- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Optimized Immunostaining of Embryonic and Early Postnatal Mouse Brain Sections
Published: Vol 11, Iss 1, Jan 5, 2021 DOI: 10.21769/BioProtoc.3868 Views: 6098
Reviewed by: Anand Ramesh PatwardhanCarmelo BellarditaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Optimizing Confocal Imaging Protocols for Muscle Fiber Typing in the Mouse Masseter Muscle
Catalina Matias [...] Jeffrey J. Brault
Apr 5, 2025 2900 Views

Development of a Novel Automated Workflow in Fiji ImageJ for Batch Analysis of Confocal Imaging Data to Quantify Protein Colocalization Using Manders Coefficient
Vikram Aditya [...] Wei Yue
Apr 5, 2025 2856 Views

Detecting the Activation of Endogenous Small GTPases via Fluorescent Signals Utilizing a Split mNeonGreen: Small GTPase ActIvitY ANalyzing (SAIYAN) System
Miharu Maeda and Kota Saito
Jan 5, 2026 482 Views
Abstract
The mammalian neocortex, the outer layer of the cerebrum and most recently evolved brain region, is characterized by its unique areal and laminar organization. Distinct cortical layers and areas can be identified by the protein expression of graded transcription factors and molecular determinants that define the identity of different projection neurons. Thus, specific detection and visualization of protein expression is crucial for assessing the identity of neocortical neurons and, more broadly, for understanding early and late developmental mechanisms and function of this complex system. Several immunostaining/immunofluorescence methods exist to detect protein expression. Published protocols vary with regard to subtle details, which may impact the final outcome of the immunofluorescence. Here, we provide a detailed protocol, suitable for both thin cryostat sections and thick vibratome sections, which has successfully worked for a wide range of antibodies directed against key molecular players of neocortical development. Ranging from early technical steps of brains collection down to image analysis and statistics, we include every detail concerning sample inclusion and sectioning, slide storage and optimal antibody dilutions aimed at reducing non-specific background. Routinely used in the lab, our background-optimized immunostaining protocol allows efficient detection of area- and layer- specific molecular determinants of distinct neocortical projection neurons.
Graphic abstract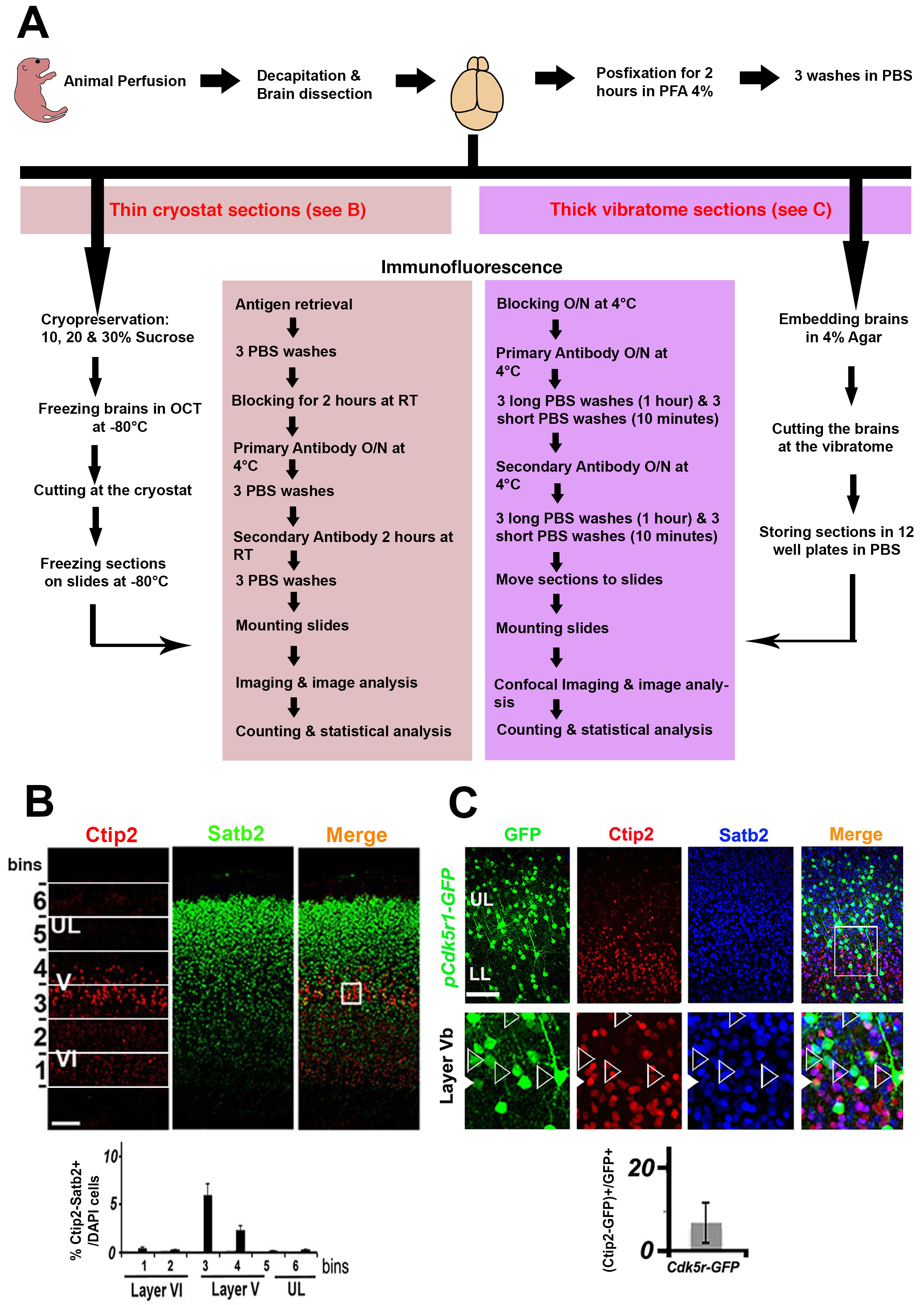
Workflow chart for the optimized immunostaining protocol of mouse brain sections. A. A flow chart for different steps of the optimized immunostaining protocol on both thin cryostat and thick vibratome sections. B. Example for immunostaining against Satb2 and Ctip2 on a thin coronal section (20 μm) at the level of the somatosensory cortex. The first column to the left shows the binning system where 6 bins can be overlaid on the image. On the bottom, an example of counting analysis showing the percentage of marker-positive cells normalized to the total number of DAPI or Hoechst-positive cells. C. Example for immunostaining against Satb2 and Ctip2 on a GFP+ thick vibratome section (200 μm). Images are taken at low magnification (10x, left) and high magnification (40x, right). The graph shows a counting of the percentage of Ctip2-positive neurons normalized to the total number of GFP-electroporated neurons on high-magnification images. Images on B and C are modified from Harb et al. (2016).
Background
The mammalian neocortex is characterized by its radial subdivision into six distinct neuronal layers. In addition, during embryonic development and with further refinement postnatally, the neocortex gets organized tangentially into distinct primary areas dedicated to the elaboration and analysis of both motor outputs and sensory inputs, a developmental process termed “arealization”. These areas within the neocortex are positioned along the antero-posterior and medio-lateral axes by graded expression of key nuclear transcription factors. The basic neocortical structure of the six layers adapts within each area, as layers are populated with specific neuronal populations, which are defined by the expression of key molecular determinants driving the area-specific acquisition of their identity, morphology and connectivity. Thus, detection of selected protein of interest in neocortical neurons is crucial for investigating the tangential and radial expression of these determinants, which in turn is critical for exploring developmental and functional mechanisms occurring in this complex structure. Widely used in biology labs to determine the tissue and cellular localization of a protein of interest, the immunohistochemistry (or immunofluorescence) is a technical procedure that assesses the presence of a specific protein or antigen by the use of a specific primary antibody, followed by a secondary antibody coupled with a fluorochrome, thus allowing indirect visualisation and examination under a microscope. Many immunostaining protocols are available in literature; however, even subtle changes in the whole procedure can affect the final outcome. Here, we present a protocol that worked in our hands for most of the antibodies aimed at detecting key determinants of neocortical development in thick vibratome or thin cryostat sections. We added exhaustive details regarding all steps, from animal sample collection until image analysis, which can help beginners in the field to easily use the protocol. Optimized for the detection of nuclear determinants in neocortical neurons of embryonic or early postnatal mouse neocortex, the protocol is easily adaptable to a wide variety of biological samples, such as other neural or non-neural tissues.
Materials and Reagents
For animal perfusion:
12-well plates (Falcon, catalog number: 353043 )
Needles, syringes
Butterfly needles:
Winged Infusion Set: 25 G 0.50 x 19mm + 30 cm tubing (Braun, catalog number: 4056370 )
Winged Infusion Set: 21 G 0.80 x 19mm + 30 cm tubing (Braun, catalog number: 4056337 )
Hypodermic needles:
18 G (Terumo Neolus, catalog number: NN-1838R)
21 G (Terumo Neolus, catalog number: NN-2138R )
25 G (Terumo Neolus, catalog number: NN-2516R )
30 G (BD Microlance 3, catalog number: 304000 )
Syringes without needle:
20 ml (Terumo, catalog number: SS+20ES 1)
50 ml (Terumo, catalog number: SS+50ES1 )
Syringes with needle:
U-100 Insulin syringe, 0.5 ml 0.33 mm (29G) (Fisher Scientific, BD Medical, catalog number: 324892 )
Ketamine
Xylazine
Paraformaldehyde (PFA) (Sigma-Aldrich, catalog number: P6148-1KG , Lot: MKCD5277)
Phosphate Buffer Saline (PBS) (Gibco, catalog number: 14190-094 ) or homemade PBS (Recipe below)
Sodium chloride (VWR, AnalaR Normapur, catalog number: 27810.295 )
Potassium chloride (Merck Millipore, catalog number: 1.04936.1000 )
Sodium phosphate dibasic heptahydrate (Sigma-Aldrich, catalog number: S9390-2.5KG )
Potassium dihydrogen phosphate (Merck Millipore, catalog number: 1.04873.1000 )
4% PFA (see Recipes)
1x PBS (see Recipes)
Tissue sample fixation and washing:
12-well plates (Falcon, catalog number: 353043 )
Paraformaldehyde (PFA) (Sigma-Aldrich, catalog number: P6148-1KG , Lot #MKCD5277)
Phosphate Buffer Saline (PBS) (Gibco, catalog number: 14190-094 ) or homemade PBS (Recipe below)
Sodium Azide (Sigma-Aldrich, catalog number: S8032-100G, Lot #0001453190 )
4% PFA (see Recipes)
1x PBS (see Recipes)
Embedding in agar for thick vibratome sections:
12-well plates (Falcon, catalog number: 353043 )
Select Agar ≥ 99.5% (GC) (Sigma-Aldrich, catalog number: S5054-250G, Lot #MKCB2702V )
Phosphate Buffer Saline (PBS) (Gibco, catalog number: 14190-094 ) or homemade PBS (Recipe below)
4% Agar (see Recipes)
Cryopreservation and Freezing for thin cryostat sections:
6 cm Petri dish
Optimal cutting medium (OCT) (Leica Tissue Freezing Medium, catalog number: 14020108926 )
Dry ice
Sucrose (Sigma-Aldrich, catalog number: S9378-1KG , Lot #BCBS5325V)
Phosphate Buffer Saline (PBS) (Gibco, catalog number: 14190-094 ) or homemade PBS (Recipe below)
1x PBS (see Recipes)
10, 20 and 30% sucrose (see Recipes)
Vibratome sectioning (thick sections):
12-well plates (Falcon, catalog number: 353043 )
Blades (Gillette "Bleue Extra")
Phosphate Buffer Saline (PBS) (Gibco, catalog number: 14190-094 ) or homemade PBS (Recipe below)
Glue (Henkel, Loctite superglue-3 )
1x PBS (see Recipes)
Immunofluorescence on thick vibratome sections:
12-well plates (Falcon, catalog number: 353043 )
Slides (Thermo Scientific, Superfrost Plus: J1800AMNZ , Lot #0180)
Cover slips (Dia path, catalog number: 0 61061 )
Nail polish
Phosphate Buffer Saline (PBS) (Gibco, catalog number: 14190-094 ) or homemade PBS (Recipe below)
Goat serum (GS) (Thermo Fisher scientific, Gibco, catalog number: 16210-064 , Lot #16711329)
Newborn calf serum (NBCS) (Thermo Fisher scientific, Gibco, catalog number: 16010-167 )
Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A-4503 or A7906-50G , Lot #SLBX7550)
Triton (Sigma-Aldrich, catalog number: T8787-250ML , Lot #SLBV4122))
N-propyl Gallate (Sigma-Aldrich, catalog number: P3130-100G , Lot #SLBD6728V)
Glycerol (VWR Chemicals, catalog number: 24388.295 , Batch: 14E050030)
Hoechst 33342, Trihydrochloride (Invitrogen, catalog number: H3570 , Lot #1116437)
Primary antibodies, example:
Secondary antibodies, Example: Alexa Fluor 488, 555, 594 and 647 anti-mouse or anti-rabbit IgG conjugates (Thermo Fisher scientific)
Blocking Solution (see Recipes)
Antibody Solution (see Recipes)
Mounting medium (see Recipes)
Antibody Solution for vibratome thick sections (see Recipes)
Cryostat sectioning (thin sections):
Slides (Thermo Scientific, Superfrost Plus: J1800AMNZ , Lot #0180)
Blades (Thermo Fisher scientific, Edge-Rite: 4280L )
Optimal cutting medium (OCT) (Leica Tissue Freezing Medium, 14020108926 )
Immunofluorescence on thin cryostat sections:
Coverslips (Dia Path, catalog number: 0 61061 )
Phosphate Buffer Saline (PBS) (Gibco, catalog number: 14190-094 ) or homemade PBS (Recipe below)
Distilled water
Trisodium Citrate Dihydrate (Sigma-Aldrich, catalog number: S1804-1KG , Lot #BCBF5684V)
Citric acid (Sigma-Aldrich, catalog number: C-0759 , Lot #34H1314)
Goat serum (GS) (Thermo Fisher scientific, Gibco, catalog number: 16210-064 , Lot #16711329)
New-born calf serum (NBCS) (Thermo Fisher scientific, Gibco, catalog number: 16010-167 )
Triton (Sigma-Aldrich, catalog number: T8787-250ML , Lot #SLBV4122)
N-propyl Gallate (Sigma-Aldrich, catalog number: P3130-100G ; Lot #SLBD6728V)
Glycerol (VWR Chemicals, catalog number: 24388.295 ; Batch 14E050030)
Hoechst 33342, Trihydrochloride (Invitrogen, catalog number: H3570 ; Lot #1116437)
Primary antibodies, example:
Nr2f1/COUP-TFI (Abcam, catalog number: ab181137 or R&D, catalog number: H8132 )
Ctip2 (Abcam, catalog number: ab18465 )
Satb2 (Abcam, catalog number: ab51502 )
Sox5 (Abcam, catalog number: ab94396 )
GFP (Thermo Fisher scientific, catalog number: A-11122 ; or: Abcam, catalog number: ab13970 )
Ski (Santa Cruz, catalog number: sc-9140 )
For other antibodies, see “Materials and Methods” in Harb et al. (2016).
Secondary antibodies, Example: Alexa Fluor 488, 555, 594 and 647 anti-mouse or anti-rabbit IgG conjugates (Thermo Fisher Scientific)
Unmasking Solution (see Recipes)
Blocking Solution (see Recipes)
Antibody Solution (see Recipes)
Equipment
Scissors, forceps, dissection and tissue handling tools
Sharp scissors (Holtex, model: IC12411 )
Fine scissors (Fine Science Tool, catalog number: 14060-11 )
45° angled tip forceps (Fine Science Tool, model: Dumont #5, catalog number: 11251-35)
Straight tip forceps (Fine Science Tool, Dumont Mini Forceps – Style 5)
Double-ended micro spatula (Fine Science Tool, catalog number: 10091-12 )
Perforated spoon (WPI, catalog number: 501997 )
Microwave
Embedding moulds:
Peel-A-Way® embedding moulds (Sigma-Aldrich, catalog number: E6032-1CS )
Peel-A-Way® Disposable Embedding Moulds (VWR Polysciences, catalog number: 18646A-1 )
Brush
Boxes for storage (Dutscher, catalog number: 0 37516 )
Glass (Leica, Insert Glass 70 mm, catalog number: 14047742497 )
Boxes for boiling (Tissue-Tek® Staining Dish, Sakura, catalog number: 4456 )
Vertical boxes for slides wash (Fisher Scientific, Fisherbrand 5-place slide mailer: HS15986 )
Shaker (Stuart 3D gyratory rocker, catalog number: SSL3 )
Horizontal Microscope slide boxes (Kartell Labware, #276 [98 mm x 83 mm], #277 [230 mm x 97 mm] and #278 [230 mm x 180 mm])
pH meter (Fisher Scientific, model: accumet AE150 )
Cryostat (Leica, model: CM3050S )
Vibratome (Leica, model: VT1000S )
Microscope (Leica, model: DM6000B +CTR6000 )
Confocal microscope (Zeiss, model: LSM 710, 2012 )
Water bath (Fisher Scientific, catalog number: Polystat 24, +5L/8662F )
Software
Adobe Photoshop CS (CS5 Extended, Version 12.0 x32)
Microsoft Excel (2005 Version)
GraphPad Prism (Version 7.00)
Zen software for confocal images (Zen Black, Version 11 SP3 HF88.1.8.484)
Leica Microsystem CMS Gmbh LASX (Version: 3.3.3.16958)
Procedure
Mouse Brain Collection and Fixation
Anesthetize the mouse with an intra-peritoneal injection of Ketamine/Xylazine mix (100 mg/kg Ketamine mixed with 10 mg/kg Xylazine in isotonic saline solution; contact local animal ethical committee for drug use and animal procedures). In our case, all mouse experiments were conducted in accordance with relevant national and international guidelines (European Union rules; 2010/63/UE), and have been approved by the local ethical committee in France (CIEPAL NCE/2014-209 and NCE/2019-548).
For postnatal/adult mice, perfuse them intracardially with 4% PFA; for embryonic stages, recover embryos on ice-cold PBS from the uterus of the anesthetized mother.
Decapitate adults/embryos and dissect the brains; for early stage embryos (embryonic age < 15.5 days), brain dissection is not mandatory, but highly recommended to reduce background signal on skin and other tissues surrounding the brain.
Keep brains in ice-cold PBS in 12-well plates during and after collection.
Post-fix brains for 2 h in 4% PFA at 4 °C (gentle agitation on shaker).
Wash the brains 3 times for 10 min each in PBS at 4 °C (on shaker).
Store brains at 4 °C until further treatment (few days); for long-term storage (few weeks), supplement PBS with 0.05% Sodium Azide.
Embedding in Agar for vibratome sectioning (thick sections: 50-200 μm)
Prepare a solution of 4% agar in PBS.
Boil in the microwave until homogenization.
Keep the 4% agar in a water bath at 50 °C.
Add 4% agar to a 12-well plate.
Remove the brain from PBS with perforated spoon and dry it on a paper towel.
Quickly immerge the brain in agar and correctly position it with forceps before agar polymerization.
Store the brain at 4 °C for short term or immediately proceed with sectioning.
Cutting vibratome thick sections
Trim the agar surrounding the brain and fix it on the vibratome with glue.
Add ice cold PBS to the vibratome and keep it cold by adding ice around the mould for cutting.
Cut 50-200 μm thick sections.
Use a brush to gently collect the sections from the vibratome before moving them to a 12-well plate; keep the plate on ice-cold PBS.
Create series of sections representing the whole cortex or brain by distributing similar sections on subsequent wells.
You can add up to 3-4 sections in each well.
Immunostaining on vibratome sections (50-200 μm)
Remove the PBS by carefully aspirating it with a pipette tip, without touching the sections (repeat for following passages).
Incubate the sections in Blocking solution (10% GS, 3% BSA, 0.3% Triton) over-night (O/N) at 4 °C by adding 1 ml for each well.
The following day add 500 μl of primary antibodies by diluting them to the appropriate concentration with the antibody solution (3% GS, 3% BSA, 0.3% Triton).
Incubate the sections with primary antibodies O/N at 4 °C.
The day after, wash the sections 3 times for 1 h each in PBS, followed by 3 short washes for 10 min each in PBS (all washes are performed at room temperature (RT), on shaker).
Prepare secondary antibodies by diluting them to 1/500 in the antibody solution (3% GS, 3% BSA, 0.3% Triton).
Incubate the sections with 500 μl of diluted secondary antibodies O/N at 4 °C by shaking.
The following day wash the sections in PBS as described in Step D5.
Incubate the sections in 1 μg/ml Hoechst in PBS for 1 h.
Carefully move the sections using a brush from the wells to a slide covered with PBS.
Aspirate the PBS with a pipette tip and let the sections dry.
Add 3 drops of mounting solution (80% glycerol, 2% N-propyl gallate, 1 μg/ml Hoechst) to each slide.
Cover the slides with cover slips and seal the edges with nail polish.
Leave the nail polish to dry for 5 min at RT.
Store the slides until imaging at -20 °C.
Cryopreservation and Freezing brains for thin sectioning at the cryostat (thin sections: 10-20 μm)
Prepare 10%, 20% and 30% sucrose in PBS.
Put the brains (dissected and post-fixed as detailed in Procedure A) in 10% sucrose, keep at 4 °C with gentle shaking. Wait to equilibrate (usually 8-12 h) until they sink to the bottom of the wells.
Repeat Step E2 with sucrose 20% and then 30%. Increasing sucrose concentration in a gradual way is key for preserving optimal histology.
Remove all sucrose from the brains by shaking them with a Pasteur pipette in OCT in a 6 cm Petri dish.
Transfer the brains in OCT in Peel-A-Way® boxes.
Snap freeze them on dry ice.
Store them at -80 °C until sectioning.
Cutting sections at the cryostat (10-20 μm)
Equilibrate the brains at -20 °C for 30 min at the cryostat.
Cut 20 μm thin sections for postnatal brains and 12-16 μm for embryonic brains.
During cutting, collect series of sections on cryostat slides.
Let the slides dry O/N at RT.
Store them at -80 °C until use.
Immunostaining on cryostat sections
Defreeze the sections for 1 h at RT.
Put the slides on a vertical slide box for boiling.
If your antibody needs unmasking, boil the slides for 15 s in an unmasking solution (0.1 M sodium citrate, pH 6).
Boil them a second time for 1 s with fresh unmasking solution (0.1 M sodium citrate, pH 6).
Cool the sections on ice for 10 min.
Move the slides to a vertical slide box for washing
Wash the sections 3 times in PBS for 10 min each at RT (on shaker).
Create a humidified chamber by adding water or PBS in a plastic box where the slides can lay horizontally.
Lay your slides horizontally and avoid the sections to dry by immediately adding the blocking solution (10% GS, 0.3% Triton in PBS).
Block the sections in blocking solution for 1 h at RT by adding 1 ml for each slide.
Prepare 200-300 μl of primary antibodies for each slide by diluting them to the appropriate concentration with the antibody solution (3% GS, 0.3% Triton in PBS).
Add 200-300 μl of the diluted primary antibodies to the slides and cover them with cover slips to homogeneously distribute the antibody.
Incubate the sections with primary antibodies (ON at 4 °C).
The following day move the slides to the vertical slide box containing PBS.
Wash the sections 3 times for 10 min each in PBS at RT while shaking.
Prepare 200-300 μl of secondary antibodies per slide by diluting them to 1/300 in the antibody solution (3% GS, 0.3% Triton).
Lay the slides horizontally again in the humidified chamber.
Add 200-300 μl of the diluted secondary antibodies to the slides and cover them with cover slips to homogeneously distribute the antibody.
Incubate the sections for 2 h at RT with diluted secondary antibodies.
Move the slides to the vertical slide box containing PBS.
Wash the sections 3 times for 10 min each in PBS at RT while shaking.
Lay the slides horizontally after taking away extra PBS.
Add 3 drops of mounting solution (80% glycerol, 2% N-propyl gallate, 1 μg/ml Hoechst) to each slide.
Cover the slides with cover slips and seal the edges with nail polish.
Leave the nail polish dry for 5 min at RT.
Store the slides at -20 °C until imaging.
Imaging
Take images of your area and layer of interest with the appropriate magnification
For thin sections:
Acquire plane images with a 10x/20x magnification using a fluorescent optical microscope.
For thick sections: Acquire Z-stack images using a confocal microscope.
Acquire Z-stack images using a confocal microscope.
Use low (10x) and high magnification (40x) images of your layer of interest.
Data analysis
Data analysis for thin cryostat sections (coronal cutting plane):
Process the 1 plane images acquired with 10x/20x magnification using Adobe Photoshop.
For quantification of specific markers based on their layer position, use a binning system in Adobe Photoshop by overlaying a grid with 6-8 bins on the neocortical area of interest, from the marginal zone until the glial surface. Six bins are typically used for embryonic or postnatal day (P) 0 brains, while 8 bins can be used from P7.
Count the cells expressing the protein of interest using Photoshop counting tool or an automated counting software like ImageJ.
Normalize the number of marker-positive cells to the total number of cells (quantified by counting Hoechst+ nuclei) in each bin.
Count at least 3 sections per animal and use at least 3 animals of each genotype for reliable statistical analysis.
To avoid variability occurring between litters, the best option is to use littermate controls to compare the expression of specific genes/proteins of interest in different genetic backgrounds.
Use two-tailed Student’s t-test on Microsoft Excel or on GraphPad Prism for statistical analysis of two independent data groups. For comparing multiple (> 2) groups, use analysis of variance (ANOVA) or other appropriate statistical tests.
Data analysis for thick vibratome sections:
Process the Z-Stack images using the correspondent software of your confocal microscope (for example, Zen lite) and make a maximum intensity projection image.
Count the cells expressing the protein of interest and normalize them to the total number of cells or to another marker depending on the purpose.
Count at least 3 sections per animal and use at least 3 animals of each genotype for statistical analysis.
Use littermate controls to compare the expression of specific genes/proteins of interest in different genetic backgrounds (example: wild-type versus knock-out) or after specific treatments, depending on your scientific question.
Use two-tailed Student’s t-test on Microsoft Excel or on GraphPad Prism for statistical analysis of two independent data groups. For comparing multiple (> 2) groups, use analysis of variance (ANOVA) or other appropriate statistical tests.
Notes
To minimize subjective bias, sample identity (e.g., genotypes) can be randomized by associating an identification number to each sample before processing.
Fixed embryos/brains/sections with clearly damaged tissue must be excluded from any further analysis/processing.
Long O/N incubation with Blocking solution BEFORE addition of antibodies, together with long and repeated PBS washes, are crucial for background reduction and signal specificity, as they contribute to mask non-specific epitope sites and to remove excess antibodies, respectively.
Troubleshooting 1: Poor tissue fixation can result from perfusion with low-quality PFA. Avoid repeated thaw-freeze cycles by freezing PFA aliquots of appropriate volumes, and by using in one-two weeks after thawing.
Troubleshooting 2: Poor histology commonly results from inefficient tissue dehydration prior OCT embedding. Be sure to cryopreserve with long incubation in increasingly concentrated sucrose solutions (see Steps E1-E3).
Troubleshooting 3: In our experience, poor adhesion of cryostat tissue sections on glass slides can happen when using expired slides. Always check the expiring date of SuperFrost slides for optimal adhesion.
Troubleshooting 4: For correct background removal, be sure to use a normal serum from the same species as the one in which the secondary antibody was generated. The use of the incorrect serum can result in non-specific signal.
Tips 1: Many of the antibodies we used for studying cortical development detect nuclear transcription factors. The easiest way to check for the specificity of the staining, is to check for proper localization of the signal inside the cell nucleus.
Tips 2: When performing multiple co-staining, use secondary antibodies associated to fluorochromes that do not overlap on the light spectrum. This largely depends on your microscope settings; check technical specifications of your microscope lasers/filters. As an example, we experienced some signal overlap when using Alexa Fluor 594 (Rhodammine) together with Alexa Fluor 647 (Far red/Cy5) on our Leica Microscope; better results (no signal overlap) were obtained by replacing Alexa Fluor 594 with Alexa Fluor 555 (Texas Red).
Recipes
4% PFA
To prepare 1 L of PFA 4%:
Dissolve 40 g of Paraformaldehyde powder (Sigma-Aldrich) in 1x PBS to reach 1 L
Heat while stirring under the chemical hood on a temperature of approximately 60 °C
When solution is transparent, let cool down then filter with Whatman paper to remove undissolved particles
Freeze PFA aliquots in 50 ml tubes at -20 °C
1x PBS
Add the following chemicals to 1 L of distilled water
8 g Sodium chloride
200 mg Potassium chloride
1.44 g Sodium phosphate dibasic heptahydrate
240 mg Potassium dihydrogen phosphate
Adjust pH to 7.4 with HCl
Autoclave it
Store it at RT
Unmasking Solution
Prepare 1 M Trisodium Citrate Dihydrate solution in distilled water (store at 4 °C)
Prepare 1 M citric acid solution in distilled water (store at 4 °C)
To prepare 1 L of unmasking solution, add 85 ml of sodium citrate 1 M to 800 ml of distilled water.
Measure pH while stirring
Adjust to pH 6 using 1 M citric acid solution
Bring the volume to 1 L with distilled water
Filter your solution
Store it at 4 °C
Agar 4%
For each brain, you need to use approximately 5 ml of Agar 4% in a 12-well plate, to prepare 100 ml of Agar 4%:
Dissolve 4 g of Select Agar ≥ 99.5% (GC) in 100 ml of PBS
Boil in the microwave to obtain a clear Agar solution with no aggregates
Incubate the agar solution in the water bath at 50 °C until mounting all the brains
10, 20 and 30% Sucrose
To prepare 50 ml of sucrose 10, 20 and 30%:
Add respectively 5 g, 10 g and 15 g of sucrose and reach 50 ml with PBS 1x
Vortex the solutions vigorously until fully dissolving the sucrose
Store the solutions at 4 °C; for long term storage, freeze them at -20 °C
Blocking Solution for cryo-sections
50 ml for immunostaining on thin cryo-sections:
Add 5 ml Goat serum or NBCS (in case one of your primary antibody is made in goat)
Add 150 μl of Triton
Reach 50 ml by adding PBS
Vortex until fully homogenizing the solution
Store it at 4 °C (for few days)
Antibody Solution for thin cryo-sections
50 ml for immunostaining on thin cryo-sections:
Add 3 ml Goat serum (or NBCS, in case one of your primary antibody is made in goat)
Add 150 μl of Triton
Reach 50 ml by adding PBS
Vortex until fully homogenizing the solution
Store it at 4 °C (for few days)
Blocking Solution for vibratome sections
50 ml for immunostaining on vibratome thick sections:
Add 5 ml Goat serum or NBCS (in case one of your primary antibodies is made in goat or sheep)
Add 150 μl of Triton 100
Add 1.5 g of BSA
Reach 50 ml by adding PBS
Vortex until fully homogenizing the solution
Store it at 4 °C (up to two weeks)
Antibody Solution for vibratome thick sections
50 ml for immunostaining on vibratome thick sections:
Add 3 ml Goat serum or NBCS (in case one of your primary antibody is made in goat)
Add 150 μl of Triton 100
Add 1.5 g of BSA
Reach 50 ml by adding PBS
Vortex until fully homogenizing the solution
Store it at 4 °C (up to two weeks)
Mounting medium
For 50 ml
Add 40 ml glycerol
Add 1 g N-propyl gallate
Reach 50 ml with distilled water
Boil in the microwave until homogenizing the solution
Add 5 μl of Hoechst 33342, Trihydrochloride
Store at 4 °C covered with aluminium foil to protect from light
Acknowledgments
This work was supported by the “Agence Nationale de la Recherche” under grant reference # ANR-13-BSV4-0011, by the “Fondation Recherche Médicale; Equipe FRM 2011” #DEQ20110421321 and by the “Fondation Jérôme Lejeune” under n° R13098AA to MS and by a postdoctoral fellowship from the city of Nice, France (“Aides Individuelles aux Jeunes Chercheurs”), to MB. KH was funded by a CNRS from Lebanon, AFM fellowship from France and an Alexander Von Humboldt fellowship from Germany. This protocol was elaborated on a previous work published in Harb et al. (2016).
Competing interests
The authors declare no competing interests.
Ethics
All animal procedures were conducted in strict accordance with relevant national and international guidelines and regulations (European Union rules; 2010/63/UE; and: Guide for the Care and Use of Laboratory Animals of the French Ministry of Research). All animal experiments were approved by the local ethical committee (CIEPAL registration numbers: NCE/2014-209 and NCE/2019-548). Standard approved housing conditions consisted in a 12 h light-dark cycle and housing with the recommended environmental enrichment (wooden cubes, cotton pad, igloo) with food and water ad libidum, three animals per cage max.
References
- Harb, K., Magrinelli, E., Nicolas, C. S., Lukianets, N., Frangeul, L., Pietri, M., Sun, T., Sandoz, G., Grammont, F., Jabaudon, D., Studer, M. and Alfano, C. (2016). Area-specific development of distinct projection neuron subclasses is regulated by postnatal epigenetic modifications. Elife 5: e09531.
Article Information
Copyright
Harb et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Harb, K., Bertacchi, M. and Studer, M. (2021). Optimized Immunostaining of Embryonic and Early Postnatal Mouse Brain Sections. Bio-protocol 11(1): e3868. DOI: 10.21769/BioProtoc.3868.
- Harb, K., Magrinelli, E., Nicolas, C. S., Lukianets, N., Frangeul, L., Pietri, M., Sun, T., Sandoz, G., Grammont, F., Jabaudon, D., Studer, M. and Alfano, C. (2016). Area-specific development of distinct projection neuron subclasses is regulated by postnatal epigenetic modifications. Elife 5: e09531.
Category
Neuroscience > Development > Immunofluorescence
Developmental Biology > Morphogenesis > Cell structure
Cell Biology > Cell imaging > Confocal microscopy
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








