- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Analysis of Isotopically-labeled Monogalactosyldiacylglycerol Molecular Species from [14C]Acetate-Labeled Tobacco Leaves
Published: Vol 10, Iss 24, Dec 20, 2020 DOI: 10.21769/BioProtoc.3864 Views: 3079
Reviewed by: Agnieszka ZienkiewiczKumiko OkazakiVenkatasalam Shanmugabalaji

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Step-by-step Protocol for Crossing and Marker-Assisted Breeding of Asian and African Rice Varieties
Yugander Arra [...] Wolf B. Frommer
Sep 20, 2024 2331 Views

Vegetative Propagation of Cannabis sativa and Resin Obtained From its Female Inflorescences
Sebastián D´Ippolito [...] Silvana L. Colman
Feb 20, 2025 1735 Views

A New Approach to Detect and Semi-quantify All Molecular Species and Classes of Anionic Phospholipids Simultaneously in Plant Samples
Manon Genva [...] Laetitia Fouillen
Apr 20, 2025 1718 Views
Abstract
Plant lipid metabolism is a dynamic network where synthesis of essential membrane lipids overlaps with synthesis of valuable storage lipids (e.g., vegetable oils). Monogalactosyldiacylglycerol (MGDG) is a key component of the chloroplast membrane system required for photosynthesis and is produced by multiple pathways within the lipid metabolic network. The bioengineering of plants to enhance oil production can alter lipid metabolism in unexpected ways which may not be apparent by static quantification of lipids, but changes to lipid metabolic flux can be traced with isotopic labeling commonly with [14C]acetate. Because lipid classes such as MGDG are composed of many different molecular species, full analysis of metabolically labeled lipids requires separation and quantification of the individually labeled molecular species which is traditionally performed by thin layer chromatography. Here we present a reverse phase HPLC method for the separation of MGDG molecular species from tobacco leaves in under 35 min. The quantification of each 14C-labeled molecular species was accomplished by an in-line flow radio detector. This method of analysis for [14C]Acetate labeled MGDG molecular species by radio-HPLC provides a rapid, high throughput, and reliable analytical approach to identify changes in MGDG metabolism due to bioengineering or other perturbations of metabolism.
Keywords: MonogalactosyldiacylglycerolBackground
Monogalactosyldiacylglycerol (MGDG) is a class of glycerolipid comprising of two fatty acid chains and a galactose moiety attached to the sn-1, -2, -3 positions of the glycerol backbone, respectively. MGDG is found as a major component in photosynthetic membranes of all plants. The complexity of MGDG lies in the chemical composition of acyl chains. Depending on the fatty acid carbon chain length and degree of unsaturation, each MGDG molecule could be further categorized into different molecular species, with different metabolic pathways leading to distinct molecular species compositions. In the research findings reported in the original research paper (Zhou et al., 2020), [14C]acetate metabolic labeling of wild-type and bioengineered oil producing leaves of tobacco was utilized to understand how oil accumulation in leaves affected fatty acid flux into essential membrane lipids. Quantification of specific 14C-labeled MGDG molecular species was utilized to estimate acyl flux through different branches of the lipid metabolic network. In this report we discuss a HPLC protocol for MGDG molecular species separation adapted from Yamauchi et al. (1982) and optimized for radio-HPLC quantification of 14C-MGDG molecular species with in-line flow liquid scintillation counting as reported in Zhou et al. (2020).
Materials and Reagents
HPLC Grade Methanol (Fisher Chemical, catalog number: A452-4 )
Water, CHROMASOLVTM, for HPLC (Honeywell Riedel-de HaenTM, Fisher Chemical, catalog number: 60-048-247 )
FlowLogic U Scintillation Cocktail (LabLogic, catalog number: SG-BXX-05 )
Equipment
HPLC Equipment: Agilent 1260 Infinity II High Performance Liquid Chromatography System
Flow Radio Detector: LabLogic β-Ram 6
Thermo Scientific Accucore C18 Column; 3 x 150 mm, 2.5 μm spherical solid core particles. (Thermo Fisher, catalog number: 17126-153030 )
Accucore C18 Guard Column; 3 x 10 mm, 2.6 μm spherical solid core particles (Thermo Fisher, catalog number: 17126-013005 )
Software
LabLogic Laura version 6.0.1.40
GraphPad Prism version 8.1.2 (332)
Procedure
Sample preparation
Isolate MGDG from the leaf lipid extract via normal phase HPLC method using a time based fraction collection method, as disclosed in Zhou et al. (2020), or by thin layer chromatography (Karki et al., 2019).
Dry collected MGDG fractions under a gentle N2 stream.
Dissolve the isolated MGDG into a small volume (e.g., 25 µl) HPLC grade methanol for analysis via Radio-HPLC. The volume of injection solvent for the sample analysis illustrated in Figure 1 is 10 μl and the total radioactivity per injection is approximately 17,500 Disintegrations Per Minute (DPM). For the analysis of MGDG molecular species, it is ideal to have at least 5,000 DPM of total MGDG radioactivity per injection of a minimal volume of solvent into the HPLC.
Transfer the MGDG in methanol to an autosampler vial with a small volume insert for HPLC analysis. Spring bottom inserts are ideal for small volumes (10-100 µl) where nearly the entire sample may be injected.
Reverse phase Radio HPLC-14C MGDG molecular species analysis
Power on the HPLC modules; pump, autosampler, multi column compartment, Ultra Violet Diode-Array Detector (UV-DAD), and flow radio detector.
The HPLC parameters and the online liquid scintillation counter are both controlled by the LabLogic Laura software.
Set up the sequence in Laura with a blank methanol as the first sample in the sequence. Allow the column to equilibrate for 20 min prior to the start of analysis. Since the method uses isocratic conditions, column equilibration is not necessary between each sample.
Once the sequence is saved, the method for the analysis will be loaded. The method conditions for the HPLC analysis are as described below:
Quaternary Pump: Solvent A: Methanol and Solvent B: Water. The method uses Isocratic elution with mobile phase–95%A/5%B. The flow is set at 0.35 ml/min and the run time is 35 min.
Autosampler: Autosampler temperature is set at 20 °C. Sample Injection solvent: Methanol. Set the injector settings in the method to as follows: Needle Draw Speed–200 μl/min, Needle eject Speed–400 μl/min, Wait Time after draw–1.2 s.
Multi Column Compartment: The temperature of the column compartment is set at 35 °C. The mobile phase is allowed to pass through a Quick-Connect Heat Exchanger prior to entering the column. A short 10 mm guard column containing same material as that of the stationary phase is installed immediately prior to the analytical column.
UV-DAD: Direct the flow of eluate from the column to UV-DAD. UV absorption at 210 nm will provide retention times of the molecular species containing unsaturated acyl chains with significant mass abundance in the sample as illustrated in Figure 1A and the original research paper (Zhou et al., 2020), Supplementary Figure S4A.
β-Ram 6 Radio Detector: Direct the eluate from UV-DAD to the radio detector. Set the type of analysis to Non-Stop Radio HPLC. The radio detector is equipped with a 500 μl adjustable volume flow cell that facilitates the radio counting process. Set the flow cell volume to 300 μl. Prior to entering the flow cell, the eluate from the UV-DAD is mixed with the scintillation cocktail whose flow is controlled by a scintillant pump. Set the scintillant flow to 1.05 ml/min (a 1:3 for eluate:scintillation cocktail ratio). The resulting residence time in the flow cell for these flow settings is 12.9 s. The recommended residence time for flow counting is above 5 s. The eluate mixed with scintillant from the radio detector is directed to the properly labeled radioactive HPLC waste container secured in a secondary waste container. In the method, adjust the detector settings of Radio 1 channel as follows: Measurement units–CPM, Dwell Time–1 s, Shift Time–0 s, Lower Limit–200, Upper Limit–9000.
After the HPLC data acquisition is completed, proceed to data analysis.
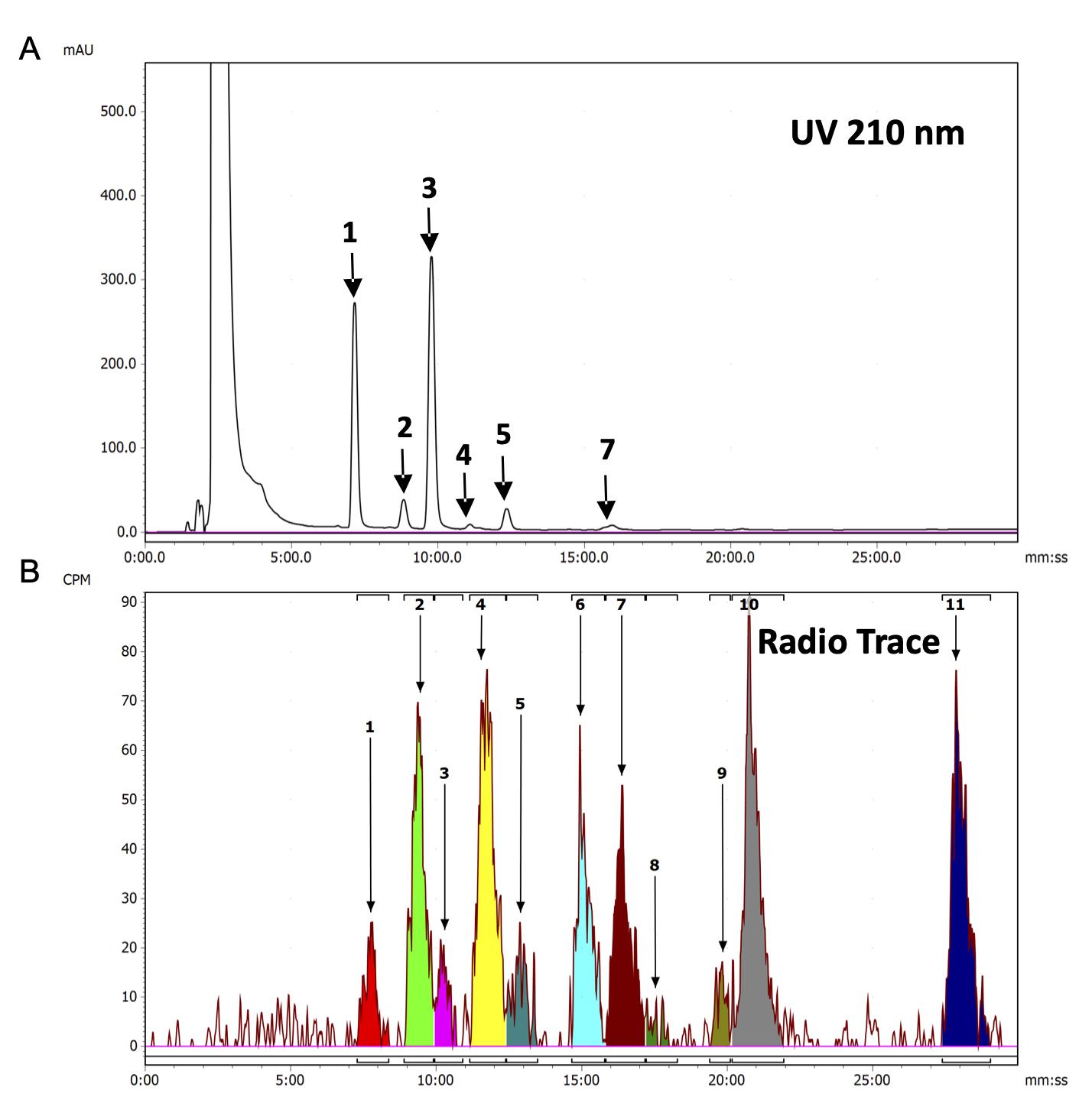
Figure 1. HPLC Analysis-MGDG molecular species from wild type tobacco leaves continuously labeled with [14C]acetate for 2 h. A. UV chromatogram at 210 nm. B. radioactivity in counts per minute (CPM). Identities of individual molecular species from B are as follows: 1-18:3/16:3, 2-18:3/16:2>18:2/16:3, 3-18:3/18:3, 4-18:2/16:2>18:3/16:1, 5-18:3/18:2, 6-18:2/16:1, 7-18:3/16:0>18:2:18:2, 8-18:3/18:1, 9-18:2/18:1, 10-18:2/16:0, 11-18:1/16:0. Figure reproduced from Zhou, X. R., Bhandari, S., Johnson, B. S., Kotapati, H. K., Allen, D. K., Vanhercke, T. and Bates, P. D. (2020). Reorganization of acyl flux through the lipid metabolic network in oil-accumulating tobacco leaves.Plant Physiol 182(2): 739-755. www.plantphysiol.org. “Copyright American Society of Plant Biologists”.
Data analysis
Analyze radio HPLC data via LabLogic Laura, and a sample chromatogram after processing is illustrated in Figure 1B and original research paper (Zhou et al., 2020) Supplementary Figure S4B.
Load the first MGDG sample in the sequence and select all the significant peaks in the order of elution.
Each peak will be designated as a region and numbers are assigned in the same elution order.
Multiple samples in the sequence via batch analysis where all the regions in a chromatogram will be selected based on the first chromatogram. It is ideal to go through each chromatogram and make sure all significant peaks are selected. This also helps to identify any peak drifts due to errors in the analysis.
The data table for the processed chromatogram provides the following information: retention time for each peak, area under the peak in CPM, % total and % region of interest (%ROI).
Export the data table to Microsoft Excel by simple copy paste function.
%Total for each identified peak in a chromatogram is used to determine the relative labeling of each molecular species in the MGDG sample. See notes for additional information on the MGDG molecular species analysis.
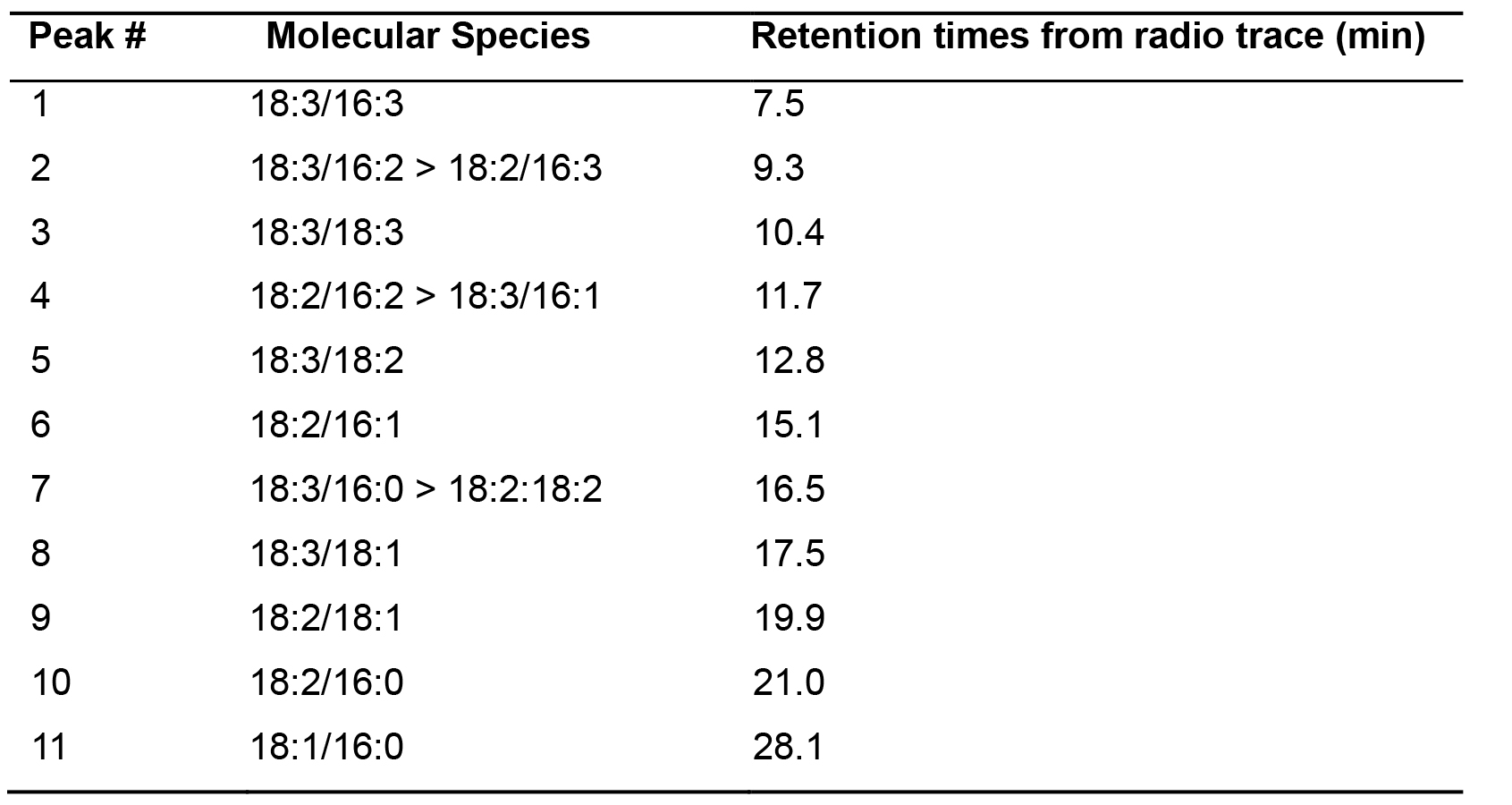
Identify the individual molecular species based on the retention times of the peaks as shown in Table 1. MGDG species are identified based on the carbon chain length and unsaturation number in the acyl chains. The data of relative %14C label between the molecular species plotted in Prism and is illustrated in original research paper (Zhou et al., 2020) Supplementary Figures S5 and S6.
Table 1. Retention times of identified 14C MGDG molecular species in WT tobacco
Notes
For the molecular species identification, it is always ideal to let the analysis go for longer than our reported method run time. This is to make sure there are no other species besides the reported ones.
If the retention times are off, then the identity of each peak needs to be determined. One procedure is to collect each 14C peak, convert to fatty acid methyl esters, and identify the 14C fatty acids via argentation TLC and phosphorimaging analysis as reported in Bates et al. (2009), this procedure was used for original identification of each labeled MGDG molecular species in Zhou et al. (2020).
Further method optimization by adjusting method parameters such as using different stationary phase (e.g., a RP column with longer carbon chain) or same column with different dimensions (e.g., increased length) might help with the separation of co-eluting molecular species of MGDG.
Acknowledgments
The research is supported by U.S. Department of Agriculture National Institute of Food and Agriculture (grant nos. 2017-67013- 26156, 2017-67013-29481, and Hatch umbrella project no. 1015621), and the National Science Foundation (grant no. 1930559).
Competing interests
The authors report no competing interests.
References
- Bates, P. D., Durrett, T. P., Ohlrogge, J. B. and Pollard, M. (2009). Analysis of acyl fluxes through multiple pathways of triacylglycerol synthesis in developing soybean embryos. Plant Physiology 150(1): 55-72.
- Karki, N., Johnson, B. S. and Bates, P. D. (2019). Metabolically distinct pools of phosphatidylcholine are involved in trafficking of fatty acids out of and into the chloroplast for membrane production. Plant Cell 31(11): 2768-2788.
- Yamauchi, R., Kojima, M., Isogai, M., Kato, K. and Ueno, Y. (1982). Separation and purification of molecular species of galactolipids by high performance liquid chromatography. Agricultural and Biological Chemistry 46(11): 2847-2849.
- Zhou, X. R., Bhandari, S., Johnson, B. S., Kotapati, H. K., Allen, D. K., Vanhercke, T. and Bates, P. D. (2020). Reorganization of acyl flux through the lipid metabolic network in oil-accumulating tobacco leaves. Plant Physiol 182(2): 739-755.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Kotapati, H. K. and Bates, P. (2020). Analysis of Isotopically-labeled Monogalactosyldiacylglycerol Molecular Species from [14C]Acetate-Labeled Tobacco Leaves. Bio-protocol 10(24): e3864. DOI: 10.21769/BioProtoc.3864.
Category
Plant Science > Plant biochemistry > Metabolite
Biochemistry > Other compound
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










