- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Efficient Transient Gene Knock-down in Tobacco Plants Using Carbon Nanocarriers
Published: Vol 11, Iss 1, Jan 5, 2021 DOI: 10.21769/BioProtoc.3897 Views: 6381
Reviewed by: Ayelign M. AdalHui Chen WuTohir A. Bozorov

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

TUNEL Assay to Assess Extent of DNA Fragmentation and Programmed Cell Death in Root Cells under Various Stress Conditions
Amit K. Tripathi [...] Sneh Lata Singla-Pareek
Aug 20, 2017 15025 Views

Faster Bacterial Gene Cloning Using the Brick into the Gateway (BiG) Protocol
Flaviani G. Pierdoná [...] Fabio T. S. Nogueira
Dec 20, 2022 2353 Views
Abstract
Gene knock-down in plants is a useful approach to study genotype-phenotype relationships, render disease resistance to crops, and enable efficient biosynthesis of molecules in plants. Small interfering RNA (siRNA)-mediated gene silencing is one of the most common ways to achieve gene knock-down in plants. Traditionally, siRNA is delivered into intact plant cells by coding the siRNA sequences into DNA vectors, which are then delivered through viral and/or bacterial methods. In this protocol, we provide an alternative direct delivery method of siRNA molecules into intact plant cells for efficient transient gene knock-down in model tobacco plant, Nicotiana benthamiana, leaves. Our approach uses one dimensional carbon-based nanomaterials, single-walled carbon nanotubes (SWNTs), to deliver siRNA, and does not rely on viral/bacterial delivery. The distinct advantages of our method are i) there is no need for DNA coding of siRNA sequences, ii) this abiotic method could work in a broader range of plant species than biotic methods, and iii) there are fewer regulatory complications when using abiotic delivery methods, whereby gene silencing is transient without permanent modification of the plant genome.
Graphic abstract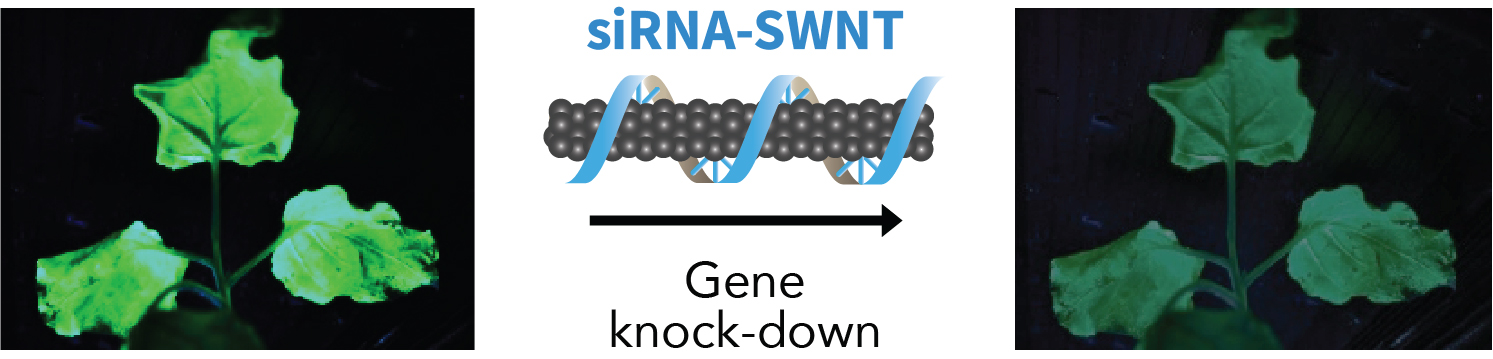
Background
Gene silencing through RNA interference (RNAi) was discovered in the early 1990s by plant researchers studying petunia flower coloring (Van der Krol et al., 1990). In RNAi, specifically in post-transcriptional gene silencing (PTGS), gene expression level is reduced through mRNA degradation caused by small RNA molecules – micro (miRNA) or small interfering (siRNA) RNA. RNAi has been a breakthrough technology, not only in plant research and biotechnology applications, but also for many other organisms, including human therapy applications (Sierakowska et al., 1996).
The first step of siRNA-mediated RNAi in plants is the delivery of siRNA molecules into plant cells. Delivery is a big bottleneck in plant biotechnology, given the presence of plant cell wall that acts as a physical barrier for the delivery of biotechnology-relevant cargoes such as DNA, RNA, and protein. In plants, siRNA delivery is most commonly accomplished through viral vector delivery via Agrobacterium tumefaciens. However, most plant viruses are limited in their host range (Silva et al., 2010) and the size of cargo they can efficiently deliver (Burch-Smith et al., 2004). Agrobacterium-mediated delivery is also limited in terms of plant host species, causes uncontrolled DNA integration into the plant nuclear genome, and results in constitutive expression of siRNA, which limits temporal control over gene silencing (Baltes et al., 2017).
Carbon nanotubes are one dimensional high-aspect-ratio nanomaterials that have many advantageous features for siRNA delivery in plants. First, given their needle-like structure with a small diameter (~1 nm), long length (~500 nm) and high stiffness, single-walled carbon nanotubes (SWNTs) have shown to transport across the plant cell wall and localize inside plant cells (Demirer et al., 2019b). Second, high surface area and diverse surface chemistry options of SWNTs enable delivery of diverse biological cargoes (Beyene et al., 2016; Del Bonis-O’Donnell et al., 2017; Demirer et al., 2020). Lastly, SWNTs have the ability to delay the intracellular degradation of biomolecular cargoes (Demirer et al., 2019a and 2020), which is especially valuable when working with fragile molecules like RNA.
Recently, we have developed a method to deliver siRNA molecules targeting the silencing of a transgenic GFP gene in Nicotiana benthamiana leaves, and an endogenous stress gene, ROQ1, using SWNTs (Demirer et al., 2020). In this approach, we first load sense and antisense strands of siRNA onto two separate SWNT nanoparticle solutions via pi-pi interactions that form between the sp2 carbon nanotube surface lattice and the aromatic bases of single stranded RNA (ssRNA). Next, we introduce an equimolar mixture of these RNA-SWNT solutions into intact plant leaves for GFP silencing. Our results demonstrate efficient silencing of GFP as assessed by confocal microscopy imaging, quantitative PCR (qPCR), and Western blotting, both for transgenic GFP and also for the endogenous ROQ1 gene, with disease-resistance applications (Demirer et al., 2020). This transient gene knock-down approach could be applied to other plant species, tissues, and target genes with minimal modifications. Additionally, the RNA loading method used in this study is not specific to siRNA, and thus, it can be adapted for the delivery of other types of nucleic acids with some optimization (e.g., guide RNA or messenger RNA for CRISPR genome editing applications).
Below, we provide a step-by-step protocol for the synthesis and characterization of siRNA loaded SWNTs, and the measurement of gene silencing efficiency in tobacco leaves through confocal imaging, qPCR and Western blotting (Figure 1).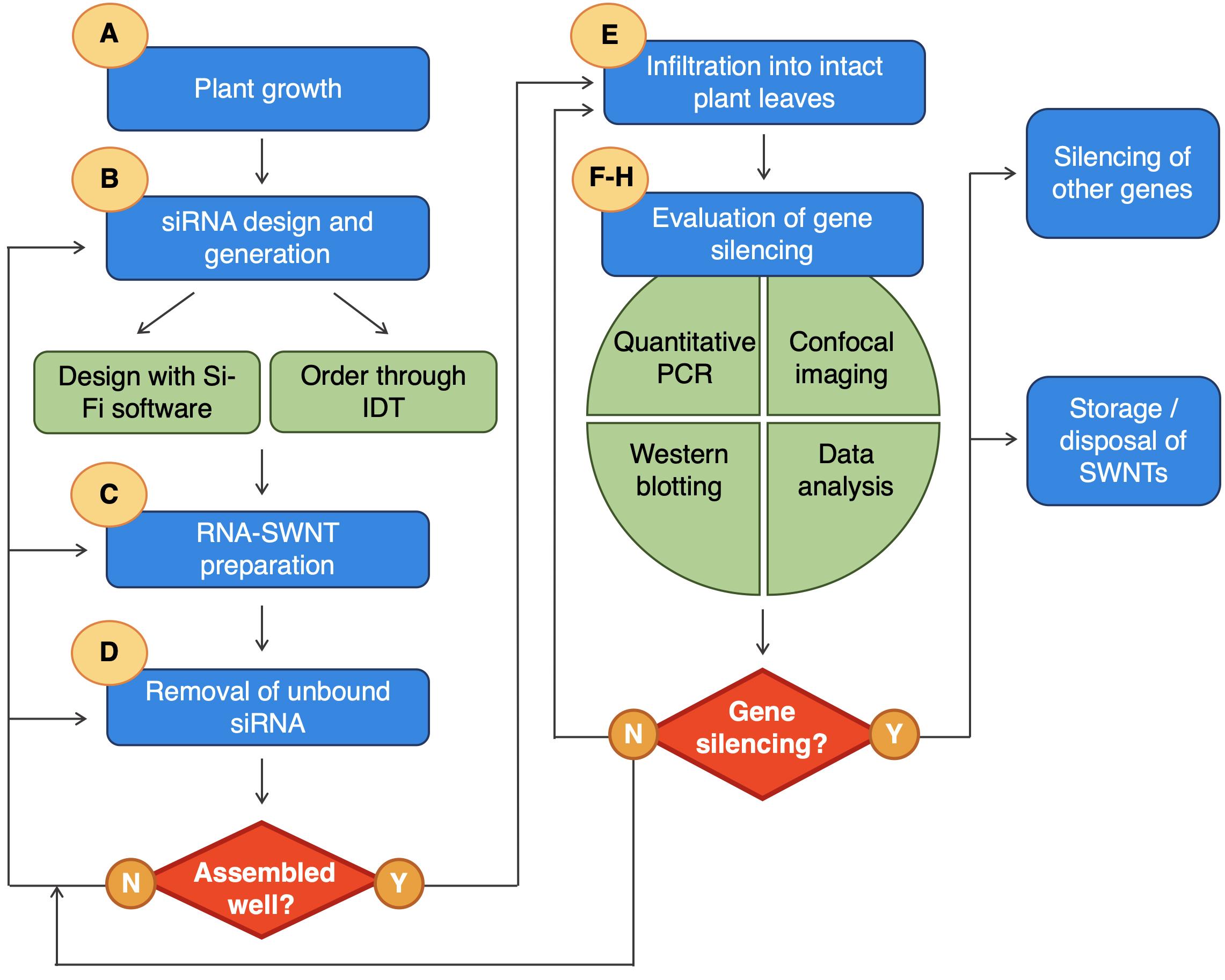
Figure 1. Overview of the siRNA-SWNT gene silencing procedure
Materials and Reagents
SunGro Sunshine LC1 Grower soil mix (SUN52128CFLP)
Delicate task wipes (Kimberly-Clark, catalog number: 06-666 )
100K MWCO Amicon spin filters (MilliporeSigma, catalog number: UFC510024 )
PVDF Membrane, Precut, 7 x 8.4 cm (Bio-Rad, catalog number: 1620174 )
Sterile syringe filter (0.45 μm; VWR, catalog number: 28145-481 )
Microcentrifuge tubes (1.5 ml; VWR, catalog number: 89000-028 )
Conical tubes (50 ml; Olympus, catalog number: 28-106 )
Pipette tips (Low retention 10 μl, 200 μl, 1,000 μl filter tips; USA Scientific, catalog numbers: 1181-3710 , 1180-8710 , 1182-1730 )
Extended-length pipette tips (1,000 μl; Eppendorf, catalog number: 00 30073614 )
#1 Microscopy cover glass (Fisher Scientific, catalog number: 12-542B )
Microscope slides (VWR, catalog number: 16004-422 )
Syringe (1 ml; BD, catalog number: 14-823-434 )
Mini Trans-Blot Filter paper (Bio-Rad, catalog number: 1703932 )
EasyStrip‚ Plus PCR Tube (Thermo Scientific, catalog number: AB2005 )
Plant seeds (mGFP5 Nicotiana benthamiana is obtained from the Staskawicz lab at UC Berkeley, mGFP5 plants constitutively express GFP targeted to the ER under the control of the Cauliflower mosaic virus 35S promoter)
Goat anti-rabbit horseradish peroxidase-conjugated antibody (Abcam, catalog number: ab205718 )
Anti-GFP antibody, ChIP Grade (Abcam, catalog number: ab290 )
HiPCO SWNTs (NanoIntegris, Super purified, catalog number: HS28-037 )
MilliQ water
Nuclease-free water (Qiagen, catalog number: 129114 )
Sodium chloride, NaCl (Sigma-Aldrich, catalog number: S9888-500G )
Hydrochloric acid, HCl (37% [vol/vol]; Sigma, catalog number: 320331 )
Single-stranded RNA oligonucleotides, including sense and antisense siRNA strands – 21 nucleotides (Integrated DNA Technologies, IDT)
Sodium dodecyl sulfate, molecular biology grade (Sigma-Aldrich, catalog number: 436143-100G )
Tris/HCl (Sigma-Aldrich, catalog number: 10812846001 )
EDTA (Sigma-Aldrich, catalog number: E9884-100G )
NP-40 (Sigma-Aldrich, catalog number: 492016-100ML )
Glycerol (Sigma-Aldrich, catalog number: G5516-500ML )
Pierce 660 nm Protein Assay (Thermo, catalog number: 22660 )
iScript cDNA synthesis kit (Bio-Rad, catalog number: 1708891 )
PowerUp SYBR green master mix (Applied Biosystems, catalog number: A25742 )
Qubit Protein Assay (ThermoFisher Scientific, catalog number: Q33211 )
RNeasy plant mini kit (QIAGEN, catalog number: 74904 )
BSA (Sigma-Aldrich, catalog number: A4737-25G )
TWEEN20 (Sigma-Aldrich, catalog number: P9416-100ML )
Ammonium persulphate, APS (Sigma, catalog number: 248614-100G )
Low range ultra agarose (Bio-Rad, catalog number: 1613107 )
ECL Prime Western Blotting System (MilliporeSigma, catalog number: GERPN2232 )
TEMED (N,N,N,N'-tetramethylethylenediamine; Sigma, catalog number: T9281 )
Glycine (Sigma, catalog number: G8898 )
Methanol (Sigma, catalog number: 179957 )
4x Laemmli sample Buffer (Bio-Rad, 10 ml, catalog number: 1610747 )
Liquid nitrogen
SYBR Gold Nucleic Acid Gel Stain (Invitrogen, catalog number: S11494 )
30% Acrylamide/Bis solution 19:1 (Bio-Rad, catalog number: 1610154 )
Protease inhibitor cocktail (Sigma, catalog number: P9599-1ML )
0.1 M NaCl (see Recipes)
10% (wt/vol) Ammonium persulphate solution (APS) (see Recipes)
10x Transfer buffer (see Recipes)
1x Transfer buffer (see Recipes)
10x Tris-Buffered Saline (TBS) buffer (1 M Tris, 1.5 M NaCl, pH 7.4) (see Recipes)
1x TBST buffer (see Recipes)
Lysis buffer (see Recipes)
Equipment
Analytical balance (Radwag, model: AS 60/220.R2 )
Ultrasonic bath (Branson, model: 15-336-100 )
Ultrasonic homogenizer with 6-mm tip (Cole-Parmer, models: UX-04711-70, UX-04712-14 )
Vortex mixer (Fisher Scientific, model: 02-215-365 )
pH meter (Spectrum, model: 242-97839 )
Orbital shaker (Waverly, model: S1CE )
NanoVue Plus spectrophotometer (GE Life Sciences, model: 28-9569-61 )
Visible spectrophotometer (Thermo Scientific, model: 14-385-445 )
Near-infrared spectrometer (Princeton Instruments IsoPlane 320 coupled to a liquid nitrogen-cooled Princeton Instruments PyLoN-IR 1D array of InGaAs pixels)
UV-Vis-NIR Spectrophotometer (Shimadzu, model: UV-3600 Plus )
Tabletop centrifuge (Eppendorf, catalog number: 5418000017 )
Centrifuge (Eppendorf, model: 5424R )
Tweezers (VWR, catalog number: 63042-518 )
Scissors (VWR, catalog number: 82027-582 )
Mortar and pestle (Cole-Parmer, catalog number: EW-63100-54 )
Pant growth chamber (HiPoint, model: 740 FHLED )
Gel image-analysis system ( Typhoon FLA 9500 , GE Healthcare Services)
Electrophoresis power supply (PowerPac basic power supply; Bio-Rad, catalog number: 1645050 )
Mini Trans-Blot Cell (Bio-Rad, catalog number: 1703811 )
Mini-Protein TGX gels (Bio-Rad, catalog number: 456-1094 )
ChemiDoc XRS+ System (Bio-Rad, catalog number: 1708265 )
Confocal Microscope (Zeiss, model: LSM 710 )
Thermal Cycler CFX96 Touch Real-Time PCR Detection System (Bio-Rad, catalog number: 1855195 )
Thermal Cycler PCR (Applied Biosystems Veriti 96-Well, catalog number: 4375786 )
Software
GraphPad Prism 7.0a (https://www.graphpad.com/scientific-software/prism/)
Fiji ImageJ 2.0.0 (https://imagej.net/Fiji/Downloads)
Zen Blue 2.6 (https://www.zeiss.com/microscopy/us/downloads.html)
Procedure
Plant growth
Germinate transgenic mGFP5 Nicotiana benthamiana seeds (see Note 1) and grow seeds in SunGro Sunshine LC1 Grower soil mix in a growth chamber for four to six weeks before experiments. Use 12-h light at 24 °C and 12-h dark at 18 °C cycle for growing plants.
Note: Different plant species may require different germination and growth conditions.
siRNA design and generation
Currently, there are many software to design gene specific siRNA sequences with minimal off-target effects. A recently developed software called “siRNA-Finder (si-Fi) Software” can be used in plants (Lück et al., 2019).
After the design of siRNA sequences, sense and antisense RNA strands can be purchased from Integrated DNA Technologies (IDT) as single-stranded oligonucleotides.
RNA-SWNT preparation
Dissolve sense and antisense siRNA strands in 0.1 M NaCl at a concentration of 100 mg/ml.
Add 1 mg dry HiPCO SWNTs to 20 μl of dissolved sense RNA, and complete the solution volume to 1 ml with 0.1 M NaCl (see Note 2).
Bath sonicate the mixture for 10 min at room temperature in Ultrasonic bath (Branson).
Probe-tip sonicate the mixture with a 3-mm tip at 50% amplitude (~7W) for 30 min in an ice bath. Renew ice bath if it starts melting during the sonication to prevent heating.
Rest the solution at room temperature for 30 min.
Centrifuge the solution at 16,100 x g for 1 h in room temperature to remove bundled SWNTs. The supernatant contains the individually suspended sense-RNA-SWNTs. Keep the supernatant and discard the SWNT pellet to the hazardous nanomaterials waste.
Repeat the same protocol for the antisense RNA strand (see Note 3). Store RNA-SWNT solutions at 4 °C.
Removal of unbound siRNA
Add 500 μl sense-RNA-SWNT and 500 μl antisense-RNA-SWNT into two separate 100K Amicon spin filters that are placed in 2 ml collection tubes. Centrifuge 4 min at 8,000 x g in room temperature.
Collect the flow-throughs from sense-RNA-SWNT and antisense-RNA-SWNT in separate tubes and place the spin filters into the same collection tubes.
Add 0.1 M NaCl into the spin filters until the volume reaches 500 μl. Repeat the wash step.
Perform a total of 8 washes to remove all unbound RNA molecules. Merge and accumulate all flow-through solutions (separately for sense-RNA-SWNT and antisense-RNA-SWNT) for later measurement of removed RNA amount.
Calculate the SWNT concentration by measuring the carbon nanotube absorbance at 632 nm using a spectrophotometer (use 2 μl for NanoVue Plus spectrophotometer or dilute to 1 ml for Thermo Scientific). Divide the absorbance value by SWNT extinction coefficient of 0.036 to obtain SWNT concentration in the unit of μg/ml (If the sample is diluted, multiply the absorbance value also by the dilution factor).
Calculate the concentration of RNA loaded on SWNTs by measuring the absorbance of collected flow-through solutions at 260 nm (use 2 μl for NanoVue Plus spectrophotometer), and subtracting the total amount of RNA removed from the total amount of RNA added (2 mg in this case).
(Optional) For additional characterization, record sense-RNA-SWNT and antisense-RNA-SWNT absorbance spectra with Shimadzu UV-3600 Plus , and fluorescence spectra with a near-infrared spectrometer (Princeton Instruments IsoPlane 320 coupled to a liquid nitrogen-cooled Princeton Instruments PyLoN-IR 1D array of InGaAs pixels). See Demirer et al., 2020 for representative examples of SWNT absorbance and fluorescence spectra.
Infiltration of leaves with RNA-SWNTs
Select healthy and fully-developed leaves from mGFP5 Nicotiana benthamiana (4-6 weeks old) plants for experiments.
Merge 100 μl of 200 nM sense-RNA-SWNTs with 100 μl of 200 nM antisense-RNA-SWNTs in a 1.5 ml Eppendorf tube (see Note 4). Mix well.
Immediately after mixing, make a small puncture on the abaxial (bottom) surface of the leaf with a pipette tip, and infiltrate ~100-200 μl of the siRNA-SWNT mixture from the hole with a 1 ml needleless syringe with caution not to damage the leaf (see Note 5).
Use a Kimwipe tissue to remove the excess siRNA-SWNT solution on the leaf surface. Mark the infiltrated area with a Sharpie pen without damaging the leaf.
Infiltrate a negative control solution, such as the free siRNA without SWNTs or scrambled RNA suspended SWNTs that does not target the gene of interest. If possible, also infiltrate a positive control solution, such as viral siRNA delivery sample (see Note 6).
Return the infiltrated plant(s) into the growth chamber until the measurement of gene silencing.
Gene silencing determination through quantitative PCR (qPCR)
24-h after infiltration, cut the infiltrated leaf areas (maximum of 100 mg leaf tissue per sample) and extract total RNA with a RNeasy plant mini kit. After cutting the leaf, immediately proceed with the first step of the RNA extraction protocol (i.e., grinding the tissue in liquid nitrogen using mortar and pestle) to make sure gene expression levels do not change after cutting. Follow the protocol of the RNeasy plant mini kit (see Note 7).
Following RNA extraction, measure the RNA concentration and purity with a spectrophotometer. Reverse transcribe 1 μg total RNA into complementary DNA (cDNA) using an iScript cDNA synthesis kit. Follow the protocol of the iScript cDNA synthesis kit.
Use PowerUp SYBR green master mix for the qPCR step with 2 μl cDNA from Step 2 and specific primers for the target and housekeeping genes. Follow the protocol of the PowerUp SYBR green kit for relative quantification of target mRNA in the siRNA-SWNT infiltrated leaf compared to negative controls of free siRNA without SWNTs or scrambled RNA-SWNTs.
Example: The target gene in our qPCR was mGFP5 (GFP transgene inserted into Nb), and EF1 (elongation factor 1) as our housekeeping (reference) gene.
Analyze the qPCR data using the ddCt method (Rao et al., 2013). See Figure 2B for representative qPCR results.
Gene silencing determination through confocal fluorescence microscopy imaging
If silencing a fluorescent protein, such as GFP, confocal fluorescence microscopy can be used to detect approximate silencing efficiency.
After infiltration, leave the plants with intact infiltrated leaves in the growth chamber for 72 h.
72-h after infiltration, cut a small flat piece (0.5-1 cm x 0.5-1 cm) of the infiltrated leaf around the infiltration point and prepare a glass slide with cover slip (thickness #1). Add 50-100 μl water in between the glass slide and cover slip for imaging. Image samples before the leaf piece dries out (optimally within 15 to 30 min).
Image the plant tissue with 488 nm laser excitation with a GFP filter cube (in the case of GFP silencing), and also capture brightfield with a transmitted light if available (see Note 8).
Capture at least 10 to 15 fields of view with same optical settings per sample, including non-treated leaf, and any negative and positive control samples.
For each sample, compare the mean fluorescence intensity value with the mean GFP fluorescence intensity of a non-treated leaf, which can be used to determine silencing efficiency of siRNA-SWNTs. Pay attention to use the same imaging parameters and quantification analyses for samples imaged on different days. See Figure 2A for representative confocal imaging results.
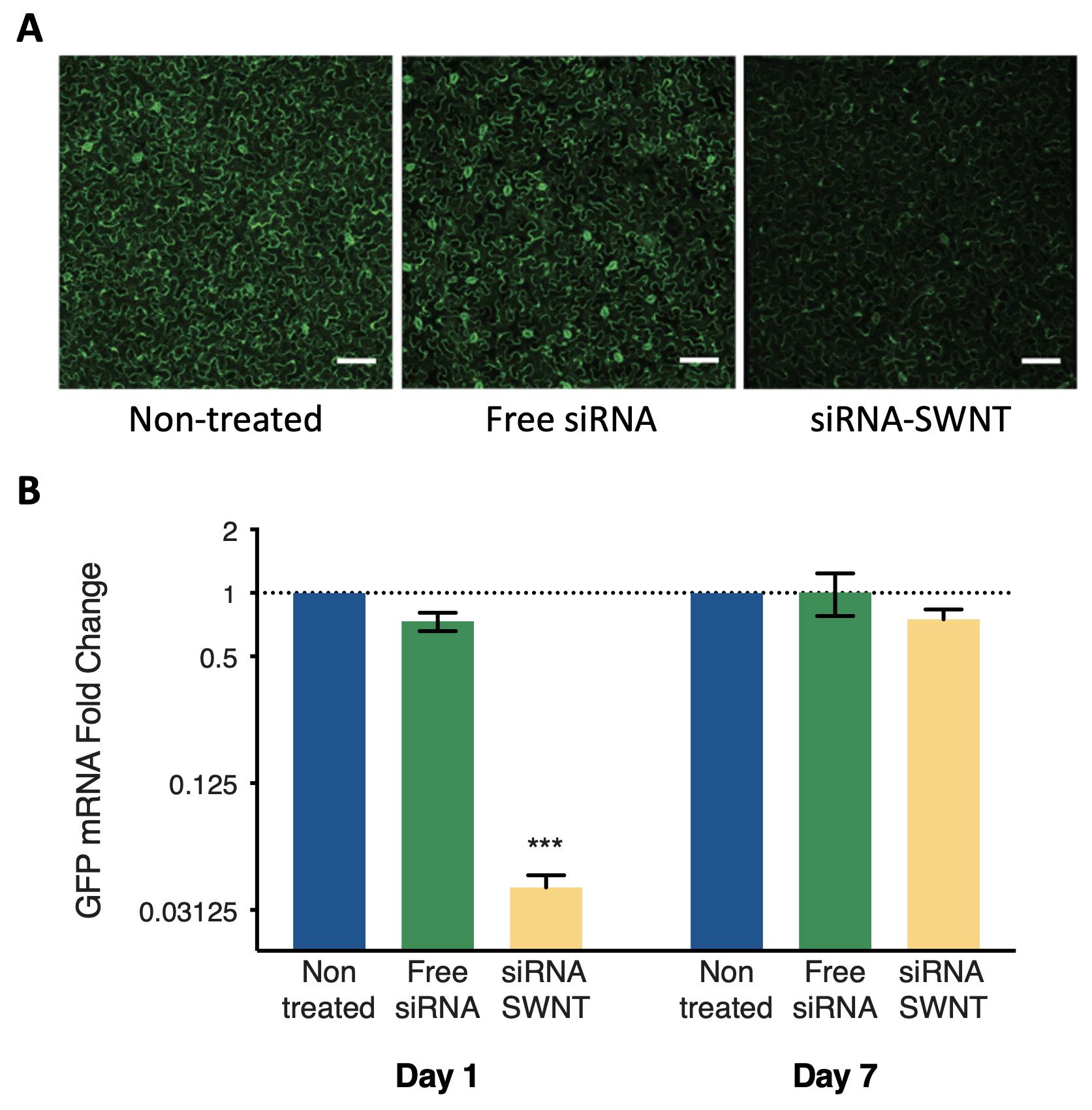
Figure 2. Representative gene silencing results. A. Confocal microscopy images of non-treated and free-siRNA treated control leaves and siRNA-SWNT infiltrated sample leaves of Nicotiana benthamiana. Imaged after 3 days post-infiltration, scale bars are 50 μm. B. Quantitative PCR analysis results for GFP gene silencing in Nicotiana benthamiana leaves with siRNA-SWNTs.
Gene silencing determination through Western blotting
72-h after infiltration, harvest infiltrated leaves and grind them in liquid nitrogen using mortar and pestle to recover dry frozen leaf powders.
Transfer the frozen leaf powder into a tube with 400 μl pre-chilled lysis buffer (see Recipes).
Lyse tissue on ice for 1 h. Centrifuge the tubes at 10,000 x g at 4 °C for 20 min. Following centrifugation, gently transfer the supernatants containing whole proteins to a new clean tube. Quantify total extracted proteins with a Pierce 660 nm Protein Assay.
Mix the samples with the appropriate loading buffer for gel electrophoresis, and boil the mixture at 95-100 °C for 5 min either using a heat block or thermal cycler. Load 0.5 µg of normalized total proteins from each sample and analyze with SDS-PAGE gel (Bio-Rad precast tris/glycine gel, 4-20% gradient). Run the gel at 120 V for 60 min.
Transfer the gel to a PVDF membrane in cold transfer buffer (see Recipes) and run at 400 mA in 1x transfer buffer with methanol for no more than 60 min in a cold room with an ice block.
Block the membrane for 1 h using 7.5% BSA in 1x TBST buffer (see Recipes) followed by overnight incubation at 4 °C with the primary GFP antibody. After extensive washing, probe the corresponding protein bands with a goat anti-rabbit horseradish peroxidase-conjugated antibody for 30 min. After washing, develop the bands by incubation with chemiluminescence (Amersham ECL prime kit) in less than 2 min and image with a ChemiDoc XRS+ System.
Quantify the intensity of GFP bands with ImageJ software.
Data analysis
For all experiments, perform at least 3 biological replicates. Biological replicates are defined as experiments consisting of independent infiltrations with RNA-SWNTs into different plants.
For quantitative analysis of confocal imaging, capture at least 10-15 non-overlapping fields of view per sample and per biological replicate, which is defined as technical replicates. Average the fluorescence intensity of these technical replicates to obtain the mean fluorescent intensity for a given biological replicate. Confocal images can be analyzed with either the ImageJ or Zen Blue software.
For the qPCR assay, perform at least 3 technical replicates, which are defined as reactions from the same isolated RNA batch obtained from the same infiltration. To analyze qPCR results for gene silencing efficiency, used the previously developed ddCt method. The details of this method can be found in Rao et al. (2013).
When comparing more than 2 samples with each other statistically for a single independent variable (e.g., silencing efficiency), use one-way ANOVA with Tukey’s multiple comparisons test. Report both the P value and F. When comparing more than 2 samples with each other statistically for multiple independent variables (e.g., silencing efficiency over multiple time points), use two-way ANOVA with Sidak’s multiple comparisons test. Report both the P value and F. When comparing two samples only, Student’s t-test can be used. Report the P value. GraphPad Prism can be used to plot the data and perform statistical significance analysis.
Notes
This protocol is provided for the silencing of GFP gene in transgenic tobacco (Nicotiana benthamiana) leaves, but can be adapted for the silencing of other genes and in other plant species, including other transgenes or endogenous genes.
RNA and SWNT amounts can be scaled up or down depending on the experimental need. In scaling, keep the mass ratio of RNA:SWNT at 2. Different final siRNA concentration may be needed for different gene targets, or in different plant species or tissues.
Typically, if one RNA strand suspends SWNTs well, its complementary strand has a lower suspension efficiency. This is expected given the complementary base pairs of the sense and antisense strands which can lead to varying adsorption ability of RNA bases to the SWNT surface. We typically obtain 20-50 μg/ml RNA-SWNT suspensions after centrifugation and purification steps.
Pay attention that 200 nM is the RNA concentration on SWNTs, and not the SWNT concentration. Depending on the suspension efficiency, this corresponds to ~2 μg/ml SWNTs.
100-200 μl infiltration is suggested for confocal imaging studies, whereas larger volume or multiple infiltrations should be performed for qPCR and Western blot to cover the full leaf area.
It is preferred to infiltrate the SWNT and control samples on the same leaf to avoid leaf to leaf variation in gene expression and imaging.
RNA work must be performed carefully on a clean bench (optimally dedicated to RNA-based experiments) to prevent RNA degradation. Wipe the surfaces and equipment with RNase solution and change gloves frequently. Keep the RNA on ice during the protocol, and transfer to -20 °C or -80 °C when the protocol is completed.
If the leaf is too thick or wavy to get the entire field of view into focus, image with z-stack and perform the silencing analysis on the stacked image. Keep the imaging parameters same between samples that will be compared with each other.
Recipes
0.1 M NaCl
Weigh 0.5844 g of NaCl and dissolve in 100 ml nuclease free water
Sterile filter with 0.22-micron syringe filter
10% (wt/vol) Ammonium persulphate solution (APS)
Add 5 g of APS to 50 ml of MilliQ water, and mix to dissolve
The solution can be stored at 4 °C for up to 3 months
10x Transfer buffer
To make 1 L of 10x Transfer buffer, add 30.3 g Tris base, 144 g Glycine to 800 ml MilliQ water, and mix to dissolve
Then add MilliQ water to a final volume of 1 L
1x Transfer buffer
To make 1 L of 1x Transfer buffer, add 100 ml 10x Transfer buffer, 200 ml methanol to 700 ml MilliQ water and mix the solution
The buffer needs to be stored at 4 °C, and it is better to prepare the buffer before running each experiment
10x Tris-Buffered Saline (TBS) buffer (1 M Tris, 1.5 M NaCl, pH 7.4)
To make 250 ml 10x TBS buffer, add 30.3 g Tris base, 21.9 g NaCl to 200 ml MilliQ water and mix to dissolve
Use HCl to adjust the pH of the solution to 7.4, then add MilliQ water to a final volume of 250 ml
The solution can be stored at 4 °C for up to 6 months
1x TBST buffer
Add 50 ml 10x TBS buffer, and 500 μl of Tween 20 (0.1%) to 500 ml MilliQ water and mix the solution
The buffer can be stored at 4 °C for at least 1 month
Lysis buffer
10 mM Tris/HCl
150 mM NaCl
1 mM EDTA
0.1% NP-40
5% glycerol
1% protease inhibitor cocktail
pH 7.5
To make 100 ml lysis buffer, add 1.21 g of Tris base, 0.877 g NaCl, 29.24 mg EDTA, 0.1 g NP-40, and 5 ml glycerol to 80 ml nuclease free water, mix to dissolve, adjust the pH to 7.5 by HCl, add 1 ml protease inhibitor cocktail, and fill the final volume to 100 ml using the nuclease free water.
Note: The lysis buffer can be stored at -20 °C for at least 6 months.
Acknowledgments
This protocol is derived from Demirer et al. (2020).
G.S.D. is supported by the Schlumberger Foundation Faculty for the Future Program and Caltech Resnick Sustainability Institute. We acknowledge support of a Burroughs Wellcome Fund Career Award at the Scientific Interface (CASI), a Stanley Fahn PDF Junior Faculty Grant with Award # PF-JFA-1760, a Beckman Foundation Young Investigator Award, a USDA AFRI award, a USDA NIFA award, the Moore Foundation, and an FFAR New Innovator Award (M.P.L). M.P.L. is a Chan-Zuckerberg Biohub investigator.
Competing interests
Authors declare no competing interest.
References
- Baltes, N. J., Gil-Humanes, J. and Voytas, D. F. (2017). Genome Engineering and Agriculture: Opportunities and Challenges. Prog Mol Biol Transl Sci 149: 1-26.
- Beyene, A. G., Demirer, G. S. and Landry, M. P. (2016). Nanoparticle-Templated Molecular Recognition Platforms for Detection of Biological Analytes. Curr Protoc Chem Biol 8(3): 197-223.
- Burch-Smith, T. M., Anderson, J. C., Martin, G. B. and Dinesh-Kumar, S. P. (2004). Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J 39(5): 734-746.
- Del Bonis-O’Donnell, J. T., Beyene, A., Chio, L., Demirer, G., Yang, D. and Landry, M. P. (2017). Engineering Molecular Recognition with Bio-mimetic Polymers on Single Walled Carbon Nanotubes. J Vis Exp 119: e55030.
- Demirer, G. S., Zhang, H., Goh, N. S., Gonzalez-Grandio, E. and Landry, M. P. (2019). Carbon nanotube-mediated DNA delivery without transgene integration in intact plants. Nat Protoc 14(10): 2954-2971.
- Demirer, G. S., Zhang, H., Goh, N. S., Pinals, R. L., Chang, R. and Landry, M. P. (2020). Carbon nanocarriers deliver siRNA to intact plant cells for efficient gene knockdown. Sci Adv 6(26): eaaz0495.
- Demirer, G. S., Zhang, H., Matos, J. L., Goh, N. S., Cunningham, F. J., Sung, Y., Chang, R., Aditham, A. J., Chio, L., Cho, M. J., Staskawicz, B. and Landry, M. P. (2019). High aspect ratio nanomaterials enable delivery of functional genetic material without DNA integration in mature plants. Nat Nanotechnol 14(5): 456-464.
- Lück, S., Kreszies, T., Strickert, M., Schweizer, P., Kuhlmann, M. and Douchkov, D. (2019). siRNA-Finder(si-Fi) Software for RNAi-Target Design and Off-Target Prediction. Front Plant Sci 10: 1023.
- Rao, X., Huang, X., Zhou, Z. and Lin, X. (2013). An improvement of the 2ˆ(–delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath 3(3): 71-85.
- Sierakowska, H., Sambade, M. J., Agrawal, S. and Kole, R. (1996). Repair of thalassemic human β-globin mRNA in mammalian cells by antisense oligonucleotides. Proc Natl Acad Sci 93(23): 12840-12844.
- Silva, A. T., Nguyen, A., Ye, C., Verchot, J. and Moon, J. H. (2010). Conjugated polymer nanoparticles for effective siRNA delivery to tobacco BY-2 protoplasts. BMC Plant Biol 10: 291.
- van der Krol, A. R., Mur, L. A., Beld, M., Mol, J. N. and Stuitje, A. R. (1990). Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell 2(4): 291-299.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Demirer, G. S. and Landry, M. P. (2021). Efficient Transient Gene Knock-down in Tobacco Plants Using Carbon Nanocarriers. Bio-protocol 11(1): e3897. DOI: 10.21769/BioProtoc.3897.
Category
Plant Science > Plant molecular biology > DNA > DNA modification
Biophysics > Bioengineering > Nanomaterials
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










