associated mitochondrial DNA redistribution in mitochondria of human cell lines. Mitochondrion 1(5): 425-435.
- Martinez-Outschoorn, U. E., Peiris-Pages, M., Pestell, R. G., Sotgia, F. and Lisanti, M. P. (2017). Cancer metabolism: a therapeutic perspective. Nat Rev Clin Oncol 14(1): 11-31.
- Mitra, K. and Lippincott-Schwartz, J. (2010). Analysis of mitochondrial dynamics and functions using imaging approaches. Curr Protoc Cell Biol Chapter 4: Unit 4 25 21-21.
- Pasto, A., Bellio, C., Pilotto, G., Ciminale, V., Silic-Benussi, M., Guzzo, G., Rasola, A., Frasson, C., Nardo, G., Zulato, E., Nicoletto, M. O., Manicone, M., Indraccolo, S. and Amadori, A. (2014). Cancer stem cells from epithelial ovarian cancer patients privilege oxidative phosphorylation, and resist glucose deprivation.Oncotarget 5(12): 4305-4319.
- Sancho, P., Burgos-Ramos, E., Tavera, A., Bou Kheir, T., Jagust, P., Schoenhals, M., Barneda, D., Sellers, K., Campos-Olivas, R., Grana, O., Viera, C. R., Yuneva, M., Sainz, B., Jr. and Heeschen, C. (2015). MYC/PGC-1alpha Balance Determines the Metabolic Phenotype and Plasticity of Pancreatic Cancer Stem Cells. Cell Metab 22(4): 590-605.
- Schieke, S. M., Ma, M., Cao, L., McCoy, J. P., Jr., Liu, C., Hensel, N. F., Barrett, A. J., Boehm, M. and Finkel, T. (2008). Mitochondrial metabolism modulates differentiation and teratoma formation capacity in mouse embryonic stem cells. J Biol Chem 283(42): 28506-28512.
- Spurlock, B., Gupta, P., Basu, M. K., Mukherjee, A., Hjelmeland, A. B., Darley-Usmar, V., Parker, D., Foxall, M. E. and Mitra, K. (2019). New quantitative approach reveals heterogeneity in mitochondrial structure-function relations in tumor-initiating cells. J Cell Sci 132(9).
- Spurlock, B., Tullet, J., Hartman, J. L. t. and Mitra, K. (2020). Interplay of mitochondrial fission-fusion with cell cycle regulation: Possible impacts on stem cell and organismal aging. Exp Gerontol 135: 110919.
- Sukumar, M., Liu, J., Mehta, G. U., Patel, S. J., Roychoudhuri, R., Crompton, J. G., Klebanoff, C. A., Ji, Y., Li, P., Yu, Z., Whitehill, G. D., Clever, D., Eil, R. L., Palmer, D. C., Mitra, S., Rao, M., Keyvanfar, K., Schrump, D. S., Wang, E., Marincola, F. M., Gattinoni, L., Leonard, W. J., Muranski, P., Finkel, T. and Restifo, N. P. (2016). Mitochondrial membrane potential identifies cells with enhanced stemness for cellular therapy. Cell Metab 23(1): 63-76.
- Toledo-Guzmán, M. E., Hernandez, M. I., Gomez-Gallegos, A. A. and Ortiz-Sanchez, E. (2019). ALDH as a Stem Cell Marker in Solid Tumors. Curr Stem Cell Res Ther 14(5): 375-388.
- Vassalli, G. (2019). Aldehyde Dehydrogenases: Not Just Markers, but Functional Regulators of Stem Cells.Stem Cells Int 3904645.
- Viale, A., Corti, D. and Draetta, G. F. (2015). Tumors and mitochondrial respiration: a neglected connection.Cancer Res 75(18): 3685-3686.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Spurlock, B., Hanumanthu, V. S. and mitra, K. (2021). Strategy of Isolating ‘Primed’ Tumor Initiating Cells Based on Mitochondrial Transmembrane Potential. Bio-protocol 11(5): e3945. DOI: 10.21769/BioProtoc.3945.
Category
Cancer Biology > Cancer stem cell > Cell biology assays
Cell Biology > Cell isolation and culture > Cell isolation > Flow cytometry
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link