- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
CENP-C Phosphorylation by CDK1 in vitro
Published: Vol 11, Iss 1, Jan 5, 2021 DOI: 10.21769/BioProtoc.3879 Views: 5040
Reviewed by: Chiara AmbrogioKuldeep Singh AttriTianjiao HuangAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

An Activity-Based Proteomics with Two-Dimensional Polyacrylamide Gel Electrophoresis (2D-PAGE) for Identifying Target Proteases in Arabidopsis Apoplastic Fluid
Sayaka Matsui and Yoshikatsu Matsubayashi
Mar 5, 2025 1976 Views

Reliable and Sensitive Detection of Carbonylated Proteins by Oxime Blot
Filip Luka Mikulić [...] Mladen Merćep
Aug 5, 2025 1241 Views

Advancing 2-DE Techniques: High-Efficiency Protein Extraction From Lupine Roots
Sebastian Burchardt [...] Emilia Wilmowicz
Oct 5, 2025 1764 Views
Abstract
Accurate chromosome segregation during mitosis requires the kinetochore, a large protein complex, which makes a linkage between chromosomes and spindle microtubes. An essential kinetochore component, CENP-C, is phosphorylated by Cyclin-B-Cyclin dependent kinase 1 (CDK1) that is a master kinase for mitotic progression, promoting proper kinetochore assembly during mitosis. Here, we describe an in vitro CDK1 kinase assay to detect CENP-C phosphorylation using Phos-tag SDS-PAGE without radiolabeled ATP. Our protocol has advantages in ease and safety over conventional phosphorylation assays using [γ-32P]-ATP, which has potential hazards despite their better sensitivity. The protocol described here can be applicable to other kinases and be also useful for analysis of phospho-sites in substrates in vitro.
Keywords: In vitro kinase assayBackground
Cyclin-B-Cyclin dependent kinase 1 (CDK1), which is a master regulator of mitosis, phosphorylates numerous targets to ensure mitotic progression (Nurse, 1990; Malumbres and Barbacid, 2005). During mitosis, chromosomes carrying the genetic information are equally divided into two daughter cells. The kinetochore is a key large protein complex ensuring the faithful chromosome segregation by bridging between chromosomes and spindle microtubules (Fukagawa and Earnshaw, 2014). Various proteins composing the kinetochore are phosphorylated by CDK1 (Gascoigne et al., 2013; Nishino et al., 2013; Hara et al., 2018b; Watanabe et al., 2019). The CDK1 phosphorylation plays critical roles in kinetochore assembly (Gascoigne et al., 2013) and also in the correct microtubule attachment (Nishino et al., 2013; Hara et al., 2018a).
In vitro kinase assays and phospho-protein analyses are important ways, which provide us understanding of how phospho-regulations are achieved in kinetochore assembly and function. A classical way to detect phospho-proteins is a radiolabeled assay using [γ-32P]-ATP. However, although the radiolabeled assay gives high sensitivity, it has potential hazards. To use radioactivity, we would need to take a radiation safety training course, and prepare controlled area and safety protection equipment. Phospho-specific antibodies can be alternative ways. However, the antibodies are not always available for targets of interest.
Here, we describe a method to phosphorylate a kinetochore protein, CENP-C, by CDK1 in vitro and to detect the phosphorylated CENP-C using Phos-tag SDS-PAGE (Kinoshita et al., 2006). Phos-tag is 1,3-bis[bis(pyridin-2-ylmethyl)amino]propan-2-olato dizinc(II) complex that has a vacancy on two metal ions and binds to phenyl phosphate dianion on the target protein via two metal ions. Phospho-proteins migrate slower in the SDS-PAGE containing Phos-tag. This method magnifies mobility shifts of the phospho-proteins, which even show no migration changes in conventional SDS-PAGE. The protocol can be applicable to detect phospho-proteins by other kinases. Given that mobility of the phospho-protein on Phos-tag SDS-PAGE changes with number of phosphorylation-sites as well as location of the phosphorylation-sites, the procedure could be also utilized for analysis of phosphorylation-sites. The protocol is easy and safe, and can be finished within a few hours including kinase reaction and detection, saving time compared with the conventional radiolabeled assays.
Materials and Reagents
Active cyclin B-CDK1 purified as described previously (Okumura et al., 1996; Watanabe et al., 2019)
Recombinant chicken MBP (maltose-binding protein)-CENP-C (aa 601-864) purified as described previously (Watanabe et al., 2019)
Xpress Micro Dialyzer MD 100 (Scienova, catalog number: 40075 )
Protein Lobind Tube 500 µl (Eppendorf, catalog number: 00 30108116 )
Super SepTM Ace, 5-20%, 17-well (FUJIFILM Wako, catalog number: 194-15021 )
CBB Stain One (Nacalai Tesque, catalog number: 04543-51 )
cOmpleteTM EDTA-free proteinase inhibitor (Roche, catalog number: 11873580001 ))
Phos-tag Acrylamide (FUJIFILM Wako, catalog number: 300-93523) (Kinoshita et al., 2006)
Tris (Trizma® base) (Sigma-Aldrich, catalog number: T1503-1KG )
MgCl2 (Nacalai Tesque, catalog number: 20909-55 )
NaCl (Nacalai Tesque, catalog number: 31320-05 )
EDTA (Ethylenediaminetetraacetic acid, Nacalai Tesque, catalog number: 15111-45 )
HCl (Nacalai Tesque, catalog number: 18321-05 )
NaOH (Nacalai Tesque, catalog number: 31511-05 )
30(w/v)%-Acrylamide/Bis Mixed Solution (29:1) (Nacalai Tesque, catalog number: 06141-35 )
ATP (Adenosine triphosphate) (Nacalai Tesque, catalog number: 01072-11 )
SDS (Sodium dodecyl sulfate) (Nacalai Tesque, catalog number: 02873-75 )
APS (Ammonium peroxodisulfate) (Nacalai Tesque, catalog number: 02627-34 )
TEMED (N,N,N',N'-Tetramethylethylenediamine) (Nacalai Tesque, catalog number: 33401-72 )
MnCl2 (FUJIFILM Wako, catalog number: 133-00725 )
Glycerol (Nacalai Tesque, catalog number: 17018-83 )
2-mercaptoethanol (SIGMA, catalog number: M3148-100ML )
Bromophenol blue (FUJIFILM Wako, catalog number: 101123 )
Glycine (Nacalai Tesque, catalog number: 17109-35 )
Precision Plus Protein Dual Color standards (Bio-Rad, catalog number: 161-0374 )
1x kinase buffer (KB) (see Recipes)
2x kinase buffer (see Recipes)
25x cOmpleteTM EDTA-free proteinase inhibitor (see Recipes)
Phos-tag 5(w/v)%-Acrylamide/Bis (29:1) gel (see Recipes)
Separation gel
Stacking gel
SDS-PAGE running buffer (see Recipes)
2x Laemmli sample buffer (see Recipes)
1x Laemmli sample buffer (see Recipes)
Equipment
NanoDrop 2000c Spectrometer (Thermo Fisher Scientific, model: NanoDropTM 2000c Spectrophotometer, catalog number: ND2000C )
SDS-PAGE gel electrophoresis chamber (BIO CRAFT, catalog number: BE-230G )
Scanner (EPSON, model: GT-X980 )
Invitroshaker (TAITEC, model: Shake-LR, catalog number: 0054809-000 )
Software
ImageJ 1.8.0_172 (Abramoff et al., 2004)
Procedure
Preparation of MBP-CENP-C (aa 601-864)
Note: Although we describe the protocol for MBP-CENP-C (aa 601-864) phosphorylation using CDK1, the protocol can be applied for other kinases and their substrates for your interests.
Purify recombinant MBP-CENP-C (aa 601-864) as described in Watanabe et al. (2019).
Note: Briefly, the MBP-CENP-C (aa601-864) was expressed in bacteria, and purified with Amylose resin and Hi-Trap SP column with Superdex 200 pg. The purified protein was dissolved with buffer containing 20 mM HEPES-NaOH (pH 7.5), 500 mM NaCl, 5% Glycerol, 1 mM EDTA-NaOH (pH 8.0), and 1 mM DTT. The purified protein was snap-frozen in liquid nitrogen and stored at -80 °C. The frozen protein should be thawed on ice just before using.
Prepare 1,000 ml of ice-cold 1x kinase buffer (listed in Recipes).
Inject MBP-CENP-C (aa 601-864) into Xpress Micro Dialyzer up to 100 µl following the product instruction manuals and dialyze the protein in 1,000 ml of 1x kinase buffer for overnight at 4 °C.
Note: Dialysis is important for removal of EDTA from your protein samples, and for adjustment of salt concentration for the next steps. Because high concentration of salt and EDTA inhibits kinase reaction and interferes phospho-protein running on Phos-tag SDS-PAGE.
Collect the dialyzed MBP-CENP-C (aa 601-864) into a Protein Lobind Tube on ice.
Analyze protein concentration using molecular absorptivity and molecular weight with NanoDrop 2000c Spectrometer (Desjardins et al., 2009).
Note: Because in the next step the substrate is added for kinase reaction at 2 mg/ml (final concentration), if concentration of the protein is less than 2 mg/ml, concentrate protein using Amicon Ultra.
Optimization of kinase reaction
Note: To get optimum phosphorylation in the kinase reaction, several conditions can be optimized: substrate concentration, kinase amount, reaction buffer, and reaction time and temperature. Here, we demonstrate an example of optimization for reaction time in the kinase assay.
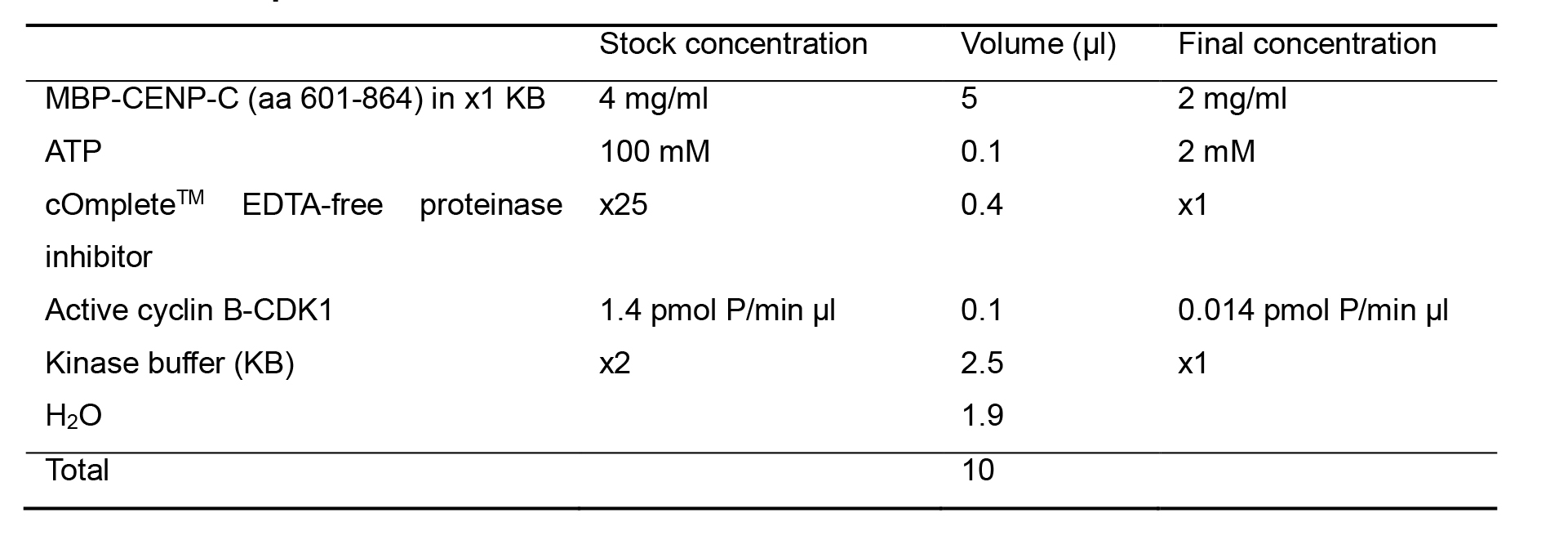
Mix the dialyzed MBP-CENP-C (aa 601-864) in kinase buffer with 100 mM ATP, 25x cOmpleteTM EDTA-free proteinase inhibitor (listed in Recipes), active cyclin B-CDK1 (relative H1 kinase activity: 1.4 pmol P/min µl), and add 2x kinase buffer (listed in Recipes) and H2O to adjust concentration of each component to final concentration in 1x kinase buffer as described in Table 1 showing our representative 10 µl-scale reaction mixture.
Notes:
You can change the volume of the kinase reaction according to your experimental design.
It is recommended to make small aliquots and store 100 mM ATP at -20 °C to avoid multiple freeze-thaw cycles.
Purification of active cyclin B-CDK1 was described in Okumura et al. (1996).
Table 1. Representative kinase reaction mix
Incubate the 10 µl reaction mixture at 25 °C. Gently mix the sample with tapping every 15 min.
Note: In this protocol, to examine optimum reaction time, several 10 µl-scale reactions are incubated for the indicated time periods in Figure 1 (0, 30, 60 and 90 min).
To stop the reaction, add 10 µl of 2x Laemmli sample buffer (listed in Recipes) to the reaction mixture. Then, add further 20 µl of 1x Laemmli sample buffer to adjust protein concentration for SDS-PAGE in the next step (listed in Recipes), and heat samples for 5 min at 96 °C.
Note: If you use the phosphorylated proteins for other in vitro assays such as a pull-down assay, stop the kinase reaction by adding 1 M EDTA-NaOH (pH 8.0) to the reaction mixture in a final concentration of 5 mM.
Detection of phosphorylated substrates using Phos-tag SDS-PAGE
Prepare a Phos-tag 5(w/v)%-Acrylamide/Bis (29:1) gel (25 µM Phos-tag, listed in Recipes).
Note: To obtain optimum separation of phosphorylated proteins in the Phos-tag SDS-PAGE, Phos-tag and acrylamide concentrations should be optimized. Assemble the gel into an electrophoresis chamber (listed in Equipment).
Add SDS-PAGE running buffer to electrophoresis chamber (listed in Recipes).
Load 0.5 µg of phosphorylated protein each lane in 1x Laemmli sample buffer, and also 5 µl of molecular weight marker on the gel.
Note: If your molecular weight marker includes salts and/or EDTA, which interfere sample running in the next lanes on Phos-tag SDS-PAGE. To avoid this problem, an empty lane should be inserted between the molecular weight marker and the next sample.
Run gels using 35 mA constant current for 55 min.
After electrophoresis, wash gels with water for 5 min using Invitroshaker (speed 60).
Remove water and fill gel with 50 ml of CBB Stain One. Warm the gel with CBB stain One by a 700 W microwave oven for 1 min (approximately 70 °C).
Shake the gel for 10 min at room temperature using Invitroshaker (speed 60).
Remove CBB stain One and wash gel with water for 1 min twice using Invitroshaker (speed 60).
Fill gel with 50 ml of new water. Warm the gel with water by a 700 W microwave oven for 1 min (approximately 70 °C).
Shake the gel with warmed water over 60 min at room temperature using Invitroshaker (speed 60).
Remove water and wash gel with water twice and acquire images of the stained gel using an image scanner.
Detection of phosphorylated substrates using regular SDS-PAGE
Note: To examine protein levels and qualities of substrates, use a conventional SDS-PAGE, because it is difficult to compare protein levels precisely and detect small amount of protein fragments on the Phos-tag SDS-PAGE. This procedure is optional, but we highly recommend it, because some proteins may be degraded during the kinase reaction.
Prepare Super SepTM Ace, 5-20%, 17-well SDS precast gel.
Assemble the gel into an electrophoresis chamber (listed in Equipment).
Add SDS-PAGE running buffer to electrophoresis chamber (listed in Recipes).
Load 0.5 µg of phosphorylated protein each lane in 1x Laemmli sample buffer, and also 5 µl of molecular weight marker on the gel.
Run gels using 35 mA constant current for 55 min.
After electrophoresis, wash gels with water for 5 min.
Remove water and fill gel with 50 ml of CBB Stain One. Warm the gel with CBB stain One by a 700 W microwave oven for 1 min (approximately 70 °C).
Shake the gel for 10 min at room temperature using Invitroshaker (speed 60).
Remove CBB stain One and wash gel with water for 1 min twice using Invitroshaker (speed 60).
Fill gel with 50 ml of new water. Warm the gel with water by a 700 W microwave oven for 1 min (approximately 70 °C).
Shake the gel with warmed water over 60 min at room temperature using Invitroshaker (speed 60).
Remove water and wash gel with water twice and acquire images of the stained gel using an image scanner.
Data analysis
In Phos-tag SDS-PAGE, if protein bands are shifted up in the kinase-treated sample compared with the untreated sample protein band, it indicates that the substrate is phosphorylated by kinase in vitro. The band shift in Phos-tag SDS-PAGE depends on the number of phosphorylated residues and their position in the substrate.
Figure 1 shows representative images of phosphor-protein analysis on Phos-tag (left) and conventional (right) SDS-PAGE gels using the protocol described in this chapter. The MBP-CENP-C (aa 601-864) treated with CDK1 clearly shows mobility shifts on Phos-tag SDS-PAGE, indicating CENP-C phosphorylation by CDK1. Importantly, the Phos-tag SDS-PAGE enable to detect the mobility shifts of phosphor-proteins, which do not change their mobility on the conventional SDS-PAGE. Phosphorylation levels of the substrate can be evaluated by quantification of the intensity of the shifted bands using ImageJ open source software (Abramoff et al., 2004).
At 30 min after CDK1 treatment, MBP-CENP-C (aa 601-864) displayed an obvious shifted band with reduced levels of unphosphorylated protein band (Figure 1, left). Subsequently, at the later time points, MBP-CENP-C (aa 601-864) showed further mobility shift, suggesting that MBP-CENP-C (aa 601-864) was phosphorylated at two sites by CDK1, but CDK1 appeared to have different phosphorylation efficiency between the two sites.
In summary, this in vitro kinase assay protocol using Phos-tag SDS-PAGE is a simple but useful method to detect kinase activity and to analyze protein phosphorylation.
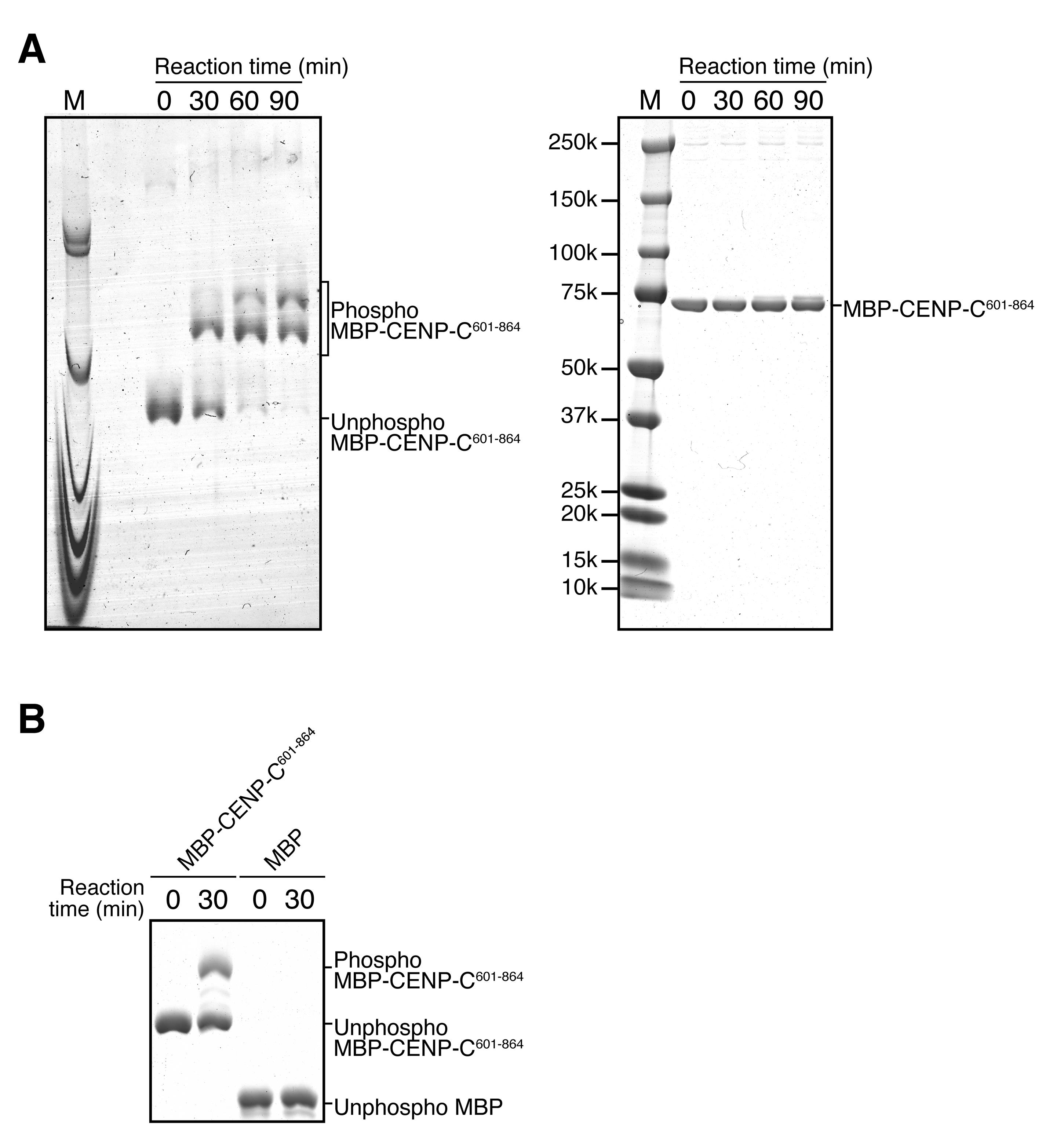
Figure 1. Analysis of CDK1-mediated CENP-C phosphorylation on Phos-tag and conventional SDS-PAGE. A. The left image shows Phos-tag SDS-PAGE analysis of MBP (maltose-binding protein)-CENP-C (aa 601-864): MBP-CENP-C601-864 was treated with CDK1 for the indicated times. Right image shows conventional SDS-PAGE analysis of same samples. M shows molecular weight marker. In the left Phos-tag SDS-PAGE analysis, MBP-CENP-C601-864 was shifted-up, indicating CENP-C phosphorylation by CDK1 in vitro. B. The image shows Phos-tag SDS-PAGE analysis of MBP-CENP-C601-864 and MBP, which were treated with CDK1 for the indicated times. MBP, which is not a CDK1 substrate, showed no mobility shifts, indicating that phosphorylated proteins can be specifically detected on Phos-tag SDS-PAGE.
Recipes
1x kinase buffer (KB)
10 mM Tris-HCl, pH 7.5
2 mM MgCl2
150 mM NaCl
2x kinase buffer
20 mM Tris-HCl, pH 7.5
4 mM MgCl2
300 mM NaCl
25x cOmpleteTM EDTA-free proteinase inhibitor
One tablet of cOmpleteTM EDTA-free proteinase inhibitor in 2 ml water
Phos-tag 5(w/v)%-Acrylamide/Bis (29:1) gel
Separation gel
5(w/v)%-Acrylamide/Bis Mixed Solution (29:1)
375 mM Tris-HCl, pH 8.8
0.1% SDS
0.1% APS
0.1% TEMED
25 µM Phos-tag Acrylamide
50 µM MnCl2
Note: It is recommended to make small aliquots and store 100 µM MnCl2 at -20 °C to avoid multiple freeze-thaw cycles. Use of an old MnCl2 stock would cause inefficient mobility changes of phospho-proteins on Phos-tag SDS-PAGE.
Stacking gel
3.5(w/v)%-Acrylamide/Bis Mixed Solution (29:1)
120 mM Tris-HCl, pH 6.8
0.1% SDS
0.2% APS
0.2% TEMED
SDS-PAGE running buffer
25 mM Tris
192 mM Glycine
0.1% SDS
2x Laemmli sample buffer
0.125 M Tris-HCl, pH 6.8
20% glycerol
4% SDS
10% 2-mercaptoethanol
0.005% bromophenol blue
1x Laemmli sample buffer
0.0625 M Tris-HCl, pH 6.8
10% glycerol
2% SDS
5% 2-mercaptoethanol
0.0025% bromophenol blue
Acknowledgments
This work was supported by JSPS KAKENHI Grant Number 17H06167, 16H06279 and 15H05972 to TF, JSPS KAKENHI Grant Number 16K18491 to MH, and JSPS KAKENHI Grant Number 18K06084 to MA. This protocol was adapted from and used in Watanabe et al. (2019).
Competing interests
The authors declare no competing interests.
References
- Abramoff, M. D., Magalhães, P. J. and Ram, S. J. (2004). Image processing with ImageJ. Biophotonics Int 11(7): 36-42.
- Desjardins, P., Hansen, J. B. and Allen, M. (2009). Microvolume protein concentration determination using the NanoDrop 2000c spectrophotometer. J Vis Exp (33): e1610.
- Fukagawa, T. and Earnshaw, W. C. (2014). The centromere: chromatin foundation for the kinetochore machinery. Dev Cell 30(5): 496-508.
- Gascoigne, K. E. and Cheeseman, I. M. (2013). CDK-dependent phosphorylation and nuclear exclusion coordinately control kinetochore assembly state. J Cell Biol 201(1): 23-32.
- Hara, M., Ariyoshi, M., Okumura, E. I., Hori, T. and Fukagawa, T. (2018). Multiple phosphorylations control recruitment of the KMN network onto kinetochores. Nat Cell Biol 20(12): 1378-1388.
- Hara, M. and Fukagawa, T. (2018). Kinetochore assembly and disassembly during mitotic entry and exit. Curr Opin Cell Biol 52: 73-81.
- Kinoshita, E., Kinoshita-Kikuta, E., Takiyama, K. and Koike, T. (2006). Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics 5(4): 749-757.
- Nishino, T., Rago, F., Hori, T., Tomii, K., Cheeseman, I. M. and Fukagawa, T. (2013). CENP-T provides a structural platform for outer kinetochore assembly. EMBO J 32(3): 424-436.
- Nurse, P., (1990). Universal control mechanism regulating onset of M-phase. Nature 344(6266): 503-508.
- Malumbres, M. and Barbacid, M. (2005). Mammalian cyclin-dependent kinases. Trends Biochem Sci 30(11): 630-641.
- Okumura, E., Sekiai, T., Hisanaga, S., Tachibana, K. and Kishimoto, T. (1996). Initial triggering of M-phase in starfish oocytes: a possible novel component of maturation-promoting factor besides cdc2 kinase. J Cell Biol 132(1-2): 125-135.
- Watanabe, R., Hara, M., Okumura, E. I., Herve, S., Fachinetti, D., Ariyoshi, M. and Fukagawa, T. (2019). CDK1-mediated CENP-C phosphorylation modulates CENP-A binding and mitotic kinetochore localization. J Cell Biol 218(12): 4042-4062.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Watanabe, R., HHara, M., Ariyoshi, M. and Fukagawa, T. (2021). CENP-C Phosphorylation by CDK1 in vitro. Bio-protocol 11(1): e3879. DOI: 10.21769/BioProtoc.3879.
- Watanabe, R., Hara, M., Okumura, E. I., Herve, S., Fachinetti, D., Ariyoshi, M. and Fukagawa, T. (2019). CDK1-mediated CENP-C phosphorylation modulates CENP-A binding and mitotic kinetochore localization. J Cell Biol 218(12): 4042-4062.
Category
Cancer Biology > Cell cycle checkpoints > Biochemical assays
Biochemistry > Protein > Electrophoresis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









