- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Triacylglycerol Measurement in HeLa Cells
Published: Vol 10, Iss 24, Dec 20, 2020 DOI: 10.21769/BioProtoc.3852 Views: 4112
Reviewed by: Chiara AmbrogioMiaowei Marwell MaoWilliam Jennings Valentine

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Metabolic Assays for Detection of Neutral Fat Stores
Jan M. Baumann [...] Douglas S. Conklin
Jun 20, 2015 15687 Views

Measurement of the Intracellular Calcium Concentration with Fura-2 AM Using a Fluorescence Plate Reader
Magdiel Martínez [...] Walter I. Silva
Jul 20, 2017 33952 Views
Abstract
Lipid droplets store triacylglycerols (triglycerides) and sterol esters to regulate lipid and energy homeostasis. Triacylglycerol measurement is often performed during the investigation of lipid droplet formation and growth. This protocol describes a reliable method using a fluorometric lipid quantification kit to measure triacylglycerols extracted from HeLa cells, which were treated with oleic acid to trigger the formation of lipid droplets. The lipid quantification kit employs a lipid-binding molecule that emits bright fluorescence only when bound to extracted triacylglycerols, whose content can be quantified by a simple fluorescence readout.
Keywords: Lipid dropletBackground
Lipid droplets (LDs) are unique organelles with characteristic functions. Different from common cellular organelles, each LD is defined by a monolayer of phospholipids, which encloses a core of neutral lipids such as triacylglycerol (TAG), also known as triglyceride, and sterol esters. As storage organelles, LDs play a major role in the homeostasis of lipids and energy, as well as other cellular events. By storing excess fatty acids in the form of TAG, LDs prevent lipotoxicity and protect cells against oxidative stress. Importantly, LDs make contacts with virtually all other organelles and regulate critical cellular processes, including membrane trafficking, protein turnover and viral infection (Olzmann and Carvalho, 2018; Gao et al., 2019). Moreover, the LD surface is decorated with various proteins that are involved in the synthesis and hydrolysis of TAG, and in the transport of lipids between LDs and other organelles (Kory et al., 2016; Kumar et al., 2018; Du et al., 2020).
Eukaryotic cells have well-established pathways to synthesize TAG stored in LDs (Coleman and Mashek, 2011). Studies on the formation and growth of LDs often involve the extraction of TAG from cells or tissues for quantification. Some sensitive methods, such as LC-MS, are available for this purpose, but requiring expensive equipment and being time-consuming. Commercially available lipid quantification kit, on the other hand, can provide a quick, accurate, and economical method to measure cellular levels of TAG. In this protocol, we describe a reliable method using a fluorometric lipid quantification kit to measure TAG extracted from HeLa cells that were grown in the presence of oleic acid to induce LD formation. The lipid quantification kit takes advantage of a lipid binding molecule that emits bright fluorescence only when bound to extracted TAG. Thus, the amount of TAG can be readily quantified by a simple fluorescence readout.
Materials and Reagents
Cell culture dish (60 x 15 mm) (Corning, catalog number: CLSE430166 )
2 ml glass vial (Agilent, catalog number: 5190-9062 )
Glass bottle, 100 ml (Therfmo Fisher Scientific, catalog number: FSBFB33144 )
Microplates for Fluorescence-based Assays, 96-well (Thermo Fisher Scientific, catalog number: M33089 )
1.5 ml microcentrifuge tubes (Sarstedt, catalog number: 72.690.001 )
Serological pipettes, 5 ml, 10 ml, and 25 ml (Corning, catalog numbers: CLS4487, 4488, 4489 )
Pipette tips with filter (Interpath Services, catalog numbers: 24750, 24900 )
Falcon tubes, 15 ml, 25 ml (Corning, catalog numbers: CLS430829, 430791 )
HeLa cell line (ATCC CCL-2TM)
Fluorometric Lipid Quantification Kit (Cell Biolabs, catalog number: STA-617 )
Dulbecco's Modified Eagle's Medium (DMEM) (Gibco, catalog number: 11995073 )
Fetal bovine serum (FBS) (Bovogen, catalog number: FFBS-500 )
Penicillin-Streptomycin-Glutamine (100x) (Gibco, catalog number: 10378016 )
DPBS, no calcium, no magnesium (Gibco, catalog number: 14190250 )
Oleic Acid-Albumin from bovine serum (Sigma-Aldrich, catalog number: O3008-5ML )
Methanol (Univar, catalog number: AJA2314-2.5LGL )
Hexane (VWR International, catalog number: VWRC24577.323 )
Chloroform (Chem-supply Pty Ltd Australia, catalog number: RP1027E-G2.5L )
Isopropanol (Unichrom, catalog number: 2323-2.5GL )
Sodium hydroxide (Univar, catalog number: AJA482-500G )
Bicinchoninic Acid Kit for protein determination (Sigma-Aldrich, catalog number: BCA1-1KT )
Sodium chloride (Chem-Supply, catalog number: SA046-500G )
TRIS (hydroxymethyl) aminomethane (Univar, catalog number: 1.08382.0500 )
Bovine serum albumin (Sigma-Aldrich, catalog number: A1933 )
Buffer A (see Recipes)
Buffer B (see Recipes)
Equipment
Biological Safety Cabinets (NuAire, model: 5437-400E )
CO2 incubator (Binder, model: CB 150)
Fume cupboard (Dynaflow, model: FC1800)
Heating block (Labnet International, model: D1100 )
Vortex mixer (Ratek, model: IC-VM1)
POLARstar Omega Microplate Reader (BMG Labtech, catalog number: 415-0704)
Water bath (Ratek, model: IC-WB1200D)
Procedure
Cell culture
Seed HeLa cells in 6-cm dishes (5 x 105 cells per dish) in 3 ml of DMEM supplemented with 10% FBS and 1x Penicillin-Streptomycin.
Incubate cells at 37 °C in a humidified incubator with 5% CO2 for 24 h.
Treat cells with 400 µM of oleic acid-albumin from bovine serum by adding 0.24 ml of the stock solution (3.3 mM) to 1.76 ml of culture medium.
Lipid extraction
Wash cells in the dishes twice with 2 ml of buffer A.
Wash cells once with buffer B.
Aspirate the buffer residues with pipette tips.
Add 2 ml of hexane-isopropanol (3:2) to each dish and incubate for 30 min at room temperature in a fume hood with the lid on.
Note: Hexane and chloroform are hazardous to breathe. Therefore, the handling of these reagents should be carried out in chemical fume hood that is safely vented to outside the laboratory.
Transfer the organic solvent from each dish to a 2 ml glass vial.
Rinse each dish briefly with 1 ml of the same solvent and combine into the same glass vial. Carefully remove all of the organic solvents and save the dish for protein determination.
Evaporate the solvent to dryness inside the fume hood by leaving the vial uncapped in the fume hood with the airflow running (takes approximately 16 h).
Protein determination
Add 1 ml of 0.1 M NaOH in water to the original cell culture dish and dissolve the cell remnants by pipetting up and down.
Remove 20 µl of aliquots in duplicate and dilute in 20 µl of water for BCA assay to determine total proteins in each dish (protein amount of each assay x 50). A typical protein yield per dish is 500-600 µg.
Lipid quantification with the Kit # STA-617 from Cell BioLabs with modifications
Mix 50 ml of methanol with 25 ml of chloroform in a 100 ml glass bottle. Store at room temperature.
Thoroughly resuspend each extracted TAG sample from Step B7 with 200 µl of methanol/chloroform mixture in capped glass vials using a vortex mixer at high speed for 30 s. Keep on ice after the vortexing.
Prepare serial dilutions of lipid standards in the methanol/chloroform mixture, with a concentration range from 0 to 5 µg/µl (Table 1). Mix each standard well before further dilution.
Table 1. Lipid standards with a serial dilution
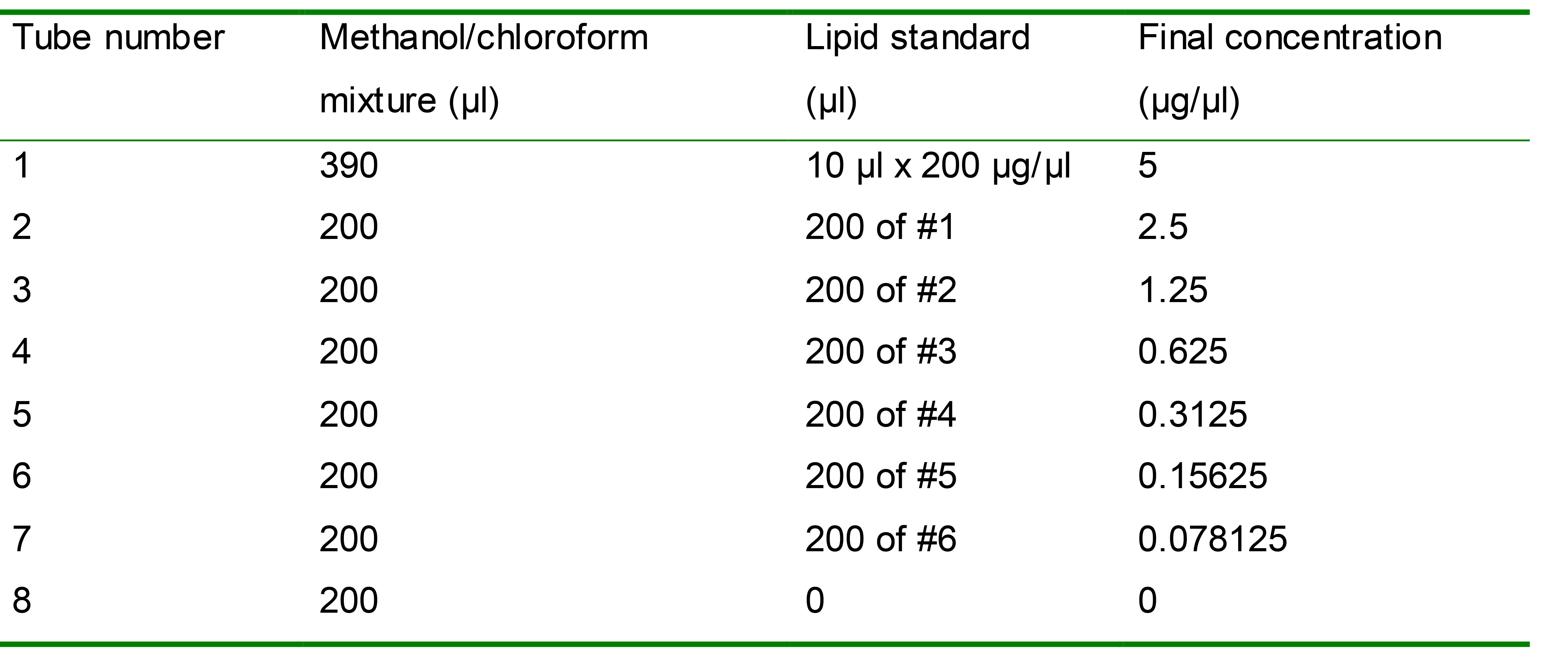
Add 40 µl of standards or TAG samples in duplicates to a 96-well microplate for fluorescence-based assays.
Evaporate the organic solvent by incubating the plate on top of a heating block set as 55 °C for 30 min or until dryness in a safety chemical hood.
Warm the 100x Fluorometric Reagent stock in a 37 °C water bath until thawed.
Prepare 1x Fluorometric Reagent by adding 0.5 ml of the thawed 100x stock to 49.5 ml of water in a 50 ml falcon tube.
Cool the 96-well plate on ice for 2 min.
Add 40 µl of isopropanol to standards or TAG samples. Pipette up and down gently to mix each well.
Add 200 µl of 1x Fluorometric Reagent to each well.
Incubate the plate in the dark for 10 min at room temperature.
Read the plate at 490 nm excitation and 585 nm emission with a POLARstar Omega Microplate Reader.
Data analysis
Generate the standard curve by plotting the amount of lipid standards against the relative fluorescent unit (RFU). An example of the standard curve with an equation is shown in Figure 1.
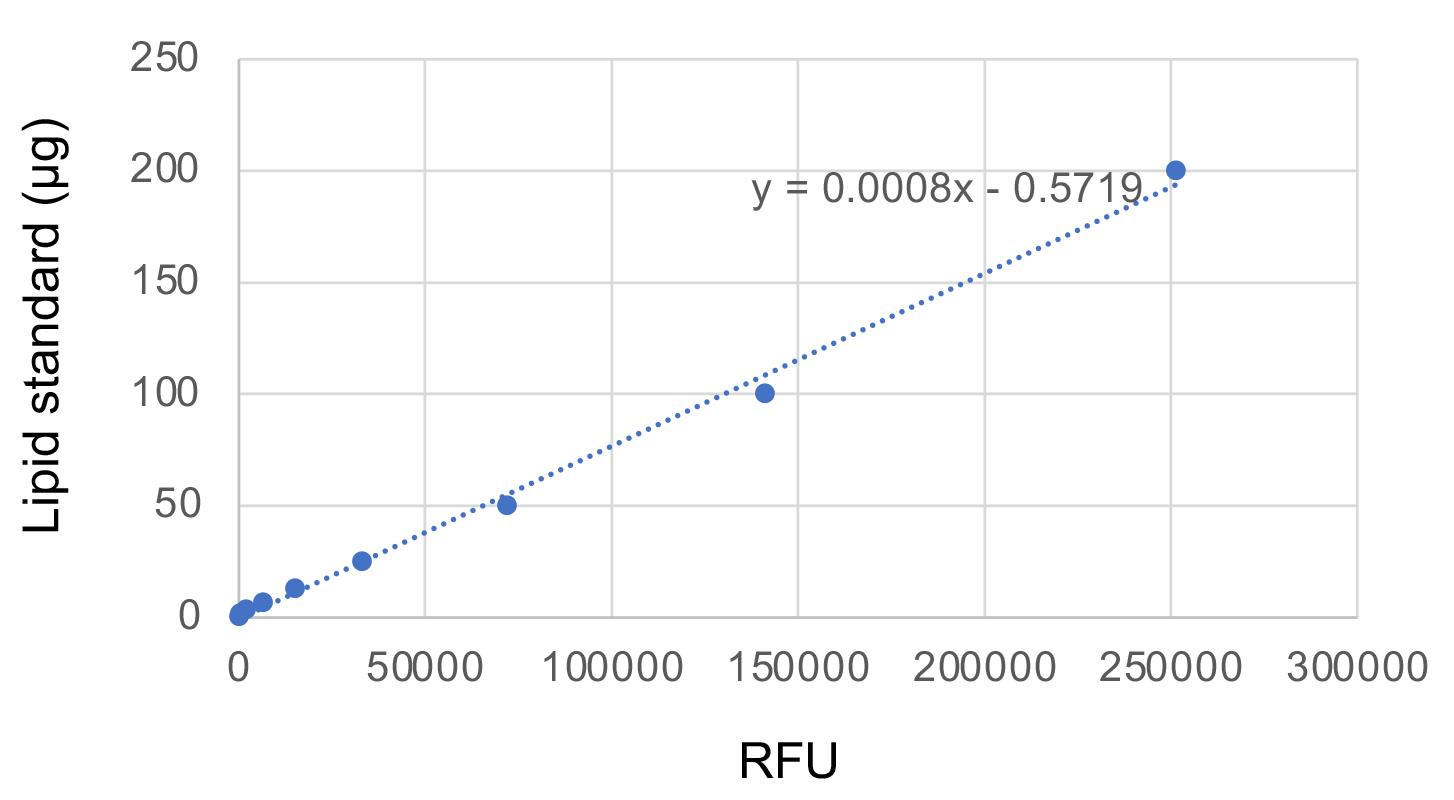
Figure 1. Fluorometric standard curve for TAG quantificationCalculate the quantity of TAG in each sample well of the 96-well plate using the equation shown in the graph of the standard curve (Figure 1).
Calculate the total quantity of TAG from each sample by multiplying the amount of TAG in the sample well with five (because 40 μl out of a 200 μl was measured per well). Typical yields of TAGs and protein per 6-cm dish of HeLa cells (~90% confluency) are shown in Table 2.
Table 2. Yields of TAGs and proteins from each dish of HeLa cells
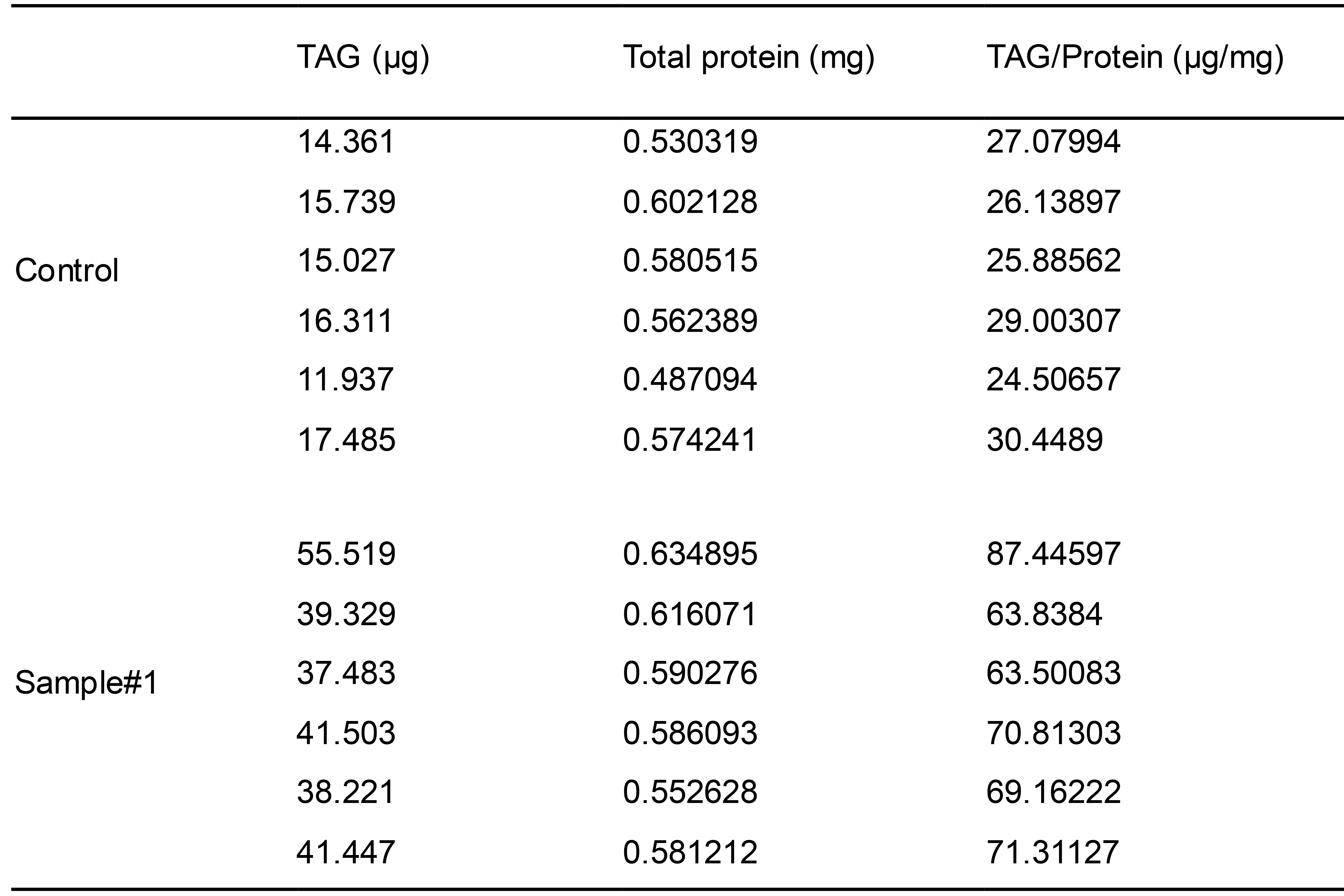
Normalize the total quantity of TAG with the total amount of proteins determined in Procedure C (Table 2). An example of final results is shown in Figure 2.
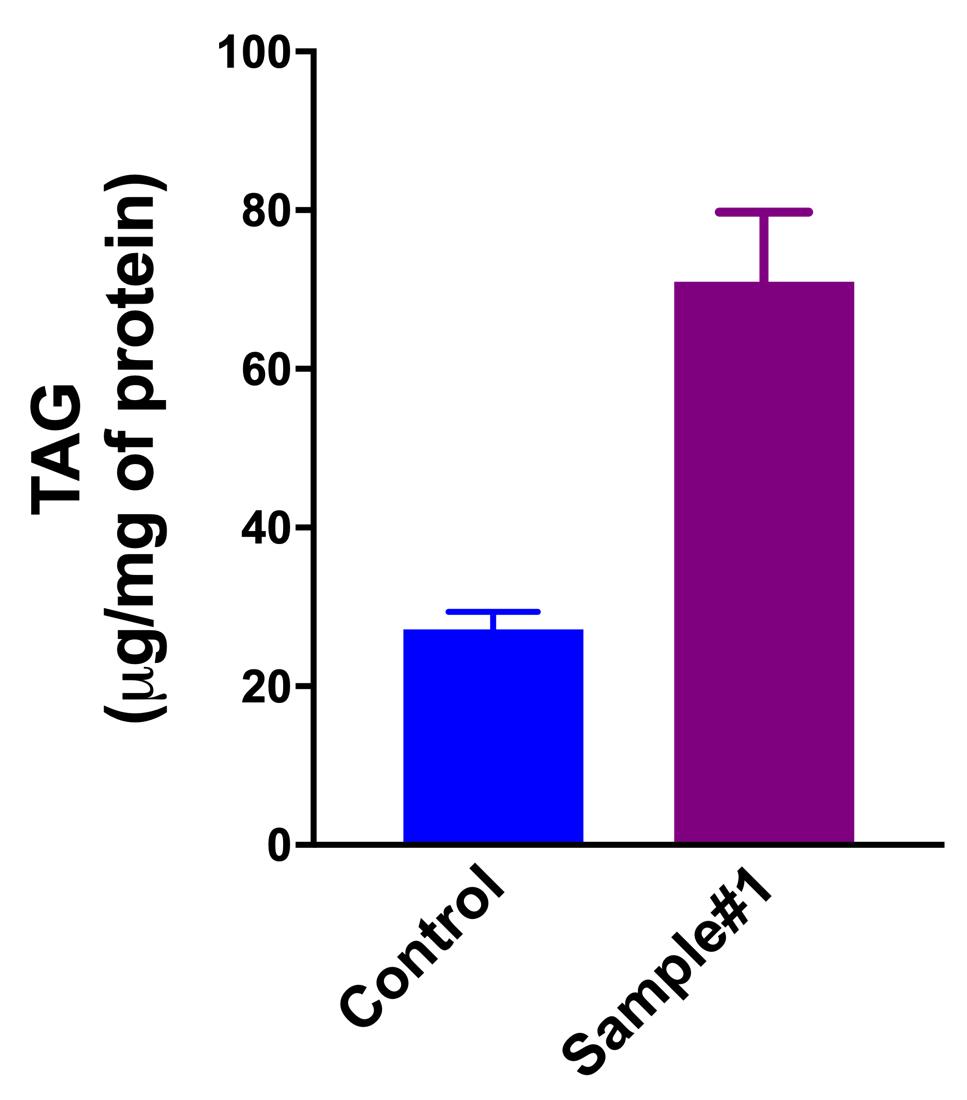
Figure 2. TAG quantities in HeLa cells (control vs. sample#1) determined by the fluorometric assay
Recipes
Buffer A
150 mM NaCl
50 mM Tris-HCl, pH 7.4
2 mg/ml Bovine serum albumin
Buffer B
150 mM NaCl
50 mM Tris-HCl, pH 7.4
Acknowledgments
This work was supported by project grants from the National Health and Medical Research Council of Australia (APP1041301, 1141939, and 114472).
TAG extraction procedure described in this protocol was adapted from a well-defined early method ( Goldstein et al., 1983 ).
Competing interests
The authors declare no competing interests.
References
- Coleman, R. A. and Mashek, D. G. (2011). Mammalian Triacylglycerol Metabolism: Synthesis, Lipolysis, and Signaling. Chem Rev 111(10): 6359-6386.
- Du, X., Zhou, L., Aw, Y. C., Mak, H. Y., Xu, Y., Rae, J., Wang, W., Zadoorian, A., Hancock, S. E., Osborne, B., Chen, X., Wu, J. W., Turner, N., Parton, R. G., Li, P. and Yang, H. (2020). ORP5 localizes to ER-lipid droplet contacts and regulates the level of PI(4)P on lipid droplets. J Cell Biol 219(1).
- Gao, M., Huang, X., Song, B. L. and Yang, H. (2019). The biogenesis of lipid droplets: Lipids take center stage. Prog Lipid Res 75: 100989.
- Goldstein, J. L., Basu, S. K. and Brown, M. S. (1983). Methods in Enzymology. Sciencedirect 98: 241-260.
- Kory, N., Farese, R. V. and Walther, T. C. (2016). Targeting Fat: Mechanisms of Protein Localization to Lipid Droplets. Trends Cell Biology 26(7): 535-546.
- Kumar, N., Leonzino, M., Hancock-Cerutti, W., Horenkamp, F. A., Li, P., Lees, J. A., Wheeler, H., Reinisch, K. M. and Camilli, P. D. (2018). VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J Cell Biol 217(10): 3625-3639.
- Olzmann, J. A. and Carvalho, P. (2018). Dynamics and functions of lipid droplets. Nat Rev Mol Cell Biol 20(3): 1.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Du, X. and Yang, H. (2020). Triacylglycerol Measurement in HeLa Cells. Bio-protocol 10(24): e3852. DOI: 10.21769/BioProtoc.3852.
- Du, X., Zhou, L., Aw, Y. C., Mak, H. Y., Xu, Y., Rae, J., Wang, W., Zadoorian, A., Hancock, S. E., Osborne, B., Chen, X., Wu, J. W., Turner, N., Parton, R. G., Li, P. and Yang, H. (2020). ORP5 localizes to ER-lipid droplet contacts and regulates the level of PI(4)P on lipid droplets. J Cell Biol 219(1).
Category
Biochemistry > Lipid > Lipid measurement
Cancer Biology > General technique > Cell biology assays > Metabolism
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










