- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Purification of Cytosolic Phospholipase A2α C2-domain after Expression in Soluble Form in Escherichia coli
(*contributed equally to this work) Published: Vol 11, Iss 3, Feb 5, 2021 DOI: 10.21769/BioProtoc.3906 Views: 3120
Reviewed by: Tao JiangArvind PandayAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Optimized Secretome Sample Preparation From High Volume Cell Culture Media for LC–MS/MS Proteomic Analysis
Basil Baby Mattamana [...] Peter Allen Faull
Dec 20, 2025 1229 Views

A One-Step Method for Efficient Purification of Functional Cas9 Protein
Xinzhi Duan [...] Aihua Mao
Feb 5, 2026 63 Views

On-Column Dual-Gradient Refolding for Efficient Recovery of Insoluble Affinity-Tagged Recombinant Proteins
Anna Vlaskina [...] Maxim Patrushev
Feb 5, 2026 38 Views
Abstract
Previous expression/purification strategies for cytosolic phospholipase A2α C2-domain in Escherichia coli have relied on refolded protein recovered from inclusion bodies and sometimes containing C-terminal Cys139Ala and Cys141Ser substitutions to eliminate potential refolding complications induced by Cys residues. The protocol presented herein describes an effective method for the expression of cytosolic phospholipase A2α C2-domain in soluble form in E. coli and subsequent purification to homogeneity. This protocol, which utilizes a cleavable 6xHis-SUMO tag, has recently been used to gain insights into the structural basis of phosphatidylcholine recognition by the C2-domain of cytosolic phospholipase A2α (Hirano et al., 2019)
Keywords: Cytosolic phospholipase A2αBackground
Phospholipase A2 (PLA2) is a member of a diverse enzyme superfamily that hydrolyzes the sn-2 acyl ester linkage of phosphoglycerides (Smith, 1989; Dennis et al., 2011; Mouchlis and Dennis, 2019). Cytosolic PLA2α (cPLA2α), a Group IV mammalian PLA2 family member, preferentially releases arachidonic acid from phosphoglycerides in a cytosolic Ca2+-concentration-dependent manner (Clark et al., 1991; Shimizu et al., 2006; Leslie et al., 2010; Vasquez et al., 2018; Astudillo et al., 2019). Arachidonic acid generated by cPLA2α serves as a precursor of pro-inflammatory eicosanoids, including certain prostaglandins and leukotrienes. Consequently, cPLA2α-mediated bioactive lipid production plays a major regulatory role in physiological and pathogenic processes (Bonventre et al., 1997; Uozumi et al., 1997; Leslie, 2015). Gaining insights into cPLA2α activation by divalent cation and lipid regulatory mediators are of great importance because arachidonic acid release by cPLA2α at the membrane surface is the rate-limiting step in eicosanoid production. Mechanistically, the membrane translocation of cPLA2α is driven primarily by its N-terminal C2-domain rather than its catalytic domain (Clark et al., 1991; Nalefski et al., 1994 and 1998; Davletov et al., 1998; Dessen et al., 1999). Complexation of two Ca2+ ions by the C2-domain neutralizes several Asp residues, facilitating protein docking and penetration into the membrane interface region (Davletov et al., 1998; Perisic et al., 1998; Xu et al., 1998; Ball et al., 1999; Bittova et al., 1999; Dessen et al., 1999; Malmberg et al., 2003; Malmberg and Falke, 2005). Among lipids, ceramide-1-phosphate is an established regulator of the cPLA2α action and C2-domain-driven membrane interaction (Pettus et al., 2004; Stahelin et al., 2007; Lamour et al., 2009; Ward et al., 2013). Previous studies of cPLA2α C2-domain have relied on expression in E. coli, recovery of inclusion bodies, followed by the laborious task of refolding of recombinant protein prior to purification. Herein we describe a direct approach for obtaining purified C2-domain in soluble form. The approach utilizes established technology involving protein fusion with a cleavable 6xHis-SUMO tag to enable expression of soluble cPLA2α C2-domain in E. coli followed by affinity purification (Malakhov et al., 2004; Marblestone et al., 2006; Yan et al. 2009; Peroutka et al., 2011). An added advantage is that removal of the 6xHis-SUMO tag relies on Ulp1 protease recognition of SUMO tertiary structure, thus avoiding unspecific cleavage associated with other proteases that target a linear protein sequence. The protocol, which has been used to facilitate successful structural characterization of chicken C2-domain complexed with dihexanoyl phosphatidylcholine, also enables successful expression and purification of mouse, zebrafish, and human phospholipase A2α C2-domains in soluble form (Hirano et al., 2019).
Materials and Reagents
Amicon Ultra Centrigual Filter (Millipore Sigma, catalog number: UFC901024 )
Escherichia coli strain BL21 star (DE3) (Thermo Fisher Scientific, catalog number: C601003 )
pET-SUMO vector
Chicken cPLA2α C2-domain cDNA (synthesized by GENEWIZ)
cOmpleteTM His-Tag Purification Resin (Millipore Sigma, catalog number: 5893682001 )
HiTrap Q HP (Millipore Sigma, catalog number: GE29-0513-25 )
Superdex 75 prep grade (Millipore Sigma, catalog number: S6657 )
Isopropyl-β-D-thiogalactopyranoside (IPTG) (Thermo Fisher Scientific, catalog number: 34060 )
Imidazole (Sigma-Aldrich, catalog number: 56748 )
Glycerol (Thermo Fisher Scientific, catalog number: BP229-1 )
Tris Base UltraPure (Research Products Intl., catalog number: T60040 )
NaCl (Sigma-Aldrich, catalog number: 71376 )
CaCl2 (Sigma-Aldrich, catalog number: C7902 )
Solution A (see Recipes)
Solution B (see Recipes)
Solution C (see Recipes)
Solution D (see Recipes)
Solution E (see Recipes)
Solution F (see Recipes)
Solution G (see Recipes)
Equipment
250 ml and 2 L Erlenmeyer culture flasks
High speed centrifuge (Beckman Coulter)
Shaker incubator (Sanyo, orbital incubator)
Sonicator (Heat Systems-Ultrasonics Inc., model: W-225 )
Benchtop centrifuge (Spectrafuge)
ÄKTA Purifier chromatography system (GE Healthcare)
Procedure
Transform E. coli strain BL21 star (DE3) cells with pET-SUMO vector containing the C2-domain open reading frame (residues 16-140) of human, chicken, mouse, or zebrafish cPLA2α.
Thaw one vial of competent cells on ice per transformation.
Add 5-10 ng of the plasmid in a volume of 1-5 μl to the cells and mix by gently tapping.
Incubate the vial(s) on ice for 30 min.
Heat shock the cells by incubating the vial(s) for exactly 30 s in a 42 °C incubator and then remove the vial(s) from the 42 °C incubator and quickly place on ice.
Add 250 μl of pre-warmed SOC medium to the vial(s) and then incubate the vial(s) at 37 °C for 1 h.
Plate the transformation reaction onto LB plates containing kanamycin (25 μg/ml) for plasmid selection. Invert the plates and incubate at 37 °C overnight.
Notes:
Chicken cPLA2α C2-domain sequences
Synthesized ORF sequence
TATAGCCATGTGTTTACCGTGACCGTGCGCAAAGCCACCAATGTGACCAAAGGCGCCATTGGCGATATGCTGGATACCCCTGATCCTTATGTGGAGCTGTTTATTCCGAGCGCACCGGATTGCCGCAAACGCACCAAGCACTTCAACAACGACGTGAACCCGGTGTGGAACGAGACCTTCGAGTTTATCCTGGACCCGAACCAGGATAACGTGCTGGAAGTGACCCTGATGGACGCCAACTATGTGATGGATGAGACCCTGGGCATGGCCACCTTTCCGATTAGCAGCCTGAAACTGGGCGAGAAGAAAGAAGTTCAGCTGACCTTCAACAACGTGACCGAGATGACCCTGGAACTGAGCCTGGAAGTTTGCAGC
Wild-type ORF sequence
TACTCACACGTATTCACTGTGACAGTAAGGAAAGCCACAAATGTCACAAAAGGAGCCATTGGCGACATGCTTGACACTCCAGACCCCTATGTAGAGCTGTTCATCCCTTCCGCTCCTGACTGCAGGAAGAGAACTAAACATTTCAATAATGATGTGAATCCTGTGTGGAATGAGACCTTCGAATTCATTCTGGACCCCAACCAGGACTCTTGGTATGGCCAACAATGTTCTTGAGGTTACTTTGATGGATGCCAACTACGTTATGGATGAGCATTTCCTATATCCTCTTTGAAACTGGGAGAGAAGAAGGAGGTCCAGTTAACTTTCAACAATGTTACAGAAATGACTTTGGAGCTGTCTCTTGAAGTGTGCTCT
Chicken C2-domain Protein sequence coded by either ORF
YSHVFTVTVRKATNVTKGAIGDMLDTPDPYVELFIPSAPDCRKRTKHFNNDVNPVWNETFEFILDPNQDNVLEVTLMDANYVMDETLGMATFPISSLKLGEKKEVQLTFNNVTEMTLELSLEVCS
In the synthesized chicken C2-domain ORF, codon usage is optimized for E. coli expression. BamHI and SalI restriction sites are used for open reading frame (ORF) ligation into a modified pET-28-SUMO vector (kanamycin-resistant). Prior to insertion, the human ORF requires mutation to remove a single BamHI internal restriction site 5’-192GGATCC197-3’ without changing the protein sequence. The corresponding GGACCC sites in the chicken sequences are underlined.
Collect 5-10 colonies and inoculate 20 ml of autoclaved LB medium (supplemented with 25 μg/ml kanamycin) in a 250 ml Erlenmeyer culture flask. Incubate overnight at 37 °C while shaking at 200 rpm.
Use 10 ml of culture from Step 2 to inoculate 1 L of fresh LB medium containing 25 μg/ml kanamycin and grow in a 2 L Erlenmeyer culture flask at 37 °C until OD600 = 0.6-1.0.
Cool the resulting culture to 20 °C and add IPTG to yield a final concentration of 0.1 mM.
After 16-20 h of induction with IPTG at 20 °C and stirring at 220 rpm, collect the cells by centrifuging the culture at 5,000 × g for 10 min.
Suspend harvested cells in 30 ml of solution A and freeze at -30 °C until use.
Thaw frozen cells and disrupt by French press or sonication. When using probe sonication, apply 6 × 20 s pulses while keeping the sample (30 ml) in an ice bath to prevent heat damage.
Transfer the lysate to a centrifuge bottle and centrifuge at 25,000 × g for 30 min at 4 °C before loading the resultant supernatant onto a 3 ml column of cOmplete His-Tag Purification Resin pre-equilibrated with Solution A.
Wash the resin with more than 10 resin-bed volumes using Solution B and then elute the protein with 10 ml of Solution C.
Add Ulp1 protease to remove the N-terminal 6xHis-SUMO tag.
Note: An advantage of the Ulp1 protease compared to other sequence-based proteases is the ability of Ulp1 to recognize SUMO tertiary structure and cleave at the C-terminal end of the conserved Gly-Gly sequence in SUMO. This ability eliminates unspecific cleavage associated with other proteases that target a linear protein sequence. SUMO cleavage by Ulp1 does leave two extra amino acids (Gly-Ser) at the N-terminal end of the C2-domain.
Dialyze the sample with 2 L of Solution D for more than 3 h at 4 °C.
Equilibrate an anion exchange chromatography column (HiTrap Q HP, 1 ml, 0.7 × 2.5 cm) with Solution E using a GE AKTA Prime Liquid Chromatography System.
Load the dialyzed solution from Step 11 onto the pre-equilibrated anion exchange column and elute using a 3 to 50% linear gradient of Solution F.
Note: The anion exchange chromatography (Steps 12 and 13) is optional. In Hirano et al. (2019), this chromatography approach was included for crystallography but not for fluorescence resonance energy transfer or surface plasmon resonance experiments since SDS-PAGE revealed similar very high purity in either case.
Combine the fractions containing cPLA2α C2-domain and load onto cOmpleteTM His-Tag Purification Resin (3 ml) pre-equilibrated with Solution B, wash the resin with 10 resin-bed volumes of Solution B and then elute the protein with 10 ml of Solution C.
Concentrate the flow-through using an Amicon Ultra-15 Centrifugal Filter (15 ml sample volume, 10 kDa cutoff) to less than 10 ml and load onto a HiLoad16/60 Superdex 75 pg gel filtration column (L x I.D.; 60 cm × 16 mm, GE Healthcare) pre-equilibrated with Solution G for final size exclusion purification of C2-domain monomer using Solution G (Figure 1). SDS-PAGE analysis shows the purity of the resulting monomeric protein (Figure 2).
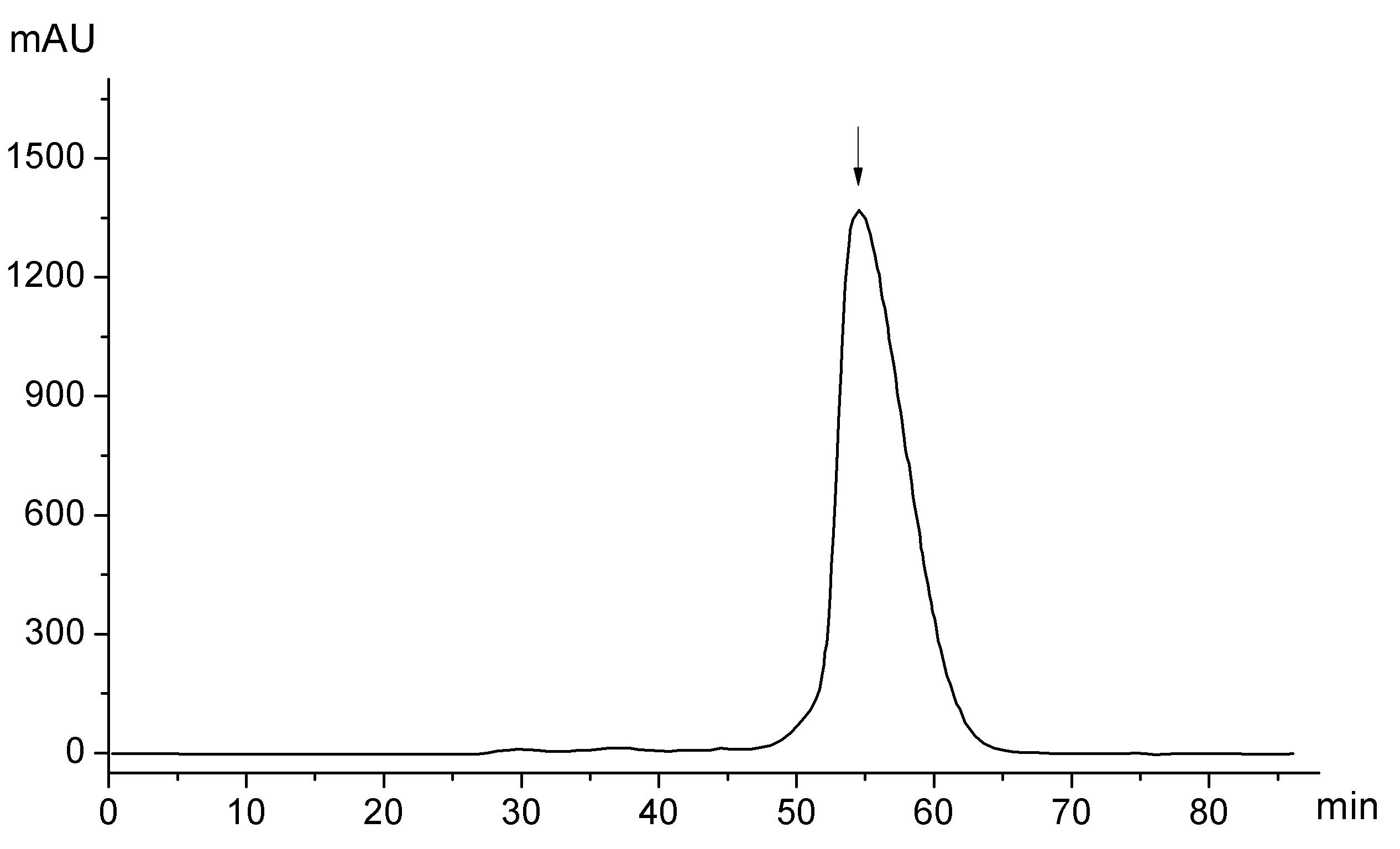
Figure 1. Chromatogram of gel filtration purification (Step 15 in the Procedure). Y-axis represents absorbance at 280 nm whereas the X-axis represents elution time. The arrow corresponds to the elution peak for monomeric cPLA2α C2-domain (16-140; 14.3 kDa).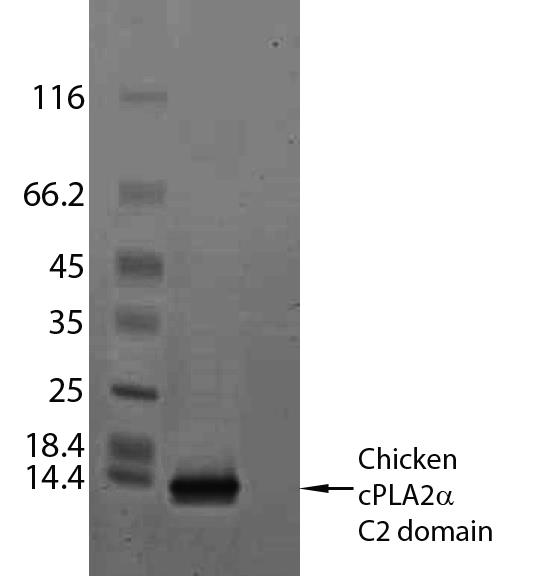
Figure 2. SDS-PAGE analysis of purified chicken cPLA2α C2-domain (16-140). The protein load for purified C2-domain equals 1-2 μg.Concentrate up to 20 mg/ml and store at -80 °C until use or add glycerol to a final concentration of 50% (v/v) and store at -20 °C. Typical yields of human C2-domain (1.3 mg) and chicken C2-domain (2.2 mg) are readily achievable from 2 L of starting culture medium.
Notes
Readers are referred to the methods section in Hirano et al. (2019) for additional details regarding application of the current approach to the purification of human cPLA2α C2-domain.
Recipes
Solution A
20 mM Tris-HCl (pH 8.0)
500 mM NaCl
2.5 mM CaCl2
Solution B
20 mM Tris-HCl (pH 8.0)
150 mM NaCl
2.5 mM CaCl2
5 mM imidazole
Solution C
20 mM Tris-HCl (pH 8.0)
150 mM NaCl
2.5 mM CaCl2
250 mM imidazole
Solution D
20 mM Tris-HCl (pH 8.0)
30 mM NaCl
2.5 mM CaCl2
Solution E
20 mM Tris-HCl (pH 8.0)
2.5 mM CaCl2
Solution F
20 mM Tris-HCl (pH 8.0)
1,000 mM NaCl
2.5 mM CaCl2
Solution G
20 mM MES-NaOH (pH 6.0)
100 mM NaCl
2.5 mM CaCl2
Acknowledgements
We are grateful for support received by the Program for Promoting the Enhancement of Research Universities from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan (YH); research grants from the National Institutes of Health via HL125353 (REB, DJP, and CEC), HD087198 (CEC) and RR031535 (CEC); the Veteran’s Administration [VA Merit Review, I BX001792 (CEC)]; a Research Career Scientist Award 13F-RCS-002 (CEC); a Memorial Sloan-Kettering Cancer Center Core Grant P30 CA008748 (DJP); the Maloris Foundation (DJP); and the Hormel Foundation (REB). The contents of this manuscript do not represent the views of the Department of Veterans Affairs or the United States Government.
Parts of the current protocol, i.e., use of SUMO tag, were adapted to enable expression of cPLA2α C2-domain in soluble form in E. coli. Original references pertaining to SUMO tag use include Malakhov et al., 2004; Marblestone et al., 2006; Yan et al. 2009 and Peroutka et al., 2011.
Competing interests
The authors declare no competing interests.
Ethics
The current study involved no use or involvement of human or animal subjects.
References
- Astudillo, A. M., Balboa, M. A. and Balsinde, J. (2019). Selectivity of phospholipid hydrolysis by phospholipase A2 enzymes in activated cells leading to polyunsaturated fatty acid mobilization. Biochim Biophys Acta Mol Cell Biol Lipids 1864(6): 772-783.
- Ball, A., Nielsen, R., Gelb, M. H. and Robinson, B. H. (1999). Interfacial membrane docking of cytosolic phospholipase A2 C2 domain using electrostatic potential-modulated spin relaxation magnetic resonance. Proc Natl Acad Sci U S A 96(12): 6637-6642.
- Bittova, L., Sumandea, M. and Cho, W. (1999). A structure-function study of the C2 domain of cytosolic phospholipase A2. Identification of essential calcium ligands and hydrophobic membrane binding residues. J Biol Chem 274(14): 9665-9672.
- Bonventre, J. V., Huang, Z., Taheri, M. R., O'Leary, E., Li, E., Moskowitz, M. A. and Sapirstein, A. (1997). Reduced fertility and postischaemic brain injury in mice deficient in cytosolic phospholipase A2. Nature 390(6660): 622-625.
- Clark, J. D., Lin, L. L., Kriz, R. W., Ramesha, C. S., Sultzman, L. A., Lin, A. Y., Milona, N. and Knopf, J. L. (1991). A novel arachidonic acid-selective cytosolic PLA2 contains a Ca2+-dependent translocation domain with homology to PKC and GAP. Cell 65(6): 1043-1051.
- Davletov, B., Perisic, O. and Williams, R. L. (1998). Calcium-dependent membrane penetration is a hallmark of the C2 domain of cytosolic phospholipase A2 whereas the C2A domain of synaptotagmin binds membranes electrostatically. J Biol Chem 273(30): 19093-19096.
- Dennis, E. A., Cao, J., Hsu, Y. H., Magrioti, V. and Kokotos, G. (2011). Phospholipase A2 enzymes: physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chem Rev 111(10): 6130-6185.
- Dessen, A., Tang, J., Schmidt, H., Stahl, M., Clark, J. D., Seehra, J. and Somers, W. S. (1999). Crystal structure of human cytosolic phospholipase A2 reveals a novel topology and catalytic mechanism. Cell 97(3): 349-360.
- Hirano, Y., Gao, Y. G., Stephenson, D. J., Vu, N. T., Malinina, L., Simanshu, D. K., Chalfant, C. E., Patel, D. J. and Brown, R. E. (2019). Structural basis of phosphatidylcholine recognition by the C2-domain of cytosolic phospholipase A2α. eLife 8: e44760.
- Lamour, N. F., Subramanian, P., Wijesinghe, D. S., Stahelin, R. V., Bonventre, J. V. and Chalfant, C. E. (2009). Ceramide 1-phosphate is required for the translocation of group IVA cytosolic phospholipase A2 and prostaglandin synthesis. J Biol Chem 284(39): 26897-26907.
- Leslie, C. C. (2015). Cytosolic phospholipase A2: physiological function and role in disease. J Lipid Res 56(8): 1386-1402.
- Leslie, C. C., Gangelhoff, T. A. and Gelb, M. H. (2010). Localization and function of cytosolic phospholipase A2α at the Golgi. Biochimie 92(6): 620-626.
- Malakhov, M. P., Mattern, M. R., Malakhova, O. A., Drinker, M., Weeks, S. D. and Butt, T. R. (2004). SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J Struct Funct Genomics 5(1-2): 75-86.
- Malmberg, N. J. and Falke, J. J. (2005). Use of EPR power saturation to analyze the membrane-docking geometries of peripheral proteins: applications to C2 domains. Annu Rev Biophys Biomol Struct 34: 71-90.
- Malmberg, N. J., Van Buskirk, D. R. and Falke, J. J. (2003). Membrane-docking loops of the cPLA2 C2 domain: detailed structural analysis of the protein-membrane interface via site-directed spin-labeling. Biochemistry 42(45): 13227-13240.
- Marblestone, J. G., Edavettal, S. C., Lim, Y., Lim, P., Zuo, X. and Butt, T. R. (2006). Comparison of SUMO fusion technology with traditional gene fusion systems: enhanced expression and solubility with SUMO. Protein Sci 15(1): 182-189.
- Mouchlis, V. D. and Dennis, E. A. (2019). Phospholipase A2 catalysis and lipid mediator lipidomics. Biochim Biophys Acta Mol Cell Biol Lipids 1864(6): 766-771.
- Mouchlis, V. D., Bucher, D., McCammon, J. A. and Dennis, E. A. (2015). Membranes serve as allosteric activators of phospholipase A2, enabling it to extract, bind, and hydrolyze phospholipid substrates. Proc Natl Acad Sci U S A 112(6): E516-525.
- Nalefski, E. A., McDonagh, T., Somers, W., Seehra, J., Falke, J. J. and Clark, J. D. (1998). Independent folding and ligand specificity of the C2 calcium-dependent lipid binding domain of cytosolic phospholipase A2. J Biol Chem 273(3): 1365-1372.
- Nalefski, E. A., Sultzman, L. A., Martin, D. M., Kriz, R. W., Towler, P. S., Knopf, J. L. and Clark, J. D. (1994). Delineation of two functionally distinct domains of cytosolic phospholipase A2, a regulatory Ca2+-dependent lipid-binding domain and a Ca2+-independent catalytic domain. J Biol Chem 269(27): 18239-18249.
- Perisic, O., Fong, S., Lynch, D. E., Bycroft, M. and Williams, R. L. (1998). Crystal structure of a calcium-phospholipid binding domain from cytosolic phospholipase A. J Biol Chem 273(3): 1596-1604.
- Peroutka Iii, R. J., Orcutt, S. J., Strickler, J. E. and Butt, T. R. (2011). SUMO fusion technology for enhanced protein expression and purification in prokaryotes and eukaryotes. Methods Mol Biol 70515-30.
- Pettus, B. J., Bielawska, A., Subramanian, P., Wijesinghe, D. S., Maceyka, M., Leslie, C. C., Evans, J. H., Freiberg, J., Roddy, P., Hannun, Y. A. and Chalfant, C. E. (2004). Ceramide 1-phosphate is a direct activator of cytosolic phospholipase A2. J Biol Chem 279(12): 11320-11326.
- Shimizu, T., Ohto, T. and Kita, Y. (2006). Cytosolic phospholipase A2: biochemical properties and physiological roles. IUBMB Life 58(5-6): 328-333.
- Smith, W. L. (1989). The eicosanoids and their biochemical mechanisms of action. Biochem J 259(2): 315-324.
- Stahelin, R. V., Subramanian, P., Vora, M., Cho, W. and Chalfant, C. E. (2007). Ceramide-1-phosphate binds group IVA cytosolic phospholipase A2 via a novel site in the C2 domain. J Biol Chem 282(28): 20467-20474.
- Uozumi, N., Kume, K., Nagase, T., Nakatani, N., Ishii, S., Tashiro, F., Komagata, Y., Maki, K., Ikuta, K., Ouchi, Y., Miyazaki, J. and Shimizu, T. (1997). Role of cytosolic phospholipase A2 in allergic response and parturition. Nature 390(6660): 618-622.
- Vasquez, A. M., Mouchlis, V. D. and Dennis, E. A. (2018). Review of four major distinct types of human phospholipase A2. Adv Biol Regul 67: 212-218.
- Ward, K. E., Bhardwaj, N., Vora, M., Chalfant, C. E., Lu, H. and Stahelin, R. V. (2013). The molecular basis of ceramide-1-phosphate recognition by C2 domains. J Lipid Res 54(3): 636-648.
- Xu, G. Y., McDonagh, T., Yu, H. A., Nalefski, E. A., Clark, J. D. and Cumming, D. A. (1998). Solution structure and membrane interactions of the C2 domain of cytosolic phospholipase A2. J Mol Biol 280(3): 485-500.
- Yan, Y., Orcutt, S. and Strickler, J. (2009). The use of SUMO as a fusion system for protein expression and purification. Chem Today 27.
Article Information
Copyright
Hirano et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Hirano, Y., Gao, Y. G., Simanshu, D. K., Stephenson, D. J., Vu, N. T., Malinina, L., Chalfant, C. E., Pate, D. J. and Brown, R. E. (2021). Purification of Cytosolic Phospholipase A2α C2-domain after Expression in Soluble Form in Escherichia coli. Bio-protocol 11(3): e3906. DOI: 10.21769/BioProtoc.3906.
- Hirano, Y., Gao, Y. G., Stephenson, D. J., Vu, N. T., Malinina, L., Simanshu, D. K., Chalfant, C. E., Patel, D. J. and Brown, R. E. (2019). Structural basis of phosphatidylcholine recognition by the C2-domain of cytosolic phospholipase A2α. eLife 8: e44760.
Category
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








