- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Differentiation of Human Induced Pluripotent Stem Cells (hiPSCs) into Osteoclasts
Published: Vol 10, Iss 24, Dec 20, 2020 DOI: 10.21769/BioProtoc.3854 Views: 5317
Reviewed by: Giusy TornilloAnthony FlamierSébastien Gillotin

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Human iPSC-Derived Neuron and Oligodendrocyte Co-culture as a Small-Molecule Screening Assay for Myelination
Stefanie Elke Chie [...] Maria Consolata Miletta
May 5, 2025 3412 Views

Isolation and Culture of Ferret Airway Stem Cells
Ziying Yan [...] Feng Yuan
Jul 20, 2025 2411 Views

Optimization of Adipogenic Differentiation Protocol for Murine and Human Cell Culture Models
Junwan Fan [...] Wenyan He
Jan 20, 2026 211 Views
Abstract
Defects in bone resorption by osteoclasts result in numerous rare genetic bone disorders as well as in some common diseases such as osteoporosis or osteopetrosis. The use of hiPSC-differentiated osteoclasts opens new avenues in this research field by providing an unlimited cell source and overcoming obstacles such as unavailability of human specimens and suitable animal models. Generation of hiPSCs is well established but efficient differentiation of hiPSCs into osteoclasts has been challenging. Published hiPSC-osteoclast differentiation protocols use a hiPSC-OP9 co-culture system or hiPSC-derived embryoid bodies (EBs) with multiple cytokines. Our three-stage protocol consists of 1) EB mesoderm differentiation, 2) expansion of myelomonocytic cells and 3) maturation of hiPSC-osteoclasts. We generate uniformly-sized EBs by culturing Accutase-dissociated hiPSCs on Nunclon Sphera microplates and promote EB mesoderm differentiation in a cytokine cocktail for 4 days. For Stage 2, EBs are transferred to gelatin-coated plates and cultured with hM-CSF and hIL-3 to expand the myelomonocytic population. By supplementing with vitamin D, hTGFβ, hM-CSF and hRANKL, cells collected at the end of Stage 2 are differentiated into mature osteoclasts (Stage 3). Compared to other techniques, our protocol does not require a co-culture system; induces EBs into mesoderm differentiation in a homogenous manner; uses less cytokines for differentiation; requires only a short time for osteoclast maturation and produces sufficient numbers of osteoclasts for subsequent molecular analyses.
Graphic abstract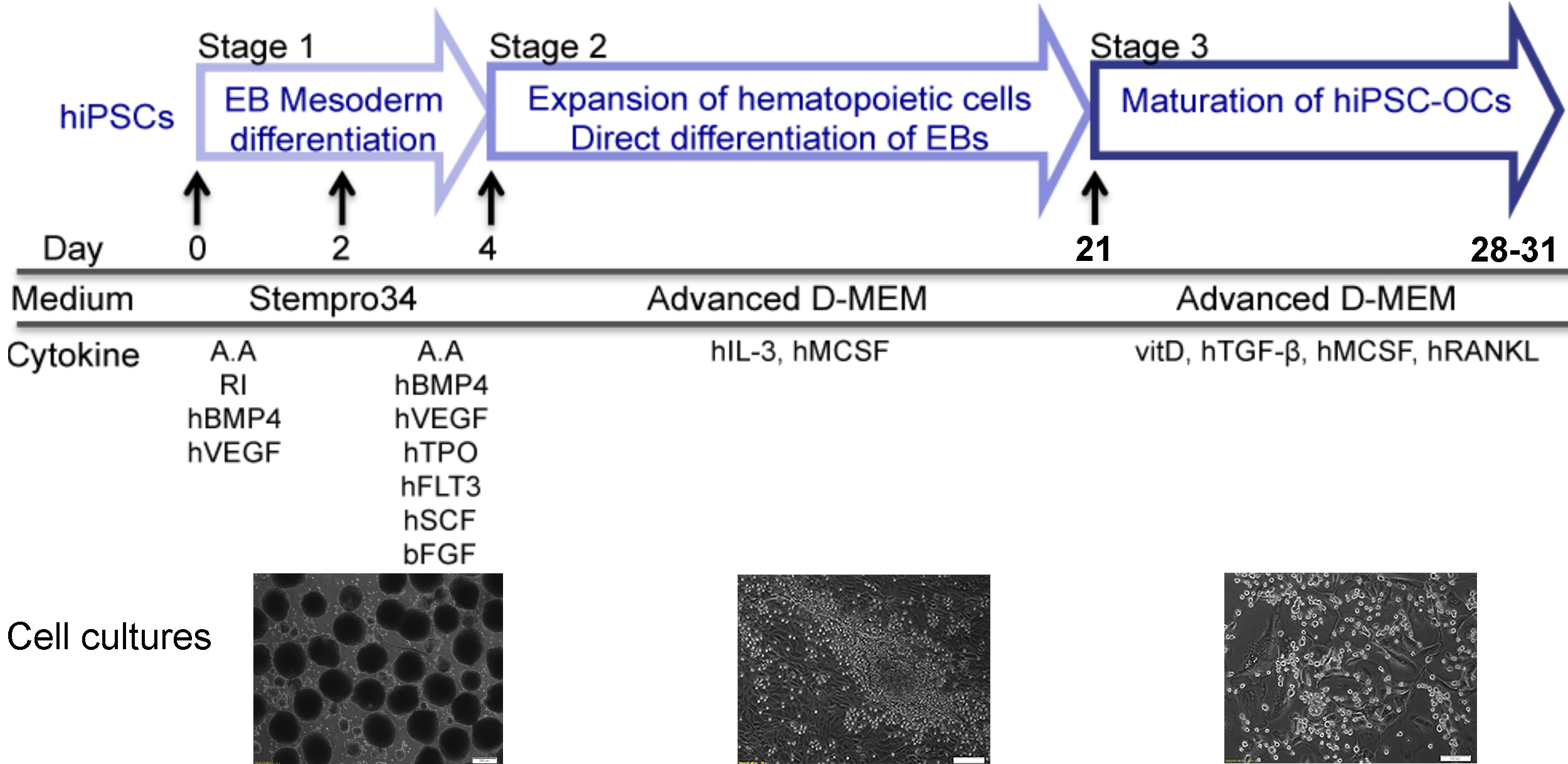
Background
The technology for generating patient-specific hiPSCs, which theoretically can be differentiated into any cell type, opens new avenues for medical research in disease modeling, including bone disorders (Deyle et al., 2012; Quarto et al., 2012a and 2012b; Cherry and Daley, 2013; Ding et al., 2013; Matsumoto et al., 2013; Chen et al., 2017). Research focusing on rare genetic bone disorders not only has the potential to find treatment for patients, but also contributes to a better understanding of skeletal biology. Many skeletal diseases involve dysfunctional osteoclasts, osteoblasts and/or osteocytes (Chen, 2014). Osteoclasts are bone resorbing cells and osteoblasts are bone forming cells. Osteocytes are derived from mature osteoblasts and are entrapped in the bone matrix that they produce (Bonewald, 2011). Bone marrow stromal cells (mesenchymal osteoblast-like cells) can be cultured from bone marrow, bone biopsies or bone excised during surgical procedures. Mesenchymal cells are proliferative and can be differentiated and passaged. The myelomonocytic population in bone marrow and peripheral blood can be differentiated into osteoclasts by culturing with human macrophage stimulating factor (M-CSF) and receptor activator of nuclear factor-kB ligand (RANKL) (Chen et al., 2011). However, once differentiated, these cells are terminally differentiated and can be used for experiments only once.
While reliable and consistent methods for reprogramming somatic cells into hiPSCs are well-established (Takahashi et al., 2007; Yu et al., 2007; Park et al., 2008; Stadtfeld et al., 2008; Sommer et al., 2009; Yu et al., 2009; Warren et al., 2010), differentiation of hiPSCs into bone cells is still more challenging. Several studies describe protocols for hiPSC differentiation into osteoblasts (Kanke et al., 2014; Kuhn et al., 2014; Ochiai-Shino et al., 2014; Kang et al., 2016), but there are relatively few protocols described for hiPSC differentiation into osteoclasts (Choi et al., 2009; Grigoriadis et al., 2010; Jeon et al., 2016). Choi et al. and Grigoriadis et al. differentiated hiPSC into osteoclasts via a hiPSC-OP9 co-culture system and through EB formation steps, respectively (Choi et al., 2009; Grigoriadis et al., 2010). A critical step for co-culture systems is to match cell densities of undifferentiated hiPSCs and OP9 cells, which can otherwise contribute to inconsistent outcomes. Generating EBs by conventional methods in cell culture dishes may cause variable EB sizes and can affect the differentiation efficiency. In addition, the use of complex cytokine cocktails is less economical. Jeon et al. (2016) reported a hiPSC-osteoblast and hiPSC-osteoclast co-culture system, which is less useful for investigating cell-autonomous osteoblast and osteoclast phenotypes in disease models.
Our protocol described here generates uniformly-sized EBs by dissociating hiPSC colonies into single cells and plating a fixed number of cells onto Nunclon Sphera microplates. EBs are stimulated to enter the mesoderm lineage in the first 4 days of differentiation and are transferred to gelatin-coated plates to expand the myelomonocytic population, which contains the osteoclast progenitors. Differentiation into osteoclasts is achieved by culturing these cells in the presence of vitamin D, hTGFβ, hM-CSF and hRANKL. The resulting mature and functional osteoclasts are tartrate-resistant acid phosphatase (TRAP)-positive multinucleated cells and are able to resorb bone.
Materials and Reagents
Materials
Pipet tips (200 μl, 1 ml)
Cell strainer 70 μm (Corning, catalog number: 352350 )
Nunclon Sphera 96-well plates, U-bottom (Thermo Fisher Scientific, catalog number: 174929 )
Tissue culture 96-well plate, polystyrene (Greiner Bio-One, catalog number: 655180 )
Tissue culture 6-well plate, polystyrene (BD, Falcon, catalog number: 08-772-1B )
Serological pipet, 5 ml (Fisher Scientific, catalog number: 13-678-11D )
Serological pipet, 10 ml (Fisher Scientific, catalog number: 13-678-11E )
15 ml polypropylene conical tubes (Thermo Scientific, catalog number: 339650 )
50 ml polypropylene conical tubes (Thermo Scientific, catalog number: 339652 )
Nalgene filter (0.2 μm PES membrane) 150 ml (Thermo Scientific, catalog number: 5650020 )
Nalgene filter (0.2 μm PES membrane) 250 ml (Thermo Scientific, catalog number: 5680020 )
Microcentrifuge tube (Fisher Scientific, catalog number: 05-408-129 )
hiPSCs generated from healthy donors (Chen et al., 2013 and 2017)
Matrigel (Corning, catalog number: 354234 )
PeproGrow embryonic stem cell (hESC) medium (Peprotech, catalog number: BM-hESC )
Accutase (Millipore Sigma, catalog number: SCR005 )
Phosphate buffered saline, no calcium, no magnesium (Thermo Fisher Scientific, catalog number: 10010049 )
DMEM/F12 medium (Thermo Fisher Scientific, catalog number: 11330032 )
Stempro-34 medium (Thermo Fisher Scientific, catalog number: 10639011 )
L-Glutamine (200 mM) (Thermo Fisher Scientific, catalog number: 25030081 )
1-Thioglycerol (MTG) (Sigma-Aldrich, catalog number: M6145 )
Ascorbic acid (Sigma-Aldrich, catalog number: A4034 )
MEM Non-essential amino acids (NEAA 100x) (Thermo Fisher Scientific, catalog number: 11140-050 )
Rock inhibitor Y-27632 (Selleck Chemicals, catalog number: S1049 )
Recombinant human BMP4 (Peprotech, catalog number: 120-05ET )
Recombinant human VEGF (Peprotech, catalog number: 100-20 )
Recombinant human TPO (Peprotech, catalog number: 300-18 )
Recombinant human Flt3-ligand (Peprotech, catalog number: 300-19 )
Recombinant human IL-3 (Peprotech, catalog number: 200-03 )
Recombinant human MCSF (Peprotech, catalog number: 300-25 )
Recombinant human RANKL (Peprotech, catalog number: 310-01 )
Recombinant human TGF-β1 (Peprotech, catalog number: 100-21C )
Recombinant human SCF (Peprotech, catalog number: 300-07 )
Recombinant human FGF basic (Peprotech, catalog number: 100-18B )
Gelatin 0.1% solution (EMD Millipore, catalog number: ES-006-B )
Advanced DMEM medium (Thermo Fisher Scientific, catalog number: 12491023 )
Alpha MEM medium (Thermo Fisher Scientific, catalog number: 12571063 )
Fetal bovine serum (Gibco, catalog number: 10437020 )
1α, 25-dihydroxyvitamin D3 (Sigma Aldrich, catalog number: D1530 )
CD14 antibody (FITC-conjugated; Biolegend, catalog number: 325604 )
CD43 antibody (APC-conjugated, Miltenyi Biotec, catalog number: 560198 )
CD45 antibody (APC-conjugated, Biolegend, catalog number: 304012 )
EDTA 0.5 M, pH 8.0 (Thermo Fisher Scientific, catalog number: 15575-038 )
TRIzol (Thermo Fisher Scientific, catalog number: 10296028 )
Direct-zol RNA (Zymo Research, catalog number: R2052 )
DNase I (Invitrogen, catalog number: 18068015 )
Superscript II reverse transcriptase (Invitrogen, catalog number: 18064071 )
SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, catalog number: 1725271 )
Glutaraldehyde 25% solution (Alfa Aesar, catalog number: A17876 )
Acid phosphatase, leukocyte (TRAP) kit (Sigma-Aldrich, catalog number: 387A )
Primers for detection of mesoderm differentiation and osteoclast maturation by qPCR (Table 1)
Matrigel coating solution (see Recipes)
hiPSC culture medium (see Recipes)
EB mesoderm differentiation medium-1 (see Recipes)
EB mesoderm differentiation medium-2 (see Recipes)
Myelomonocytic expansion medium (see Recipes)
FACS buffer (see Recipes)
Osteoclast differentiation medium (see Recipes)
TRAP staining solution (see Recipes)
Reconstitution of hBMP4 (see Recipes)
Reconstitution of other cytokines (see Recipes)
Table 1. Sequences of qPCR primers to detect marker gene expression for mesoderm differentiation and osteoclastogenesis

Equipment
Pipettes
Water bath
AirClean 600 PCR workstation (AirClean Systems, model: AC648A )
Stereomicroscope (Carl Zeiss, model: Stemi 508 )
Cell culture incubator (Thermo Fisher Scientific, model: Heracell 240i, catalog number: 51026332)
Eppendorf refrigerated centrifuge (Eppendorf, model: 5810R)
CFX96 Touch Real-time PCR Detection System (Bio-Rad, catalog number: 1855196)
MACSQuant Analyzer 10 (Miltenyi Biotech, catalog number: 130-096-343)
Upright microscope (Carl Zeiss, model: Axio Imager.D2m)
Tabletop scanning electron microscope (Hitachi, model: TM1000)
Software
CFX manager software (Bio-Rad, 18450000)
FlowJoTM software (BD Bioscience, https://www.flowjo.com/)
GraphPad Prism (GraphPad, https://www.graphpad.com/)
Fiji ImageJ (NIH, image.nih.gov)
ZEN microscope software (Zeiss, https://www.zeiss.com/microscopy/us/products/microscope-software/zen-lite.html)
Procedure
Maintenance of hiPSCs cultures
Prepare Matrigel-coated 6-well plate by adding 1 ml of coating solution (see recipe Table 4) to each well and leave the plate at room temperature for 1 h before use.
Maintain hiPSCs in PeproGrow hESC medium (see recipe Table 5) and change medium every other day.
Passage undifferentiated hiPSCs every 4-5 days.
Aspirate old medium and add 2 ml fresh PeproGrow hESC medium to each well.
Scrape undifferentiated hiPSC colonies into small pieces using 200 μl pipet tips under a stereomicroscope within an AirClean PCR Workstation. Depending on the size, a hiPSC colony can be broken up into 15-40 pieces.
Aspirate the Matrigel coating solution from the freshly prepared plates and transfer the lifted hiPSC fragments to new Matrigel-coated plates.
Mesoderm differentiation of hiPSC-derived embryoid bodies (EBs) (Stage 1)
Examine hiPSC cultures and ensure hiPSCs are undifferentiated. Lift and remove the differentiated hiPSCs, if any, using a 200 μl pipet tip under a microscope in the PCR Workstation.
Prewarm an aliquot of Accutase solution in 37 °C water bath for 10 min.
Aspirate hiPSC culture medium. Wash hiPSCs with pre-warmed PBS (2 ml per well) twice. Add 1 ml of Accutase solution to each well and incubate culture plates in 37 °C incubator for 10 min.
Add 2 ml of DMEM/F12 medium to each well. Pipette the cell suspension with 10 ml pipette gently up and down to dislodge the hiPSCs. Transfer the contents to 50 mL conical tubes.
Centrifuge the cells at 340 x g for 7 min at 4 °C. Resuspend cells in EB basal medium-1 (see recipe Table 6). Filter the cells using a 70 μm strainer. Count cells.
Plate 15,000 cells to each well in 150 μl EB mesoderm differentiation medium-1 (see recipe Table 6) on a Nunclon Sphera 96-well plate. Keep the plates at 37 °C in a 5% CO2 and 5% O2 incubator for 2 days. Each well forms one EB.
Change half medium by pipetting out 75 μl of EB mesoderm differentiation medium-1 and adding 75 μl of EB mesoderm differentiation medium-2 (see recipe Table 7).
Keep the plates at 37 °C in a 5% CO2 and 5% O2 incubator for 2 more days.
Analyze mesoderm differentiation by examining the expression levels of mesoderm marker genes in EBs cultured for 1, 2, 3, and 4 days by qPCR (Figure 1).
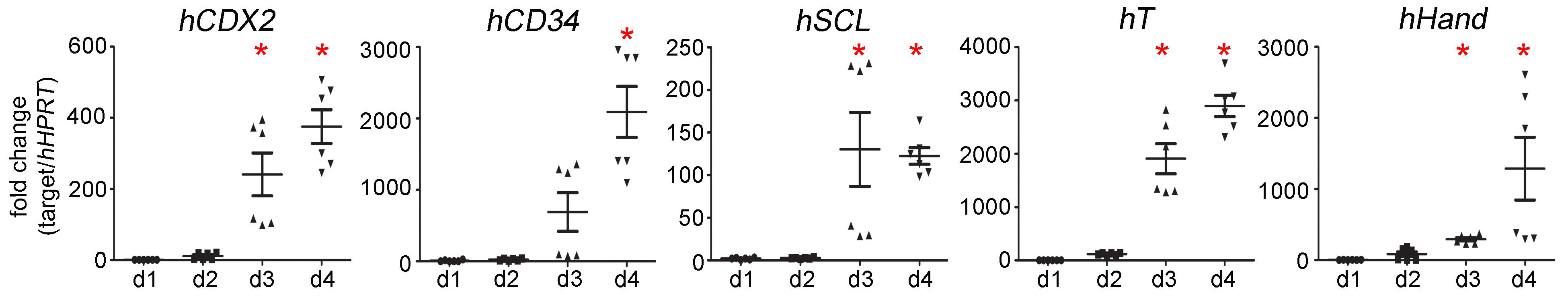
Figure 1. Expression levels of mesoderm marker genes hCDX2, hCD34, hSCL, hT, hHand, in EBs cultured for 1, 2, 3, and 4 days (d1, d2, d3 and d4). * P < 0.05 by one-way ANOVA compared to day 1 samples. Data was relative quantification and presented as mean ± SEM.
Expansion of myelomonocytic population released from EBs (Stage 2)
Prepare gelatin-coated plate by adding 1 ml of 0.1% gelatin solution to each well of a 6-well plate and leave at room temperature for 1 h before use.
Collect EBs from Nunclon Sphera 96-well plate by suctioning out the medium with EBs using a 1 ml pipet tip and transfer these EBs to a 50 ml conical tube.
Let EBs sink down by gravity for 3-5 min.
Aspirate medium with a glass Pasteur pipette carefully without removing EBs in the bottom.
Resuspend EBs in myelomonocytic expansion medium (see recipe Table 8) according to numbers of EBs. Seed 40-50 EBs in one well of 6-well plate with 4 ml of myelomonocytic expansion medium. For example: EBs collected from a full 96-well plate can be resuspended in 8 ml medium and plated in 2 wells of a gelatin-coated 6-well plate.
Keep the plates at 37 °C in a 5% CO2 and 5% O2 incubator.
EBs attach to the plates. Some cells are released from EBs. The floating non-adherent cells are the myelomonocytic population.
Change expansion medium after 4 days with the same medium but increase hMCSF concentration from 50 ng/ml to 100 ng/ml.
Continue culture for another 13 days (medium changes at days 4, 8, 12, and 16).
Floating cells released from EBs at days 10, 13, 17, 21 are collected and stained with primary antibodies against CD14 (1:20), CD43 (1:50), and CD45 (1:50) in FACS buffer (see recipe Table 9) for 30 min on ice. Protect from light. Table 2 shows FACS analysis data.
Table 2. CD14, CD43, and CD45 expression in cells collected at day 10, 13, 17, and 21 days in Stage 2 analyzed by flow cytometry
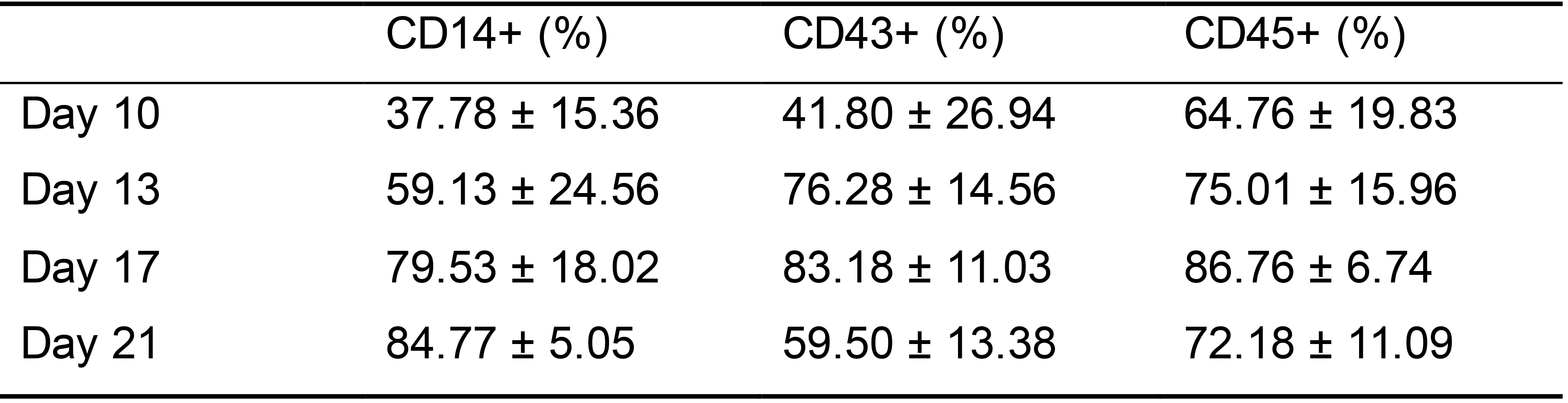
Data presented: mean ± SD. Data collected from 3 hiPSC lines. Each line had 3-4 technical repeats.
Promotion of osteoclast maturation (Stage 3)
Collect floating cells (17 days of Stage 2) to a 15 ml or 50 ml conical tube. Typically, 0.2-0.45 million cells can be expected from EBs grown on one well of a gelatin-coated 6-well plate.
Centrifuge the cells at 340 x g for 7 min at 4 °C. Count cells.
Plate cells at a density of 10,000 cells per well in a 96-well plate and on bone chips.
Culture osteoclast progenitors in osteoclast differentiation medium (see recipe Table 10). Change medium every 2-3 days.
Multinucleated mature osteoclasts form in 7-10 days in osteoclast differentiation medium (Figure 2A).
Fix hiPSC-OCs at day 10-12 for TRAP staining (Figures 2B-2C).
Remove culture medium. Wash gently with PBS once.
Fix cells in 2.5% glutaraldehyde (150 μl per well) at room temperature for 10 min.
Prepare TRAP staining solution (see recipe Table 11).
Add 100 μl of TRAP solution in each well of 96-well plate.
Incubate culture plate at 37 °C for 45 min to 1 h.
Remove the TRAP solution. Rinse plates with distilled water.
Dry the plate before taking images.
Fix TRAP stained cells on bone chips at day 14. Take images of resorption pits with TM1000 tabletop scanning electron microscope (Figure 2D).
Confirm differentiation status of osteoclasts by examining expression levels of osteoclast marker genes by qPCR (Table 3).
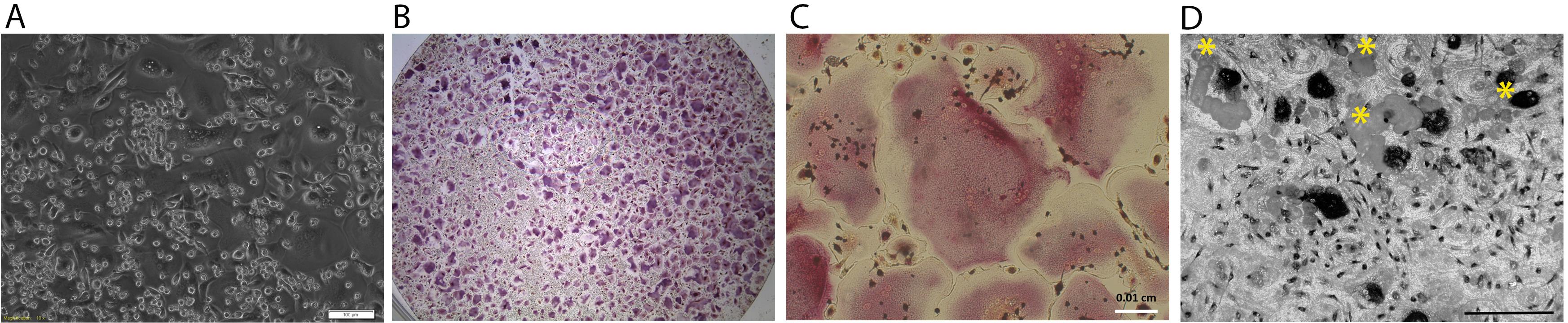
Figure 2. hiPSC-OC cultures. A. Bright field of hiPSC-OC culture at day 10 in OC differentiation medium. Scale bar = 100 μm; B. TRAP staining of one well of 96-well plate. Purplish large sized cells are TRAP positive multinucleated osteoclasts; C. High magnification of TRAP-positve multinucleated cells. Scale bar = 100 μm; D. Resorption pits (indicated by yellow asterisks) on bone chip. Large black spots are TRAP positive osteoclasts. Scale bar = 200 μm.Table 3. qPCR data of expression levels of osteoclast marker genes in hiPSC-osteoclasts

Data presented: mean ± SD. Data collected from 3 hiPSC lines. Each line had 3 technical repeats. *P < 0.05 by Student’s t-test. OC progenitors are the floating cells collected at day 17 of stage 2 and cultured in osteoclast differentiation medium without 1α,25-dihydroxyvitamin D3, hTGFβ-1, and hRANKL.
Data analysis
qPCR data were analyzed using CFX manager software. The significant difference was determined by Student’s t-test or one-way ANOVA followed by Tukey’s post-hoc test using GraphPad Prism software.
Flow cytometry data were analyzed using FlowJo software.
Notes
Differentiation efficiency of stem cells can vary based on the size of EBs (Moon et al., 2014, Ng et al., 2015). The step of forming uniform-sized EBs is important to achieve consistant efficiency of hiPSC-OCs differentiation.
We determined the duration of Stage 2 as 17 days based on flow cytometry data for the expression levels of CD14, CD43, and CD45, which are expressed highest during these 17 days with little variability in comparison to other time points.
The non-specific/spontaneous differentiation of hiPSCs can be identified by their heterogeneous morphology within a hiPSC colony. Differentiation usually starts in the center of a large hiPSC colony and appears as a dark field under a brightfield microscope.
Isogenic hiPSCs have identical genetic background except for a genetic variant that has been introduced. Thus isogenic hiPSCs are the best controls to use when researchers aim to determine the impact of a specific disease mutation.
Recipes
Matrigel coating solution (Table 4)
Thaw frozen Matrigel in refrigerator overnight and aliquot Matrigel in ice-cold microcentrifuge tubes (60 μl for one 6-well plate)
Thaw Matrigel aliquots on ice and transfer Matrigel to ice-cold DMEM/F12 medium. For one 6-well plate, add 60 μl Matrigel to 6 ml DMEM/F12 medium
Table 4. Matrigel coating solution

hiPSC culture medium (Table 5)
Centrifuge the vial with lyophilized Peprotech growth factor prior to opening and reconstitute with sterile water. Use 500 μl water for each 500 ml PeproGrow hESC basal medium kit
Add the reconstituted growth factor to the basal medium aseptically and mix well by swirling or pipetting. Store at 2-8 °C and use within 2 weeks
Table 5. hiPSC medium

EB mesoderm differentiation medium-1 (Table 6)
Prepare 100 ml EB mesoderm differentiation medium as shown in Table 6
Sterilize EB basal medium (including Stempro-34, L-glutamine, MTG) by filtering through a 0.2 μm PES membrane filter and keep at 2-8 °C for up to 1 month
Add hBMP4, hVEGF, Y-27632, and ascorbic acid in EB basal medium just before use
Table 6. EB mesoderm differentiation medium-1

EB mesoderm differentiation medium-2 (Table 7)
Prepare 100 ml EB mesoderm differentiation medium as shown in Table 7
Sterilize EB basal medium (including Stempro-34, L-glutamine, MTG) by filtering through a 0.2 μm PES membrane filter and keep at 2-8 °C for up to 1 month
Add 2x concentration of hBMP4, hVEGF, Y-27632, and ascorbic acid in EB basal medium just before use
Table 7. EB mesoderm differentiation medium-2
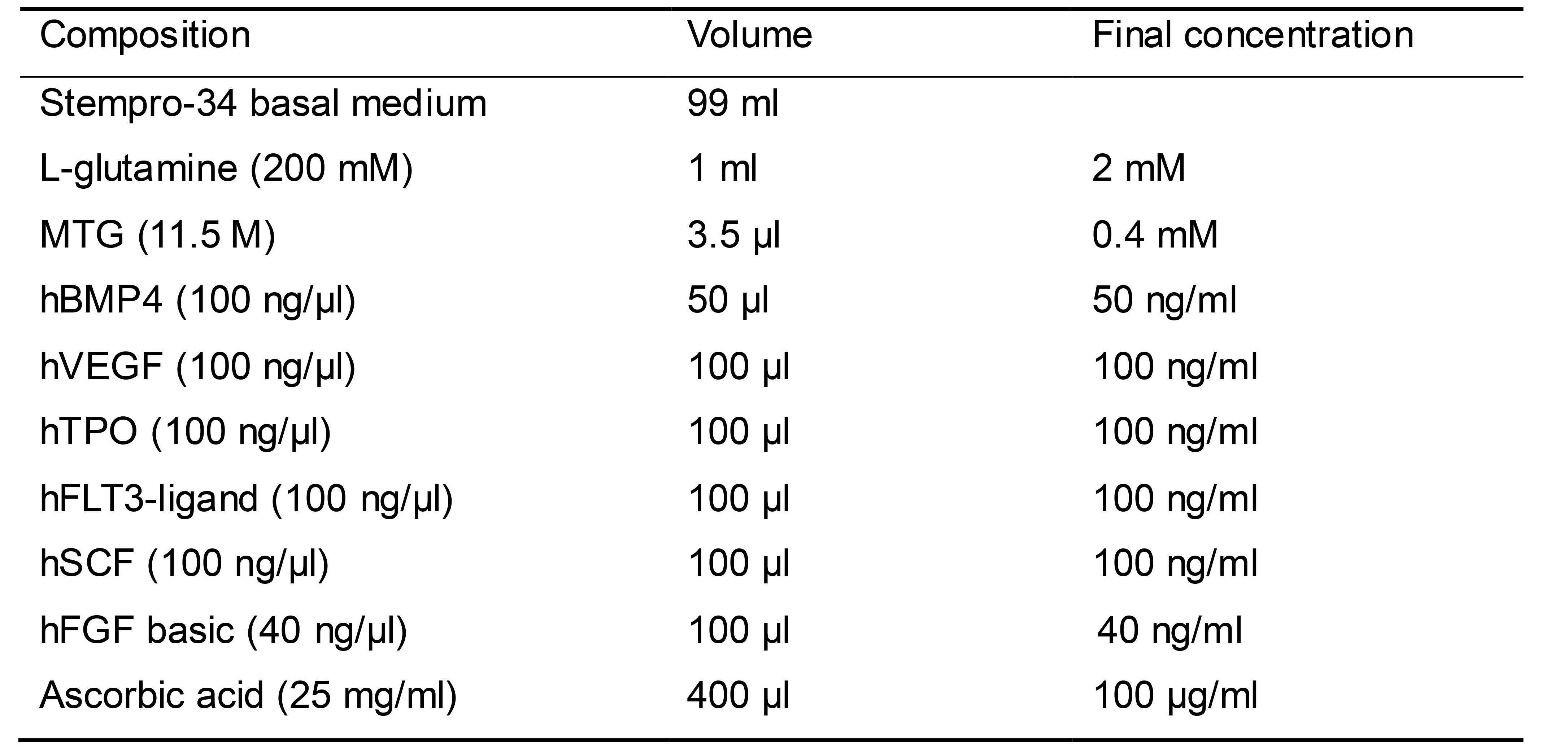
Myelomonocytic expansion medium (Table 8)
Prepare 100 ml myelomonocytic expansion medium as shown in Table 8
Sterilize myelomonocytic expansion medium (including advanced DMEM, FBS, L-glutamine, β-mercaptoethanol) by filtering through a 0.2 μm PES membrane filter and keep at 2-8 °C for up to 1 month
Add hIL-3 and hM-CSF aseptically only before use. Increase hM-CSF concentration from 50 to 100 ng/ml starting from day 4 medium change
Table 8. Myelomonocytic expansion medium

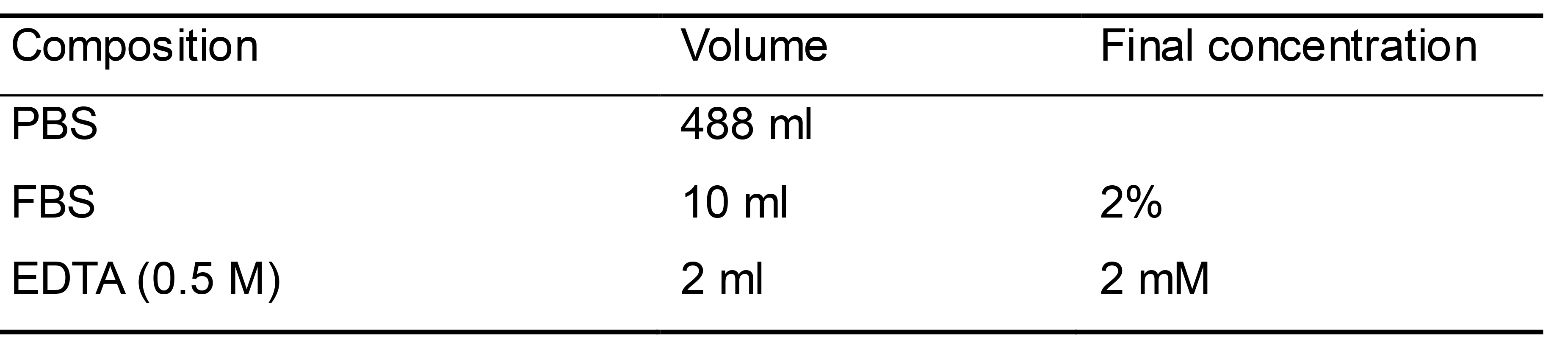
FACS buffer (Table 9)
Prepare 500 ml of FACS buffer as shown in Table 9
Store at 2-8 °C for up to 6 months
Table 9. FACS buffer
Osteoclast differentiation medium (Table 10)
Prepare 100 ml osteoclast differentiation medium as shown in Table 10
Sterilize osteoclast differentiation basal medium (including alpha-MEM, FBS) by filtering through a 0.2 μm PES membrane filter and keep at 2-8 °C for up to 1 month
Add vitamin D, hTGFβ, hMCSF, and hRANKL aseptically only before use
Table 10. Osteoclast differentiation medium

TRAP staining solution (Table 11)
All reagents are included in the acid phosphatase, leukocyte (TRAP) kit
Premix Fast Garnet GBC and sodium nitrate
Add the rest of reagents after 2 min
Table 11. TRAP staining solution

Reconstitution of hBMP4
Centrifuge the vial prior to opening
Reconstitute in 5 mM HCl, pH 3.0 to a concentration of 0.1-1.0 mg/ml
Do not vortex
Further dilute in a buffer containing 0.1% BSA as carrier protein and store in working aliquots at -80 °C
Reconstitution of other cytokines
Centrifuge the vial prior to opening
Reconstitute in water to a concentration of 0.1-1.0 mg/ml
Do not vortex
Further dilute in a buffer containing 0.1% BSA as carrier protein and store in working aliquots at -80 °C
Acknowledgments
The work was supported by funds from National Institute of Health (K99/R00 DE021442 and R01 DE025664). We adapted published protocols using M-CSF and IL-3 for myeloid differentiation (Panicker et al., 2012; Jeon et al., 2016).
Competing interests
The author declares no conflict of interest.
Ethics
The hiPSC lines from peripheral blood were generated as previous described (Chen et al., 2013; Chen et al., 2017). This work was in accordance with guidelines of the Institutional Review Board of the University of Connecticut Health (IRB protocol 09-199).
References
- Bonewald, L. F. (2011). The amazing osteocyte. J Bone Miner Res 26(2): 229-238.
- Chen, I. P. (2014). The use of patient-specific induced pluripotent stem cells(iPSCs) to identify osteoclast defects in rare genetic bone disorders. J Clin Med 3(4): 1490-1510.
- Chen, I. P., Fukuda, K., Fusaki, N., Iida, A., Hasegawa, M., Lichtler, A. and Reichenberger, E. J. (2013). Induced pluripotent stem cell reprogramming by integration-free Sendai virus vectors from peripheral blood of patients with craniometaphyseal dysplasia. Cell Reprogram 15(6): 503-513.
- Chen, I. P., Luxmi, R., Kanaujiya, J., Hao, Z. and Reichenberger, E. J. (2017). Craniometaphyseal dysplasia mutations in ANKH negatively affect human induced pluripotent stem cell differentiation into osteoclasts. Stem Cell Reports 9(5): 1369-1376.
- Chen, I. P., Wang, L., Jiang, X., Aguila, H. L. and Reichenberger, E. J. (2011). A Phe377del mutation in ANK leads to impaired osteoblastogenesis and osteoclastogenesis in a mouse model for craniometaphyseal dysplasia(CMD). Hum Mol Genet 20(5): 948-961.
- Cherry, A. B. and Daley, G. Q. (2013). Reprogrammed cells for disease modeling and regenerative medicine. Annu Rev Med 64: 277-290.
- Choi, K. D., Vodyanik, M. A. and Slukvin, II (2009). Generation of mature human myelomonocytic cells through expansion and differentiation of pluripotent stem cell-derived lin-CD34+CD43+CD45+ progenitors. J Clin Invest 119(9): 2818-2829.
- Deyle, D. R., Khan, I. F., Ren, G., Wang, P. R., Kho, J., Schwarze, U. and Russell, D. W. (2012). Normal collagen and bone production by gene-targeted human osteogenesis imperfecta iPSCs. Mol Ther 20(1): 204-213.
- Ding, Q., Lee, Y. K., Schaefer, E. A., Peters, D. T., Veres, A., Kim, K., Kuperwasser, N., Motola, D. L., Meissner, T. B. and Hendriks, W. T. (2013). A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell 12(2): 238-251.
- Grigoriadis, A. E., Kennedy, M., Bozec, A., Brunton, F., Stenbeck, G., Park, I. H., Wagner, E. F. and Keller, G. M. (2010). Directed differentiation of hematopoietic precursors and functional osteoclasts from human ES and iPS cells. Blood 115(14): 2769-2776.
- Jeon, O. H., Panicker, L. M., Lu, Q., Chae, J. J., Feldman, R. A. and Elisseeff, J. H. (2016). Human iPSC-derived osteoblasts and osteoclasts together promote bone regeneration in 3D biomaterials. Sci Rep 6: 26761.
- Kang, H., Shih, Y. R., Nakasaki, M., Kabra, H. and Varghese, S. (2016). Small molecule-driven direct conversion of human pluripotent stem cells into functional osteoblasts. Sci Adv 2(8): e1600691.
- Kanke, K., Masaki, H., Saito, T., Komiyama, Y., Hojo, H., Nakauchi, H., Lichtler, A. C., Takato, T., Chung, U. I. and Ohba, S. (2014). Stepwise differentiation of pluripotent stem cells into osteoblasts using four small molecules under serum-free and feeder-free conditions. Stem Cell Reports 2(6): 751-760.
- Kuhn, L. T., Liu, Y., Boyd, N. L., Dennis, J. E., Jiang, X., Xin, X., Charles, L. F., Wang, L., Aguila, H. L., Rowe, D. W., Lichtler, A. C. and Goldberg, A. J. (2014). Developmental-like bone regeneration by human embryonic stem cell-derived mesenchymal cells. Tissue Eng Part A 20(1-2): 365-377.
- Matsumoto, Y., Hayashi, Y., Schlieve, C. R., Ikeya, M., Kim, H., Nguyen, T. D., Sami, S., Baba, S., Barruet, E., Nasu, A., Asaka, I., Otsuka, T., Yamanaka, S., Conklin, B. R., Toguchida, J. and Hsiao, E. C. (2013). Induced pluripotent stem cells from patients with human fibrodysplasia ossificans progressiva show increased mineralization and cartilage formation. Orphanet J Rare Dis 8: 190.
- Ochiai-Shino, H., Kato, H., Sawada, T., Onodera, S., Saito, A., Takato, T., Shibahara, T., Muramatsu, T. and Azuma, T. (2014). A novel strategy for enrichment and isolation of osteoprogenitor cells from induced pluripotent stem cells based on surface marker combination. PLoS One 9(6): e99534.
- Panicker, L. M., Miller, D., Park, T. S., Patel, B., Azevedo, J. L., Awad, O., Masood, M. A., Veenstra, T. D., Goldin, E., Stubblefield, B. K., Tayebi, N., Polumuri, S. K., Vogel, S. N., Sidransky, E., Zambidis, E. T. and Feldman, R. A. (2012). Induced pluripotent stem cell model recapitulates pathologic hallmarks of Gaucher disease. Proc Natl Acad Sci U S A 109(44): 18054-18059.
- Park, I. H., Arora, N., Huo, H., Maherali, N., Ahfeldt, T., Shimamura, A., Lensch, M. W., Cowan, C., Hochedlinger, K. and Daley, G. Q. (2008). Disease-specific induced pluripotent stem cells. Cell 134(5): 877-886.
- Quarto, N., Leonard, B., Li, S., Marchand, M., Anderson, E., Behr, B., Francke, U., Reijo-Pera, R., Chiao, E. and Longaker, M. T. (2012). Skeletogenic phenotype of human Marfan embryonic stem cells faithfully phenocopied by patient-specific induced-pluripotent stem cells. Proc Natl Acad Sci U S A 109(1): 215-220.
- Quarto, N., Li, S., Renda, A. and Longaker, M. T. (2012). Exogenous activation of BMP-2 signaling overcomes TGFβ-mediated inhibition of osteogenesis in Marfan embryonic stem cells and Marfan patient-specific induced pluripotent stem cells. Stem Cells 30(12): 2709-2719.
- Sommer, C. A., Stadtfeld, M., Murphy, G. J., Hochedlinger, K., Kotton, D. N. and Mostoslavsky, G. (2009). Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells 27(3): 543-549.
- Stadtfeld, M., Nagaya, M., Utikal, J., Weir, G. and Hochedlinger, K. (2008). Induced pluripotent stem cells generated without viral integration. Science 322(5903): 945-949.
- Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K. and Yamanaka, S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131(5): 861-872.
- Warren, L., Manos, P. D., Ahfeldt, T., Loh, Y. H., Li, H., Lau, F., Ebina, W., Mandal, P. K., Smith, Z. D., Meissner, A., Daley, G. Q., Brack, A. S., Collins, J. J., Cowan, C., Schlaeger, T. M. and Rossi, D. J. (2010). Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7(5): 618-630.
- Yu, J., Hu, K., Smuga-Otto, K., Tian, S., Stewart, R., Slukvin, II and Thomson, J. A. (2009). Human induced pluripotent stem cells free of vector and transgene sequences. Science 324(5928): 797-801.
- Yu, J., Vodyanik, M. A., Smuga-Otto, K., Antosiewicz-Bourget, J., Frane, J. L., Tian, S., Nie, J., Jonsdottir, G. A., Ruotti, V., Stewart, R., Slukvin, II and Thomson, J. A. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318(5858): 1917-1920.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Chen, I. (2020). Differentiation of Human Induced Pluripotent Stem Cells (hiPSCs) into Osteoclasts. Bio-protocol 10(24): e3854. DOI: 10.21769/BioProtoc.3854.
Category
Stem Cell > Pluripotent stem cell > Cell differentiation
Cell Biology > Cell isolation and culture > Cell differentiation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








